Abstract
Background
Nuclear transfer has been utilized as a means of selectively modifying the mammalian genome. One possible consequence of this technology is that the oocytes used in nuclear transfer may provide additional antigens via cytoplasmic inheritance of maternally derived, mitochondrial DNA. These studies examine the potential consequences of such inheritance in a large animal transplantation model.
Methods
Renal transplants were performed between MHC-identical animals differing only in the source of their maternally derived cytoplasmic DNA, using a protocol which uniformly leads to tolerance within standard MHC-inbred lines. In an attempt to correlate transplant results with a putative marker for disparities in cytoplasmically inherited minor histocompatibility antigens, we examined one hypervariable region of mitochondrial DNA (mtDNA), designated HV1.
Results
The mtDNA sequence of the HV1 region was found to be invariant among MGH miniature swine of different haplotypes, despite twenty years of selective breeding of the sublines of this colony. In contrast, swine derived by nuclear transfer into outbred oocytes differed in the HV1 region sequence from each other and from MGH swine. Renal transplants from standard, inbred MGH swine to their MHC-identical knockout counterparts derived from outbred oocytes were rejected within two weeks, while transplants in the reverse direction were accepted for over 30 days.
Conclusions
The HV1 sequence of mtDNA may serve as a marker for the level of diversity of mtDNA. These transplant data are consistent with the existence of mtDNA-encoded mitochondrial minor antigens with a similar level of diversity that can influence the outcome of renal transplantation.
Keywords: transplantation, mitochondria, transplantation antigens, tolerance, rejection
Introduction
Nuclear transfer technology has been used as a means of deleting the expression of single genes in the mammalian genome(1). The gene to be modified is first “knocked-out” using a targeted insertion to disrupt its coding sequence in the chromosomal DNA. The genetically engineered nucleus containing the modified gene is then transferred into an enucleated oocyte and the resulting embryo is implanted into the uterus of a surrogate animal. While the genome of the resulting “knockout” (KO) should be identical to that of the source nucleus except for expression of the targeted gene, the cytoplasmic contents of the embryo could differ due to the use of an oocyte from an unrelated animal. Since mtDNA is a component of the cytoplasm that can potentially be expressed, one unintentional result of nuclear transfer technology could be the maternal inheritance(2) of transplantation antigenic differences contributed by the oocyte cytoplasm. Indeed, mitochondrial encoded proteins were among the first known minor histocompatibility antigens(3). Such antigens (minor H antigens) were reported to be encoded by both the NADH dehydrogenase and the Cytochrome C oxidase subunits of mouse mitochondrial DNA(4,5).
MGH miniature swine that have been intentionally inbred to homozygosity at the major histocompatibility complex (MHC), have served as a large animal model for numerous studies of transplantation biology(6-8). We have previously shown in this model that MHC-matched, vascularized kidney transplants to recipients treated with 12 days of cyclosporine are uniformly accepted long-term (greater than 60 days)(9). To facilitate the potential use of these animals as donors for xenotransplantation, we have recently developed GalT-KO animals in which the gene encoding galactosyl-alpha-1,3-galactosyltransferase (GalT) has been disrupted zin order to eliminate expression of the ubiquitous alpha-1,3-galactose antigen on the pig cell surface(1,10). These animals were derived from the fully inbred SLAdd line of MGH miniature swine(11) using nuclear transfer technology. Therefore, as explained above, although these animals remain of the SLAdd genotype, the cytoplasmic and mtDNA content of subsequent maternally derived offspring would be expected to be inherited from the outbred oocytes used for nuclear transfer. We have now performed renal allografts between these animals and the highly inbred MGH swine from which they originated and the results reported here are consistent with a role for mitochondrial antigens in determining the outcomes of these transplants.
Materials and Methods
Animals
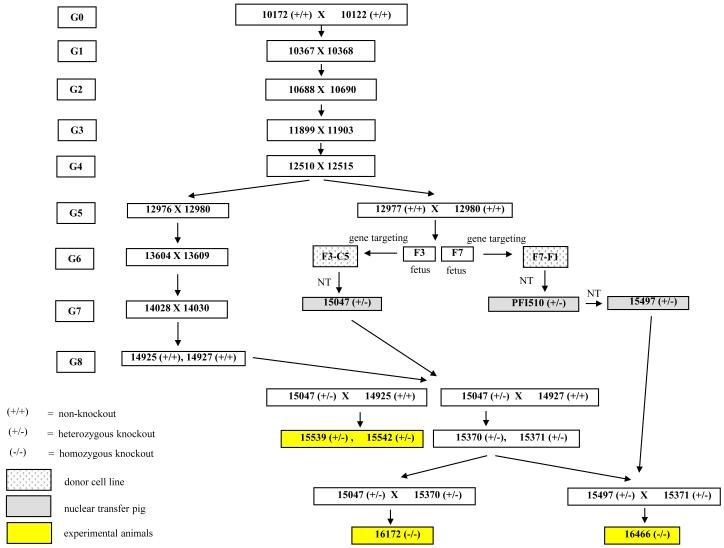
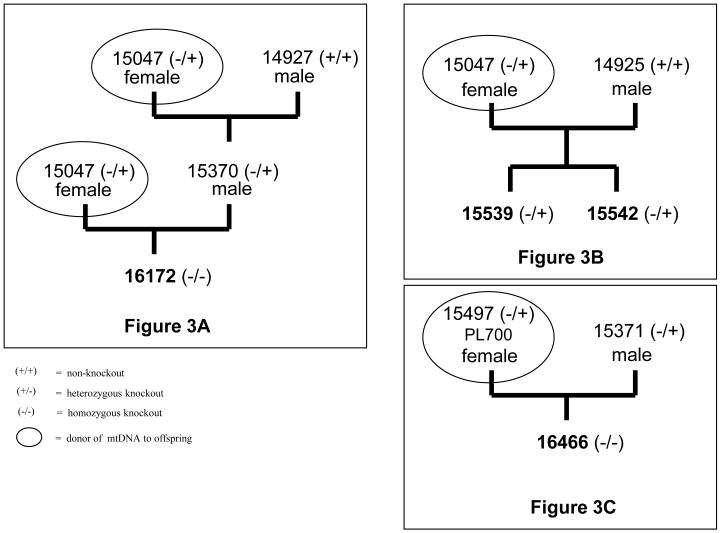
The standard MGH miniature swine have been inbred to homozygosity at the major histocompatibility locus, producing three MHC defined haplotypes (SLAdd, SLAaa, SLAcc), as described previously(6-8) (Figure 1). A subline of the standard SLAdd animals was further inbred to produce a line of highly inbred histocompatible swine, as described previously(11). The initial GalT-KO (subsequently referred to as ‘knockout’) swine were produced from this highly inbred SLAdd line by gene-targeting of fibroblasts, followed by transfer into enucleated oocytes derived from outbred domestic swine(1,10). The knockout animals used in these experiments were the offspring derived from several of these initial knockout animals (15047, 15497) (Figure 2). The heterozygous knockout animals (15539, 15542) were produced from the mating of a nuclear transfer derived heterozygous knockout female (15047) to a non-knockout G8 highly inbred male (14925). The homozygous knockout animals (16172, 16466) were produced from the mating of nuclear transfer derived heterozygous knockout females (15047, 15497, respectively) to heterozygous knockout males derived by breeding (15370, 15371, respectively) (Figures 2, 3).
Figure 1.
MGH miniature swine haplotypes. Three strains of MGH swine homozygous for class I and class II MHC (SLAdd, SLAaa, and SLAcc) were derived by selective breeding. The highly inbred line of MHC homozygous SLAdd animals was derived from the SLAdd strain by further sequential brother-sister matings.
Figure 2.
Pedigree of the knockout animals. The knockout animals were derived by nuclear transfer technology from the G5 generation of highly inbred SLAdd animals (Fig. 1). Donor oocytes for the nuclear transfer were derived from non-MGH swine. The heterozygous knockout animals (15539, 15542) were offspring, and siblings, of a nuclear transfer derived heterozygous knockout female (15047) mated with a non-knockout G8 inbred animal. The homozygous knockout animals (16172, 16466) were offspring from the mating of heterozygous knockout animals. Different nuclear transfer derived knockout females (15407, 15497, respectively) were used in the matings to derive the offspring 16172 and 16477.
Figure 3.
Partial pedigree and mtDNA inheritance of the knockout animals. A) the mtDNA of the homozygous knockout 16172 was derived from the nuclear transfer derived knockout female (15047), B) the heterozygous knockouts 15539 and 15542 were siblings and the mtDNA was derived from the same nuclear transfer derived knockout female (15047) as 16172, C) the mtDNA of the homozygous knockout (16466) was derived from a different nuclear transfer female (15497) than knockouts 16172, 15539, and 15542.
Surgery and post-transplant monitoring of graft
The surgical procedures used for kidney transplants have been described in detail previously(12). Briefly, animals were subjected to a bilateral nephrectomy via a paramedian incision, followed by an orthotopic renal transplantation utilizing a Carrel patch to implant the renal artery end-to-side into the recipient’s aorta. The renal vein was anastomosed end-to-side to the recipient’s vena cava, and a vesico-ureteral anastomosis was performed. Total cold ischemic time was kept to less than 60 min. No blood transfusions were required. All recipients were treated with 12 days of cyclosporine, 10-15 mg/kg, i.v., starting on the day of transplant, with dosing to achieve blood levels of 400-800 ng/ml. Graft status was monitored in all animals by daily serum creatinine level and platelet count for at least 31 days post transplant. A normal stable serum creatinine and platelet count indicated acceptance of the graft. A precipitous rise in creatinine (>6.0) and a simultaneous decrease in platelet count were defined as rejection.
Kidney Biopsy and Pathology
Wedge kidney biopsies were performed through flank incisions as described(12). The biopsy tissues were stained with hematoxylin and eosin (H&E) and periodic acid-Schiff. Allograft rejection was scored by standard pathologic criteria according to the Cooperative Clinical Trials in Transplantation criteria (13).
Swine Hypervariable mtDNA Sequence
Corresponding positions of two known human mitochondrial hypervariable segments HVS1 (16,024 - 16,383bp) and HVS2 (57 – 372bp) (www.mitomap.org; Genbank accession number NC_001807) were correlated with the complete swine mitochondrial genome (Genbank accession number AJ002189)(14,15). Appropriate primers were selected and amplification of the corresponding swine hypervariable regions was confirmed by agarose gel electrophoresis (data not shown). The region of the swine mitochondrial sequence, corresponding to the human HVS1 region, was designated hypervariable region I (HV1) and chosen for this study. The HV1 region corresponds to nucleotides 15,370 through 15,730 of the swine mitochondrial genome and is a noncoding variable portion of the swine control region(14).
PCR Amplification of the HV1 Mitochondrial Sequence
Peripheral blood lymphocytes (PBL) were isolated from heparinized blood by Ficoll gradient centrifugation. The DNA was isolated from the PBL with a DNeasy tissue kit (Qiagen, Valencia CA). A 346bp fragment of the swine mtDNA HV1 region was amplified by polymerase chain reaction (PCR) using forward primer 463 (15,428bp) (TTCCCTGCAACCAAAACAAG) and reverse primer 464 (15,773bp) (TCACGCGGCATGGTAATTAA). The reaction consisted of 1× buffer A (Fisher Scientific Co., Pittsburgh, PA), 1uM of each primer, 80uM each dNTP and 2.5U Taq polymerase (Fisher Scientific Co., Pittsburgh, PA) in a final volume of 50ul. The PCR cycle conditions were twenty cycles of 95°C for 30 seconds, 55°C for 30 seconds and 72°C for 45 seconds followed by 10 minutes at 72°C. The PCR product was then sequenced by the MGH Core Sequencing Facility.
Results
Pedigree of experimental transplant animals
Two heterozygous* knockout animals (15539, 15542) were used in these experiments either as kidney donors or recipients. These animals were the offspring of a female heterozygous knockout animal (15047) mated with a male non-knockout animal (14925) (Figure 2). The female heterozygous knockout (15047) was derived from a nuclear transfer in which the nucleus from a heterozygous knockout fibroblast cell line (F3-C5), isolated from a G5 highly inbred SLAdd animal (fetus #3 from 12977 × 12980), was transferred into an outbred oocyte(1). The outbred oocyte was derived from an animal unrelated to the MGH herd. The highly inbred SLAdd line, from which the knockout fibroblast cell line was derived, was produced by sequential brother-sister matings from two founder SLAdd animals (10172, 10122) of the standard MGH SLAdd herd. The non-knockout male (14925) used in the mating was from the eighth generation of this same inbred SLAdd line.
Due to the maternal inheritance of mitochondrial DNA, the female used in the mating (15047) would determine the mitochondrial content of the offspring (15539, 15542) (Figure 3B). The mitochondrial content of the nuclear transfer derived heterozygous knockout (15047) was expected to be from the outbred animal unrelated to the MGH herd, since mitochondrial DNA is cytoplasmically derived. Thus, since the heterozygous knockout animals 15539 and 15542 were sibling offspring derived from the same mating, the mitochondrial DNA content was from the same nuclear transfer and expected to be identical in sequence.
Sequence of the mitochondrial HV1 region as a marker of cytoplasmic inheritance
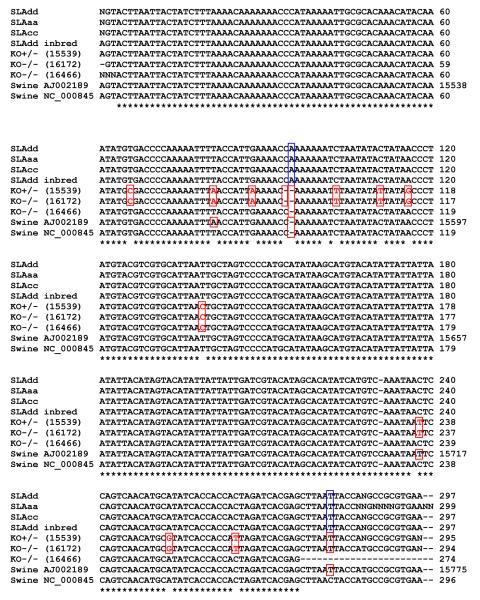
Since the HV1 region represents a region of higher variablility relative to the remainder of the mitochondrial sequence(15), it was chosen as a marker of diversity for tracing cytoplasmic inheritance. The mitochondrial HV1 hypervariable region of animals from several different MHC haplotypes of the standard MGH miniature swine and from representatives of the most highly inbred line, from which the knockouts were produced, were determined (Figure 1). All of these animals demonstrated identical mtDNA sequences (Figure 4). Thus, there appeared to be no differences in this marker of mitochondrial genomic inheritance among the existing haplotypes of MGH miniature swine. This sequence was however different in two positions (+A at 15,573bp, and a C - > T at 15,616bp of the published swine mtDNA sequence (Genbank accession number AJ002189). The swine used to obtain the published sequence was derived from a slaughterhouse animal of unknown breed(14).
Figure 4.
Sequence of the mitochondrial HV1 region in animals from the MGH herd and in knockout animals. The HV1 sequence was determined to be identical in the MGH animals of the three haplotypes (SLAdd, SLAaa, SLAcc) and inbred SLAdd. The HV1 sequence of the heterozygous knockout animals (KO+/−) and homozygous knockout (KO−/−) animals differed from the MGH herd. Knockout animals that were offspring of the same knockout female had identical HV1 sequences. HV1 sequence variation was observed between different breeds of swine (Genbank accession numbers AJ002189, NC_000845).
Further confirmation of this lack of heterogeneity was obtained by comparing the HV1 sequences in samples of stored peripheral blood lymphocyte DNA from animals born twenty-five years ago (SLAaa, SLAcc) to those from animals born more recently (SLAdd, inbred SLAdd). As seen in Figure 4, the HV1 sequences were found to be identical in all of these animals, suggesting that this mitochondrial genomic sequence has not changed in the past twenty-five years.
In contrast, marked differences were observed between these sequences and those from offspring of female knockout animals. The knockout animals (16172, 15539) were offspring derived from the same nuclear transfer derived heterozygous knockout mother (15047), thus from the same original nuclear transfer. Both animals had the identical mtDNA sequence in the HV1 region, indicative of the maternal inheritance of mtDNA (Figure 3A, B), and showing twelve nucleotide differences from the MGH miniature swine (Figure 4).
The homozygous knockout animal (16466) was the offspring of the heterozygous knockout mother (15497) which was originally derived from a different nuclear transfer than animal 15047 (Figure 3C). The HV1 mitochondrial sequence of this knockout likewise differed both from the standard animals and from knockouts 16172 and 15539. This knockout animal differed from the MGH miniature swine at two nucleotide positions (Figure 4).
Heterozygous knockout kidney transplants into standard inbred MGH swine
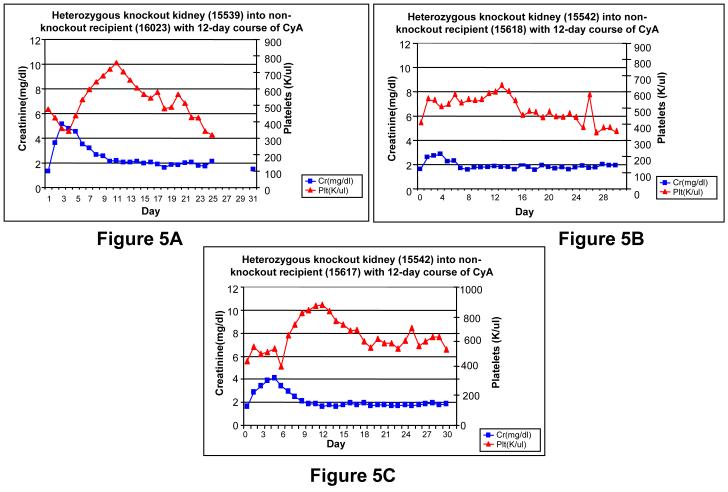
Three standard non-knockout SLAdd MGH miniature swine (16023, 15617, 15618) received kidney transplants from a heterozygous knockout animal (15539 or 15542), were treated with 12 days of cyclosporine as previously described(9) and monitored for 31 days post-operatively. All three non-knockout recipients accepted their kidney transplants (Figure 5A-C). Animal 16023 experienced an initial transient bump in creatinine of 5.17 at post-operative day (POD) 2, likely due to acute tubular necrosis (ATN) secondary to operative ischemia time. The creatinine returned to a baseline level of 2.0 by POD 9 and remained stable until POD 30. A decrease in platelet count, which correlates with rejection, was not observed in this animal. Indeed, the animal experienced a transient increase in platelet count to a peak of 757,000 per ul on POD 10, with return to baseline thereafter. Thus, the stable creatinine and a platelet count, at or above the pre-transplant baseline, indicated acceptance of the transplant. A kidney biopsy performed on POD 7 showed no acute cellular rejection (ACR), and a biopsy at POD 30 demonstrated focal mononuclear cell infiltrates, and no glomerulonephritis or endothelialitis, consistent with a mild ACR type I.
Figure 5.
Heterozygous knockout kidney transplants into non-knockout recipients. Kidneys were transplanted using the standard tolerance-inducing regimen, consisting of a 12-day course of cyclosporine. A) The MGH non-knockout recipient (16023) experienced an initial transient elevation in both creatinine and platelets, which returned to baseline by POD 30, B-C) The MGH non-knockout animals (15617, 15618) were siblings and each received a knockout kidney from the same donor (15542). Both recipients experienced an initial transient elevation in creatinine and platelets, which returned to baseline by POD 30.
The standard inbred MGH animals (15617, 15618), which were siblings, each received a kidney from the same heterozygous knockout animal (15542) and were monitored post-operatively (Figure 5B-C). Both animals experienced a transient increase in creatinine. In animal 15617, the creatinine peaked at 4.0 on POD 5, returned to a baseline value of 2.0 by POD 8 and remained stable until POD 30. In animal 15618 the creatinine peaked at 2.31 at POD 5, returned to a baseline value of 2.0 by POD 6.0 and remained stable until POD 30. In both animals the transient rise in creatinine was again likely due to ATN secondary to operative ischemia time. The platelet count remained relatively stable in both animals. In animal (15617) the platelet count peaked at 782,000 per ul on POD 11 from its pre-transplant value of 416,000 per ul, and remained stable at 550,000 per ul from PODs 18 to 31. In animal (15618) the platelet count peaked at 598,000 per ul on POD 13, from its pre-transplant value of 403,000 per ul, with a gradual downward trend until POD 29 of 350,000 per ul which is near pre-transplant values. Both the stable creatinine and platelet counts are strong indicators of acceptance in both transplants. Kidney biopsies performed on POD 30 for both 15617 and 15618 demonstrated focal mononuclear cell infiltrates with tubulitis, and no glomerulonephritis or endothelialitis, consistent with a mild ACR type I.
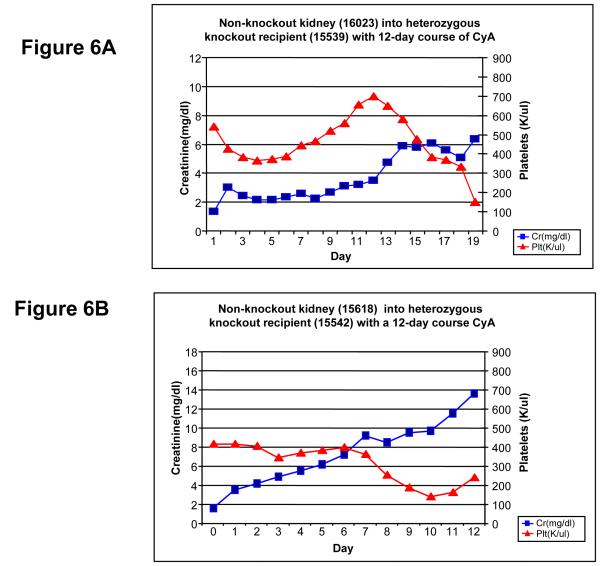
Standard inbred MGH kidney transplants into heterozygous knockout swine
Two heterozygous knockout animals (15539, 15542) received kidney transplants from standard non-knockout SLAdd MGH miniature swine (Figure 6A-B). Both animals rejected their kidney transplants at PODs 18 and 12 respectively. After an initial creatinine level of 1.5 at POD 0, animal 15539 experienced a brief period of stable creatinine of 2.0 from PODs 3 to 8. This brief period of stable creatinine was then followed by a steady rise in creatinine to a level of 6.34 by POD 18. The platelet count dropped post-transplant to 367,000 per ul by POD 3 from its initial value of 540,000 per ul, but eventually peaked at 700,000 per ul by POD 11. After this peak at POD 11, the platelets steadily dropped to 152,000 per ul on POD 18, which was well below the initial post-transplant levels. Given the steady rise in creatinine and precipitously decline in platelets, both markers of rejection, animal 15539 was determined to have rejected the transplant. A kidney biopsy performed on POD 18 demonstrated moderate mononuclear cell infiltrates with glomerulonephritis, endothelialitis, and fibrinoid necrosis of small arteries. In addition, interstitial edema and focal hemorrhage were present, consistent with ACR type III.
Figure 6.
Non-knockout kidney transplants into heterozygous knockout recipients. Kidneys were transplanted using the standard tolerance-inducing regimen, consisting of a 12-day course of cyclosporine.A-B) Both knockout recipients were siblings and each experienced a steady increase in creatinine post-transplant. Recipient (15539) rejected the transplant by POD 19, and recipient (15542) rejected the graft by POD 12.
Animal 15542 experienced a continual rise in creatinine from its initial post-operative level of 1.6 to 13.6 by POD 12 (Figure 6B). In addition, the platelets steadily declined post-transplant from an initial level of 416,000 per ul to a final value of 239,000 per ul by POD 12. Both the rise in creatinine level and drop in platelets were indicative of rejection of the transplanted graft by POD 12. A kidney biopsy at this time demonstrated moderate mononuclear cell infiltrates with glomerulonephritis, endothelialitis, and fibrinoid necrosis of small arteries. Interstitial edema and focal interstitial hemorrhage with neutrophilic infiltration were also present, consistent with ACR type III.
Discussion
This study was designed to study the effects of mitochondrial derived antigens on transplant outcome. Nuclear transfer technology used to create knockout animals was an ideal method for isolating mitochondrial antigenic differences for these studies. In the case of the knockout swine, the mitochondrial DNA of these animals had no relationship to the MGH herd. Yet the nuclear DNA, and thus MHC antigens, were still of MGH miniature swine origin. The effects of these minor antigen differences on transplant outcome were studied with a full class I and class II matched kidney transplant treated with a 12 day course of cyclosporine, since universal acceptance of the graft results when standard MGH miniature swine are used in this model(9).
In order to assess potential differences in mitochondrial DNA (mtDNA), the HV1 hypervariable mtDNA region was chosen since this would be expected to be the most sensitive target for determining sequence variations between animals. We reasoned that if there were no sequence differences within this region, then it would be highly unlikely that there would be disparities in more conserved regions of the mitochondrial genome, and thus unlikely that mitochondrial antigenic differences would contribute to transplant outcome. Conversely, observing polymorphic mtDNA variation in hypervariable regions would suggest that variation might exist in other regions of the mtDNA, specifically coding regions, and could contribute to minor antigenic variation. In fact, there were polymorphic variations among published HV1 regions of different swine breeds (Genbank accession numbers AJ002189, NC_000845), as indicated in Fig. 4.
The gene structure and organization of the swine mitochondrial genome is similar to the human (www.mitomap.org), making it relatively easy to identify the corresponding human hypervariable regions in the swine. The HV1 region, like the human hypervariable region (HVS1), is 3′ to the gene for tRNA proline and is present within the swine mitochondrial control region(16). Alignment of the human HSV1 region with the corresponding swine sequence demonstrated a homology of only 48%, as expected for inter-species homologies of hypervariable regions. In comparison, there was about a 70% homology between the entire human and swine mitochondrial sequence.
The mitochondrial HV1 region was found to be identical in all three lines of MGH miniature swine. In addition, the mtDNA from two of these lines, SLAaa and SLAcc, was derived from animals born twenty-five years ago and the sequence was identical to mtDNA isolated from SLAdd animals born within the past three years, suggesting relative stability of this sequence within these inbred lines. Similarly, the mitochondrial HV1 region in one of the highly inbred SLAdd animals had also remained identical. This uniformity of mtDNA in our miniature swine may be due to the fact that all of these lines were originally derived from two founder animals(6). Since mtDNA is maternally inherited, the mtDNA in the single female founder is likely to have survived throughout more than 30 years of subsequent breeding. These data suggest a lower rate of mutation and loss of homogeneity of mtDNA than might have been expected from previous studies of mtDNA inheritance during early oogenesis(2,17,18).
In contrast, all of the knockout animals we have examined differed from the MGH miniature swine at the HV1 sequence. These differences are undoubtedly due to the fact that the mtDNA of the knockout animals was derived from non-MGH swine. In addition, the enucleated oocytes used in each nuclear transfer were isolated from different non-MGH swine. The mitochondrial HV1 sequences of the knockout swine were identical in offspring produced from matings in which all of the females were maternal descendents from the same original nuclear transfer, again indicative of the relative stability of maternally inherited mtDNA during the time frames of these breedings. Taken together, these data suggest that if mitochondrial antigens are encoded by mtDNA with similar or less diversity than that of the HV1 region, then the effects of these antigens should be readily discernible from the outcomes of transplants between animals of the MGH herd and the knockout animals.
The rejection that was observed when kidneys were transplanted from MGH swine into heterozygous knockout swine suggest that the mtDNA antigens of our inbred lines can serve as relatively strong minor antigens, compared to other minor antigens that are not capable of causing rejection using the short course of CyA immunosuppression used here(19). The fact that heterozygous kidneys were not rejected by 30 days in MGH swine may indicate that there are additional mtDNA antigens in MGH swine, possibly expressed at low levels, that are similar enough to the mtDNA antigens of the outbred animals from which the heterozygous animals were derived to tolerize and avoid rejection. Since these grafts were only followed for 30 days before the animals were euthanized for use in a different experiment, we cannot say for certain whether the MGH animals would also have rejected the heterozygous kidney grafts at a later time. Clearly, minor antigens can differ in the strength of the rejection they cause(20).
Another consideration that may support this explanation for transplant outcomes could be the existence of more than one sequence in either the MGH or outbred oocyte donors or both, since mtDNA is present in multiple copies per cell (roughly 1 × 103 copies per cell)(21) and not all of these copies are necessarily identical. Our sequence data would not allow us to determine the true homogeneity of the mtDNA in these animals, since we utilized PCR-amplified HV1 region DNA for the sequencing. The standard PCR techniques used in these studies are biased towards the amplification of the most abundant signal and thus may not identify rare sequences. Should rare sequences also code for minor antigens, then it is possible that either the MGH animals or the heterozygous animals might be tolerant to more “self” mtDNA antigens than the other.
In any case, these results indicate that mitochondrial hypervariable sequence differences can be found between the MGH herd and knockout animals, therefore supporting the possibility that similar differences may exist in those mtDNA antigens that encode expressed proteins that can be detected as minor histocompatibility antigens. Thus, the existence and inheritance pattern of mitochondrial-encoded minor antigens should be considered in any transplant experiments involving animals produced by nuclear transfer.
Footnotes
Since, except for sequencing data, only +/− heterozygous knockout animals (which, like +/+ non-knockout animals, express Gal) were used in these studies, we will refer henceforth to these animals as “knockouts”.
Contribution: J.S.H., K.Y., and D.H.S. designed the research; J.S.H., M.O., A.T., and K.Y. performed the research and collected data; J.S.H. and D.H.S. analyzed the data; S.A. provided the animals; J.S.H. and D.H.S. wrote the paper.
Reference List
- 1.Lai L, Kolber-Simonds D, Park K, et al. Production of a-1,3-Galactosyltransferase Knockout Pigs by Nuclear Transfer Cloning. Science. 2002;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 2.Wagner RP. The role of maternal effects in animal breeding. II. Mitochondria and animal inheritance. J Anim Sci. 1972;35:1280–1287. doi: 10.2527/jas1972.3561280x. [DOI] [PubMed] [Google Scholar]
- 3.Simpson E. Minor transplantation antigens: animal models for human host-versus-graft, graft-versus-host, and graft-versus-leukemia reactions. Transplantation. 1998;65:611–616. doi: 10.1097/00007890-199803150-00002. [DOI] [PubMed] [Google Scholar]
- 4.Morse MC, Bleau G, Dabhi VM, et al. The COI mitochondrial gene encodes a minor histocompatibility antigen presented by H2-M3. J Immunol. 1996;156:3301–3307. [PubMed] [Google Scholar]
- 5.Dabhi VM, Lindahl KF. CTL respond to a mitochondrial antigen presented by H2-Db. Immunogenetics. 1996;45:65–68. doi: 10.1007/s002510050168. [DOI] [PubMed] [Google Scholar]
- 6.Sachs DH, Leight G, Cone J, Schwartz S, Stuart L, Rosenberg S. Transplantation in miniature swine. I. Fixation of the major histocompatibility complex. Transplantation. 1976;22:559–567. doi: 10.1097/00007890-197612000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Pennington LR, Lunney JK, Sachs DH. Transplantation in miniature swine. VIII. Recombination within the major histocompatibility complex of miniature swine. Transplantation. 1981;31:66–71. [PubMed] [Google Scholar]
- 8.Sachs DH. MHC Homozygous Miniature Swine. In: Swindle MM, Moody DC, Phillips LD, editors. Swine as Models in Biomedical Research. Iowa State University Press; Ames, Iowa: 1992. pp. 3–15. [Google Scholar]
- 9.Gianello P, Fishbein JM, Sachs DH. Tolerance to primarily vascularized allografts in miniature swine. Immunol Rev. 1993;133:19–44. doi: 10.1111/j.1600-065x.1993.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 10.Kolber-Simonds D, Lai L, Watt SR, et al. Production of a-1,3-galactosyltransferase null pigs by means of nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc Natl Acad Sci U S A. 2004;101:7335–7340. doi: 10.1073/pnas.0307819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mezrich JD, Haller GW, Arn JS, Houser SL, Madsen JC, Sachs DH. Histocompatible miniature swine: an inbred large-animal model. Transplantation. 2003;75:904–907. doi: 10.1097/01.TP.0000054839.43852.BF. [DOI] [PubMed] [Google Scholar]
- 12.Utsugi R, Barth RN, Lee RS, et al. Induction of transplantation tolerance with a short course of tacrolimus (FK506): I. Rapid and stable tolerance to two-haplotype fully mhc-mismatched kidney allografts in miniature swine. Transplantation. 2001;71:1368–1379. doi: 10.1097/00007890-200105270-00003. [DOI] [PubMed] [Google Scholar]
- 13.Colvin RB. The renal allograft biopsy. Kidney Int. 1996;50:1069–1082. doi: 10.1038/ki.1996.410. [DOI] [PubMed] [Google Scholar]
- 14.Ursing BM, Arnason U. The complete mitochondrial DNA sequence of the pig (Sus scrofa) J Mol Evol. 1998;47:302–306. doi: 10.1007/pl00006388. [DOI] [PubMed] [Google Scholar]
- 15.Alves-Silva J, da Silva SM, Guimaraes PE, et al. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons TJ, Muniec DS, Sullivan K, et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 17.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 18.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 19.Rosengard BR, Ojikutu CA, Guzzetta PC, et al. Induction of specific tolerance to class I disparate renal allografts in miniature swine with cyclosporine. Transplantation. 1992;54:490–497. doi: 10.1097/00007890-199209000-00020. [DOI] [PubMed] [Google Scholar]
- 20.Fuchimoto Y, Gleit ZL, Huang CA, et al. Skin-specific alloantigens in miniature swine. Transplantation. 2001;72:122–126. doi: 10.1097/00007890-200107150-00024. [DOI] [PubMed] [Google Scholar]
- 21.Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31:e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]