Abstract
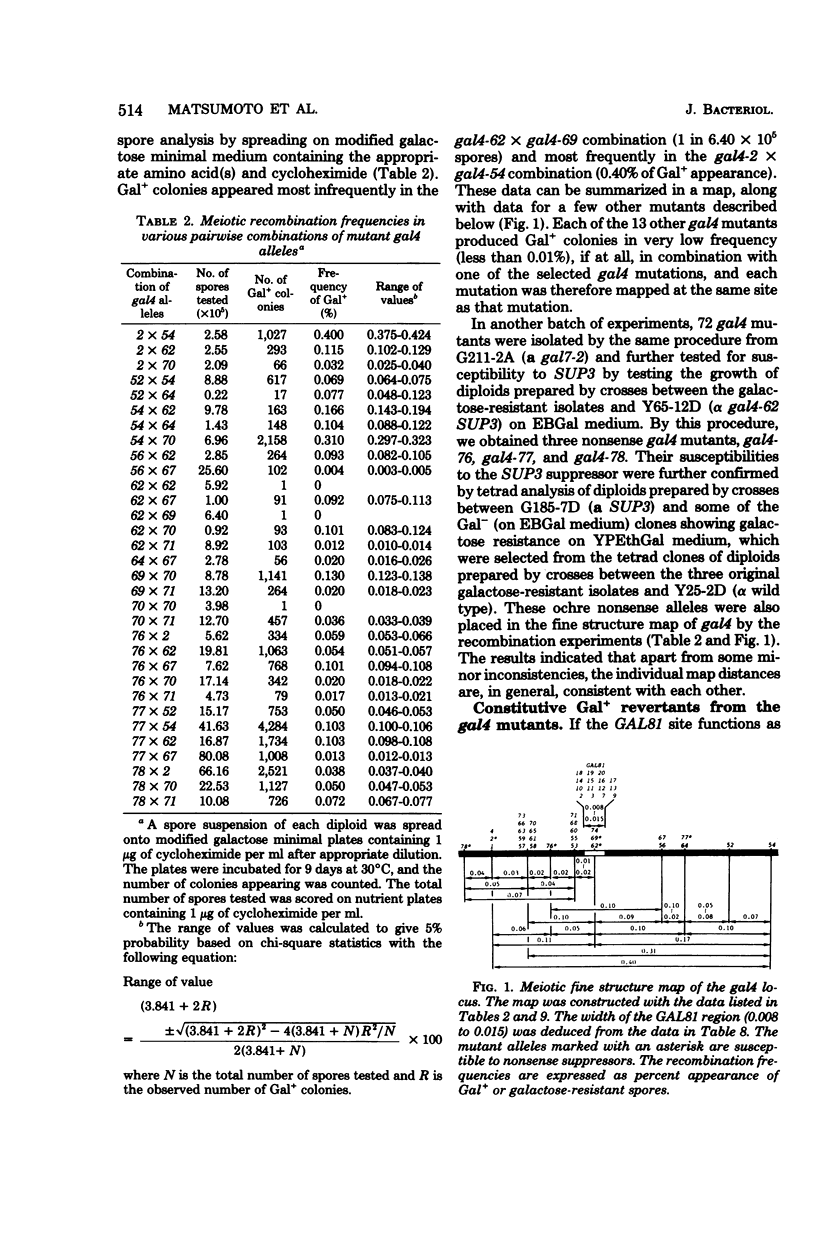
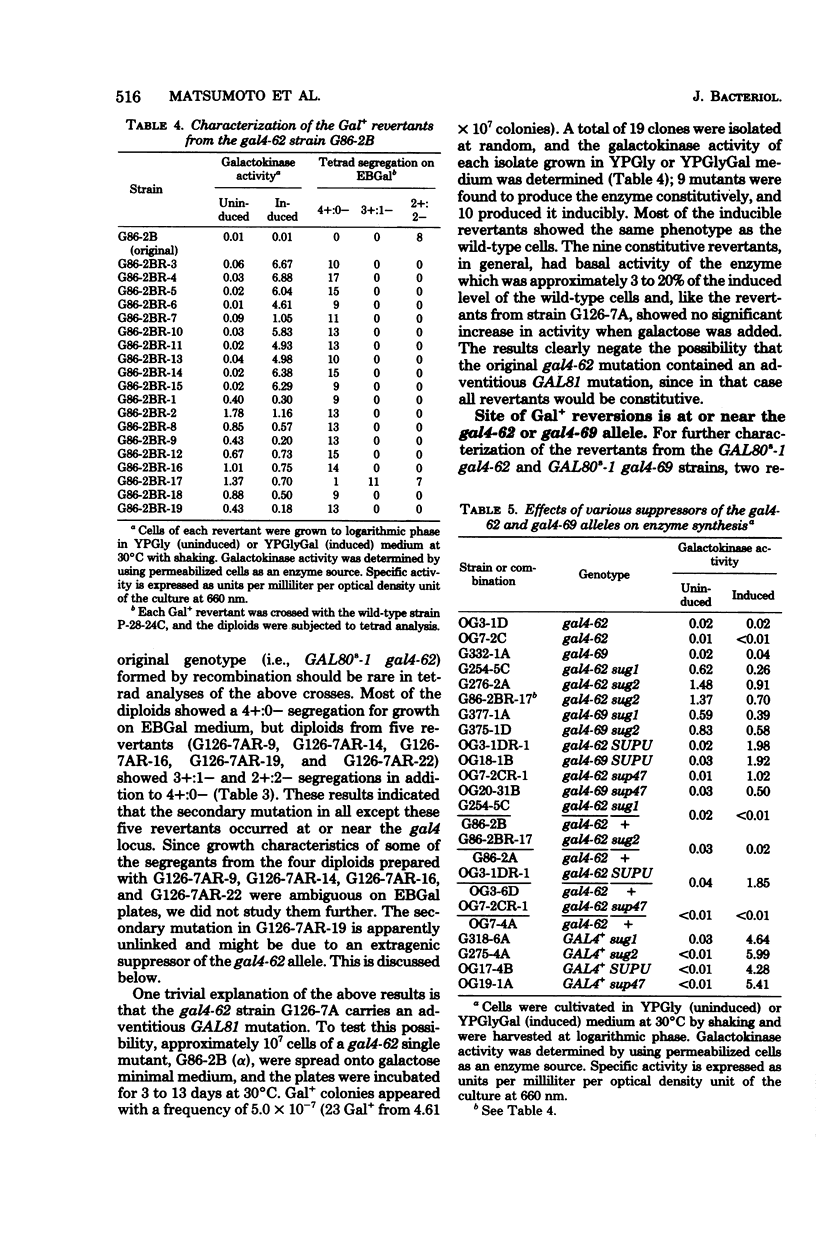
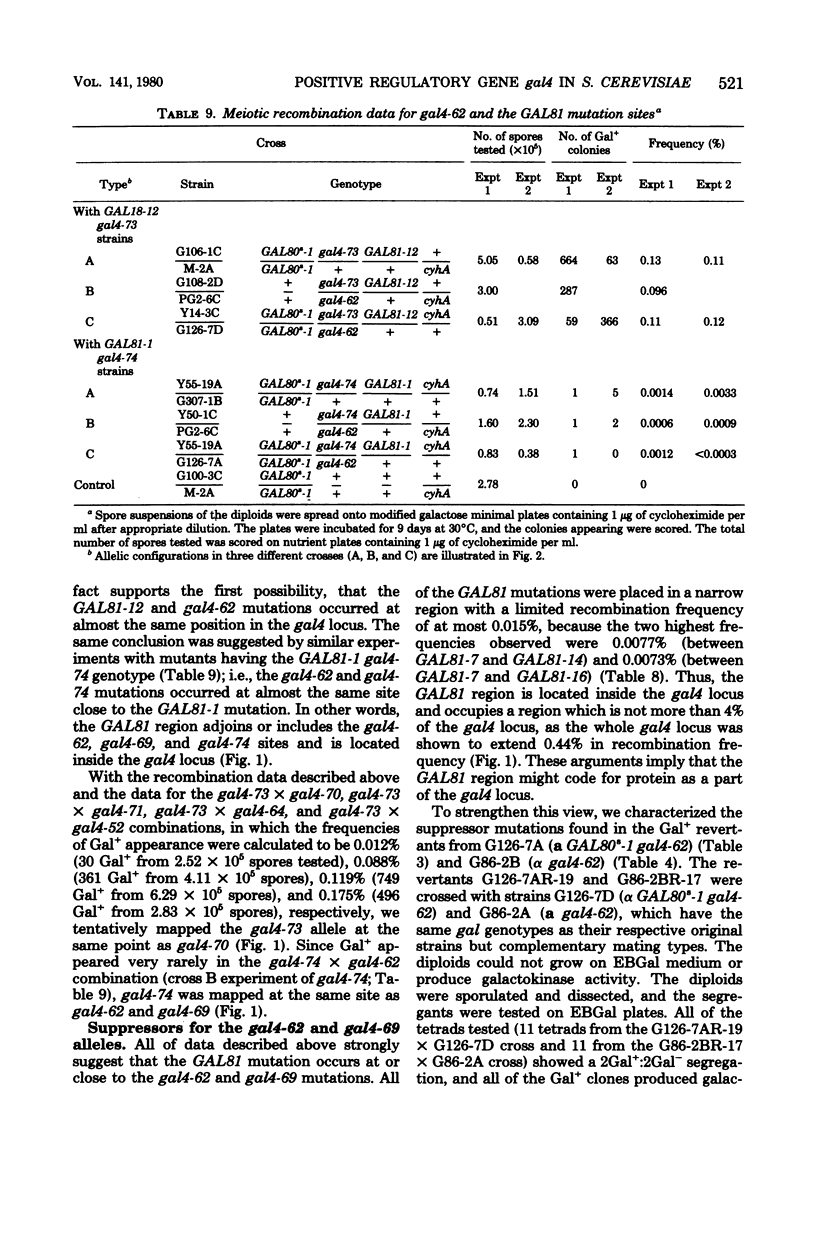
A meiotic fine structure map of the gal4 locus was constructed, which extended over 0.44 units on the chromosome (units in percent frequency of supposed recombination). Several nonsense gal4 mutations (four UAA and two supposed UGA [gal4-62 and gal4-69]) were placed at various sites on the map. In reversion experiments with 20 independently isolated gal4 mutants, only the gal4-62 and gal4-69 alleles, which are located at the same site on the map, could revert to overcome the superrepression of gal80s-1 spontaneously with a frequency of approximately 4 x 10(-7). Secondary mutations in the revertants occurred in the region of gal4-62 or were due to unlinked suppressors. A total of 15 GAL81 mutations in 19 isolates were found to be located in the same region as gal4-62 by three-point crosses with the aid of gal4 mutants; the other four could not be analyzed. The reverted gal4 gene and GAL81 mutations were semidominant over the wild-type GAL4+ allele and fully dominant over a nonsense gal4 mutation. Four suppressors (one dominant and three recessive) effective against gal4-62 and gal4-69 were isolated. The dominant suppressor was also effective against three independent, authentic auxotrophic UGA nonsense mutations, and one of the three recessive suppressors were effective against the authentic auxotrophic UAA and UAG mutations. These results strongly support the idea that the gal4 locus is expressed constitutively and codes for a regulatory protein. The GAL81 site mapped inside the locus codes for a part of the gal4 protein but does not work as an operator.
Full text
PDF
















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassel J., Mortimer R. Genetic order of the galactose structural genes in Saccharomyces cerevisiae. J Bacteriol. 1971 Oct;108(1):179–183. doi: 10.1128/jb.108.1.179-183.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGLAS H. C., HAWTHORNE D. C. ENZYMATIC EXPRESSION AND GENETIC LINKAGE OF GENES CONTROLLING GALACTOSE UTILIZATION IN SACCHAROMYCES. Genetics. 1964 May;49:837–844. doi: 10.1093/genetics/49.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne D. C. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics. 1966 Sep;54(3):911–916. doi: 10.1093/genetics/54.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Rowe L. B. Molecular expression and regulation of the galactose pathway genes in Saccharomyces cerevisiae. Distinct messenger RNAs specified by the Gali and Gal7 genes in the Gal7-Gal10-Gal1 cluster. J Biol Chem. 1978 Oct 25;253(20):7566–7569. [PubMed] [Google Scholar]
- Klar A. J., Halvorson H. O. Effect of GAL4 gene dosage on the level of galactose catabolic enzymes in Saccharomyces cerevisiae. J Bacteriol. 1976 Jan;125(1):379–381. doi: 10.1128/jb.125.1.379-381.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Halvorson H. O. Studies on the positive regulatory gene, GAL4, in regulation of galactose catabolic enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1974;135(3):203–212. doi: 10.1007/BF00268616. [DOI] [PubMed] [Google Scholar]
- Littlewood B. S., Chia W., Metzenberg R. L. Genetic control of phosphate-metabolizing enzymes in Neurospora crassa: relationships among regulatory mutations. Genetics. 1975 Mar;79(3):419–434. doi: 10.1093/genetics/79.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Toh-e A., Oshima Y. Genetic control of galactokinase synthesis in Saccharomyces cerevisiae: evidence for constitutive expression of the positive regulatory gene gal4. J Bacteriol. 1978 May;134(2):446–457. doi: 10.1128/jb.134.2.446-457.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., Sherman F. Role of DNA sequences in genetic recombination in the iso-1-cytochrome c gene of yeast. I. Discrepancies between physical distances and genetic distances determined by five mapping procedures. Genetics. 1975 Mar;79(3):397–418. doi: 10.1093/genetics/79.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Matsumoto K., Toh-e A., Oshima Y. Interaction of super-repressible and dominant constitutive mutations for the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1977 Apr 29;152(3):137–144. doi: 10.1007/BF00268810. [DOI] [PubMed] [Google Scholar]
- Perlman D., Hopper J. E. Constitutive synthesis of the GAL4 protein, a galactose pathway regulator in Saccharomyces cerevisiae. Cell. 1979 Jan;16(1):89–95. doi: 10.1016/0092-8674(79)90190-9. [DOI] [PubMed] [Google Scholar]
- Singh A., Manney T. R. Genetic analysis of mutations affecting growth of Saccharomyces cerevisiae at low temperature. Genetics. 1974 Aug;77(4):651–659. doi: 10.1093/genetics/77.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To-E A., Ueda Y., Kakimoto S. I., Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973 Feb;113(2):727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-E A., Nakamura H., Oshima Y. A gene controlling the synthesis of non specific alkaline phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1976 Mar 25;428(1):182–192. doi: 10.1016/0304-4165(76)90119-7. [DOI] [PubMed] [Google Scholar]
- Toh-E A., Oshima Y. Characterization of a dominant, constitutive mutation, PHOO, for the repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1974 Nov;120(2):608–617. doi: 10.1128/jb.120.2.608-617.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Kakimoto S. Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae. Mol Gen Genet. 1975 Dec 30;143(1):65–70. doi: 10.1007/BF00269421. [DOI] [PubMed] [Google Scholar]
- Toh-e A., Kobayashi S., Oshima Y. Disturbance of the machinery for the gene expression by acidic pH in the repressible acid phosphatase system of Saccharomyces cerevisiae. Mol Gen Genet. 1978 Jun 14;162(2):139–149. doi: 10.1007/BF00267870. [DOI] [PubMed] [Google Scholar]
- Tsuyumu S., Adams B. G. Dilution kinetic studies of yeast populations: in vivo aggregation of galactose utilizing enzymes and positive regulator molecules. Genetics. 1974 Jul;77(3):491–505. doi: 10.1093/genetics/77.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., To-E A., Oshima Y. Isolation and characterization of recessive, constitutive mutations for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Jun;122(3):911–922. doi: 10.1128/jb.122.3.911-922.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valone J. A., Jr, Case M. E., Giles N. H. Constitutive mutants in a regulatory gene exerting positive control of quinic acid catabolism in Neurospora crassa. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1555–1559. doi: 10.1073/pnas.68.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKIE D., LEE B. K. GENETIC ANALYSIS OF ACTIDIONE RESISTANCE IN SACCHAROMYCES CEREVISIAE. Genet Res. 1965 Feb;6:130–138. doi: 10.1017/s0016672300003992. [DOI] [PubMed] [Google Scholar]


