Abstract
Werner syndrome and Bloom syndrome result from defects in the RecQ helicases Werner (WRN) and Bloom (BLM), respectively, and display premature aging phenotypes. Similarly, XFE progeroid syndrome results from defects in the ERCC1-XPF DNA repair endonuclease. To gain insight into the origin of cellular senescence and human aging, we analyzed the dependence of sister chromatid exchange (SCE) frequencies on location [i.e., genomic (G-SCE) vs. telomeric (T-SCE) DNA] in primary human fibroblasts deficient in WRN, BLM, or ERCC1-XPF. Consistent with our other studies, we found evidence of elevated T-SCE in telomerase-negative but not telomerase-positive backgrounds. In telomerase-negative WRN-deficient cells, T-SCE—but not G-SCE—frequencies were significantly increased compared with controls. In contrast, SCE frequencies were significantly elevated in BLM-deficient cells irrespective of genome location. In ERCC1-XPF-deficient cells, neither T- nor G-SCE frequencies differed from controls. A theoretical model was developed that allowed an in silico investigation into the cellular consequences of increased T-SCE frequency. The model predicts that in cells with increased T-SCE, the onset of replicative senescence is dramatically accelerated even though the average rate of telomere loss has not changed. Premature cellular senescence may act as a powerful tumor-suppressor mechanism in telomerase-deficient cells with mutations that cause T-SCE levels to rise. Furthermore, T-SCE-driven premature cellular senescence may be a factor contributing to accelerated aging in Werner and Bloom syndromes, but not XFE progeroid syndrome.
Keywords: T-SCE, WRN, BLM, ERCC1-XPF, Monte Carlo
Understanding the molecular processes that drive cellular replicative senescence is critical to understanding human longevity. Much attention has focused on telomere length as a determinant of replicative capacity (1). Telomeres are nucleoprotein structures composed of species-specific tandemly repeated, G-rich DNA sequences that cap and protect chromosome ends from nucleolytic attack and degradation (2). Telomeres also conceal linear chromosome ends from inappropriate attempts at double-strand break (DSB) repair that might otherwise join chromosomes end-to-end. The preservation of natural chromosome ends and the rejoining of broken DNA ends, both of which are essential for preserving genomic integrity, rely on a common subset of proteins (3), and both decline with advancing age (4, 5).
The G-rich nature of telomeric DNA renders it susceptible to G-quadruplex formation, oxidative damage, and alkylation by electrophiles (6). Telomeres are also unique in their chromatin composition, being bound by the shelterin complex and associated proteins, their T-loop structure, and their nucleosomal organization (7, 8). Telomeres protect genetic information by providing a buffer against the end-replication problem, but their terminal position at the ends of linear DNA poses a special challenge to their own replication (9). Telomerase is a ribonucleoprotein complex consisting minimally of RNA template and catalytic reverse transcriptase subunits responsible for de novo synthesis of telomeric repeats (10). In the absence of telomerase, telomeres shorten with each cell division (11). Critically short telomeres fail to form proper protective end structures, which are sensed as DSBs and trigger a permanent cell-cycle arrest known as senescence (12). Most human tissues lack sufficient telomerase activity to maintain telomere length throughout life, limiting cell division potential. The majority of cancers circumvent this tumor-suppressor mechanism by reactivating telomerase (13), thus removing telomere shortening as a barrier to continuous proliferation. In some situations, a recombination-based mechanism known as “alternative lengthening of telomeres” (ALT) maintains telomere length in the absence of telomerase (14).
Chromosome orientation (CO)-FISH (15) is a strand-specific modification of standard FISH capable of providing information not obtainable by any other means, including detection of sister chromatid exchange (SCE) recombination between telomeres, events termed T-SCE (16). T-SCE are emerging as important features of certain telomerase-negative backgrounds, such as ALT (17) and early embryogenesis before activation of telomerase (18). In the context of combined Werner (WRN) helicase and telomerase deficiency in the mouse, both central features of human Werner syndrome (WS) pathogenesis, elevated T-SCE was observed, with higher rates associated with greater immortalization potential (19). Conditional deletion of the “protection of telomeres” (Pot)1a single-stranded telomere-binding protein has been shown to elicit a DNA damage response at mouse telomeres, as well as aberrant homologous recombination manifested as increased T-SCE (20). Human POT1 stimulates the RecQ helicases WRN and Bloom (BLM) to unwind telomeric DNA (21). Furthermore, in the absence of WRN, leading- and lagging-strand DNA synthesis are uncoupled at replication forks, and POT1 is required for efficient replication of C-rich telomere strands (22). Such studies imply a relationship between T-SCE rate and proliferative potential. However, the complexity of the process—which entails a large number of telomeres, an ongoing exchange process throughout colony growth, exponentially increasing cell number, and independent assortment of chromatids into daughter cells during cell division—precludes a simple analytical approach to understanding this relationship. We therefore turned to theoretical methods in the search for a causal connection between T-SCE and cell proliferation.
Homologous recombination provides the most plausible mechanism by which telomeric DNA could be exchanged between sister telomeres, and in the case of T-SCE, homology can be found at many points on the sister chromatid. Thus, T-SCE need not exchange equal quantities of DNA. In an unequal T-SCE, one sister telomere becomes longer at the expense of the other. Initially, we suggested that in telomerase-deficient backgrounds unequal T-SCE might confer a proliferative growth advantage to a subset of cells that stochastically acquired longer telomeres, enabling their escape from cellular senescence (16). Conversely, those cells inheriting shorter telomeres from an unequal exchange would be at risk for triggering senescence prematurely. Using a theoretical model and Monte Carlo simulations (23), we investigated the possibility that ALT arises from elevated T-SCE levels and concluded it was unlikely unless there was an additional mechanism, such as has been proposed by others (14, 24), through which longer telomeres segregate preferentially into the same daughter cell. This study (23) strongly suggested that in ALT cells, T-SCE coexists with an as yet unknown telomere-lengthening mechanism.
SCE is a relatively common event in normal human cells that occurs along the entire length of a chromosome. Although recognized for over 50 y (25), their biological significance remains neither fully understood nor appreciated. SCE are induced by DNA-damaging agents that block DNA replication (26), and can lead to deletions, translocations, and loss of heterozygosity (27), manifestations of genomic instability and precursors of carcinogenesis. These adverse outcomes likely originate from an inability to resolve replication forks stalled at sites of DNA damage, whereupon SCE provides a means to “bypass” the damage (28). Interestingly, per unit length of DNA, T-SCE are much more frequent than genomic (G)-SCE (16, 29, 30).
We investigated the contribution of three enzymes, each colocalizing with telomeric DNA and linked to accelerated aging and increased cancer risk, to regulation of T- and G-SCE frequencies. WS is a rare, autosomal recessive premature aging disease caused by truncation mutations of WRN, a member of the RecQ helicase family. WRN is the only RecQ helicase to also possess a 3′- to 5′-exonuclease domain, the exact function of which is unclear, but which may assist in forming “reverse chicken-foot structures” at stalled replication forks (31).
Bloom syndrome (BS) is also a rare, autosomal recessive disease associated with segmental, or tissue-specific, premature aging and a high risk of developing a broad spectrum of cancers, primarily carcinomas (32). The defective protein in BS, BLM, is also a RecQ helicase. Extraordinarily elevated levels of G-SCE are diagnostic of the disease and BS cells are exquisitely sensitive to treatment with the interstrand cross-linking agent mitomycin C, supporting the concept that BS patients suffer from an inability to resolve complex DNA structures that stall DNA replication. The helicase activities of both WRN and BLM facilitate resolution of such DNA structures (e.g., crosslinks and G-quadruplexes), thus facilitating progression of the replication machinery (33).
XFE progeroid syndrome is caused by mutations in XPF, which result in reduced expression of ERCC1-XPF (34). ERCC1-XPF is an endonuclease required for nucleotide excision repair and repair of DNA interstrand crosslinks (35). In the absence of ERCC1-XPF, replication of cross-linked DNA leads to an accumulation of DSBs and increased chromosomal aberrations rather than elevated SCE. This suggests that although BLM normally functions to suppress homologous recombination and SCEs, ERCC1-XPF promotes them in response to at least some types of replication stress. Of particular interest, ERCC1-XPF colocalizes with the shelterin protein TRF2 and promotes end-fusion of critically short telomeres (36).
Investigation of cells defective in WRN, BLM, and ERCC1-XPF affords a unique opportunity to decipher the contributions of T- and G-SCE to cellular senescence. Herein we provide evidence that increased T-SCE frequency can dramatically accelerate cellular senescence of telomerase-negative cells, and that this mechanism likely functions as a potent tumor-suppressor.
Results
WRN Suppresses SCE Recombination Specifically in Telomeric DNA.
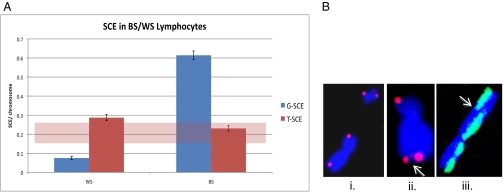
Human EBV-immortalized WS lymphoblasts (telomerase-positive) did not display elevated levels of G-SCE (0.08 G-SCE per chromosome); shaded regions on the graph in Fig. 1 represent our established normal human background levels. WS lymphoblasts also displayed normal background T-SCE frequencies (0.29 T-SCE per chromosome) (Fig. 1).
Fig. 1.
G- and T-SCE frequencies in immortalized human WS and BS lymphoblasts (telomerase-positive). (A) G-SCE frequencies were determined using fluorescence-plus-Giemsa staining, and T-SCE frequencies were determined using CO-FISH. The shaded region represents range of typical frequencies seen in other human cell lines. Error bars were calculated using SEM. (B) Illustrative examples of (i) telomere CO-FISH, characteristic strand-specific (single-sided) signals; (ii) T-SCE, CO-FISH telomere signal split between sister chromatids (arrow); and (iii) G-SCE, color switch between sister chromatids (arrow).
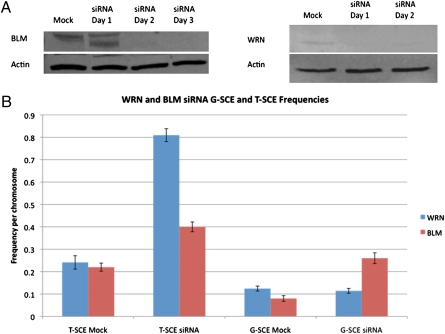
To evaluate the consequence of WRN and BLM deficiency on T-SCE frequencies in the absence of telomerase, we used an RNA interference siRNA strategy to knock down their expression in normal human primary fibroblasts (telomerase-negative). The WRN-depleted cells displayed no significant increase in G-SCE frequencies vs. the mock control (0.12 vs. 0.11) (Fig. 2), although a statistically significant (P < 0.05) increase in T-SCE was observed (0.81 vs. 0.25) (Fig. 2). These results are consistent with our previous study using cells derived from Wrn−/−,Terc−/− double-knockout mice (19).
Fig. 2.
Small interfering RNA knockdown of WRN and BLM in normal human fibroblasts (telomerase-negative). (A) Western blot analyses of protein levels demonstrate efficient knockdown following siRNA transfection. (B) Evaluation of G- and T-SCE frequencies in mock- and siRNA-transfected cells revealed elevated T-SCE upon depletion of either WRN or BLM, although only BLM-depletion resulted in an elevation of G-SCE frequency.
BLM Suppresses SCE Recombination Globally.
In contrast to WS lymphoblasts, human EBV-immortalized BS lymphoblasts (telomerase-positive) displayed a dramatic elevation of G-SCE (0.61) (Fig. 1). Similar to WS lymploblasts, BS lymphoblasts displayed background T-SCE frequencies (0.23) (Fig. 1). Shaded regions on the graph in Fig. 1 represent our established normal human background levels for both G- and T-SCE.
We hypothesized that BLM may perform a reciprocal function to WRN, in that BLM may be restricted to suppressing G-SCE, but WRN involvement is in suppressing T-SCE. However, siRNA knockdown of BLM in normal human fibroblasts (telomerase-negative) demonstrated a role for BLM in suppressing both G- and T-SCE. Depletion of BLM in the absence of telomerase again revealed a significant increase in G-SCE frequency vs. the mock control (0.26 vs. 0.08) (Fig. 2), as well as a significant increase in T-SCE (0.40 vs. 0.22) (Fig. 2).
ERCC1-XPF Does not Influence SCE.
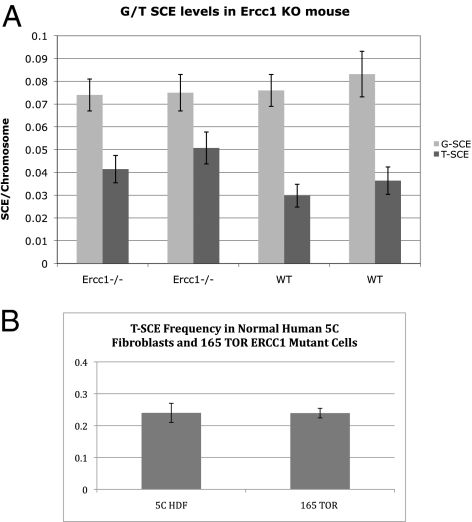
To evaluate the role of ERCC1-XPF in SCE regulation, we used four congenic primary mouse embryonic fibroblast (MEF) cell lines (telomerase-positive), two of which were wild-type and two Ercc1−/−. No significant difference in G-SCE frequencies was observed between the wild-type (0.07, 0.12) (Fig. 3) and the Ercc1−/− (0.07, 0.07) (Fig. 3) MEFs. T-SCE frequencies were assessed and no significant difference was detected between wild-type (0.029, 0.036) (Fig. 3) and Ercc1−/− (0.041, 0.050) (Fig. 3). These results highlighted the necessity of analyzing ERCC1’s T-SCE role in a telomerase-negative background.
Fig. 3.
G- and T-SCE frequencies with Ercc1 deficiency. (A) G- and T-SCE frequencies in two independent Ercc1−/− and congenic wild-type primary MEF lines (telomerase-positive). No significant difference in G- or T-SCE frequencies between the two genotypes was observed. (B) T-SCE frequencies in ERCC1-deficient human patient primary fibroblasts (165TOR). Even in this telomerase-negative background, no difference in T-SCE frequencies was observed as compared with normal human fibroblasts. G-SCE frequencies were not determined.
We attempted siRNA knockdown of ERCC1 in normal human fibroblasts (telomerase-negative). Although similar knockdown of either WRN or BLM was successful, ERCC1 proved to be problematic. ERCC1 siRNA-transfected human primary fibroblasts ceased growth almost immediately and metaphases could not be obtained for analysis. Similarly, deletion of Ercc1 by transfecting Ercc1−/cond mouse embryonic stem cells with a plasmid expressing Cre was not achieved in over 1,000 clones screened. As further testament to the requirement for ERCC1 for survival, only one patient with a mutation in ERCC1 has been reported to date (37).
We sought to evaluate the role of ERCC1 in T-SCE recombination in primary cells from the ERCC1 patient, 165TOR, who despite mutations in the gene, maintained diminished but detectable ERCC1-XPF nuclease activity, allowing survival until 14 mo of age (37). The 165TOR cells grew poorly, even in a low-oxygen environment, but sufficient metaphase spreads for T-SCE analysis were obtained. The ERCC1-deficient cells did not display a T-SCE phenotype; background T-SCE frequencies were identical to those of normal human dermal fibroblast (5C) controls (0.24 vs. 0.24) (Fig. 3). The 165TOR cells did not grow well enough for G-SCE analysis. However, G-SCE levels determined in Ercc1−/− MEFs (Fig. 3) and ES cells (38) revealed no significant difference compared with wild type, and G-SCE frequencies are not influenced by telomerase status.
Senescence in Silico.
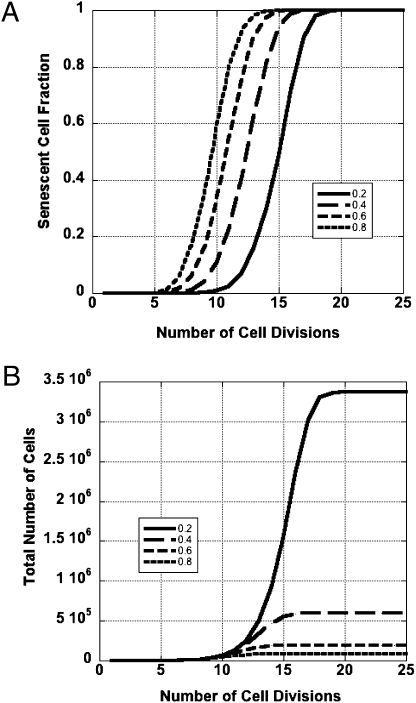
To understand the role T-SCE play in replicative senescence, we developed a theoretical model of cell proliferation in the presence of telomere recombination (23) and studied in silico the dependence of proliferative potential on T-SCE rate. Although a simple idealization of the model in which cells have one telomere is analytically tractable (39), in our experiments human cells had 92 telomeres. Because the model is analytically intractable when many telomeres participate in exchanges, we studied it using Monte Carlo simulations. In all cases we assumed that a single, critically short telomere triggers senescence, which is consistent with experimental data (40). Two possibilities were considered: one in which only specific chromosomes participated in T-SCE, and one in which T-SCE were randomly distributed among all telomeres. In the former case, at each cell division, only the chosen telomeres may exchange (quenched case), but in the latter case the chromosomes that exchange telomere material are “chosen” randomly (annealed) from the ensemble of all telomeres in the cell. In the quenched case, if only a few telomeres participate and these are the short telomeres in the cell, the model indicates that increasing T-SCE rates will prolong the proliferative potential of cells growing into colonies (23, 24). However, T-SCE rates observed in this study are sufficiently high so as to indicate that many telomeres participate in T-SCE during each cell division, and although not specifically evaluated, there was no indication of preferential participation of a subset of chromosomes. To address this situation we studied the growth of hypothetical cells with 46 chromosomes into colonies. In the model, the length of each telomere shortened with each cell division, just as occurs in real cells. Whether or not particular sister telomeres engage in T-SCE is chosen at random, with a probability determined by the T-SCE rate at each cell division. To simulate unequal T-SCE, the two breakpoints of each exchange were chosen at random locations within the two sister telomeres. Each exchange is always balanced such that for a pair of sister telomeres, there is no net gain or loss of telomeric DNA. Sister chromatids are randomly segregated into daughter cells. If a daughter cell inherited or, because of the end-replication problem, obtained one or more telomeres shorter than a critical length, the cell “senesced.” A senescent cell contributed to the number of cells in a colony but could not undergo further cell divisions. Through each simulated cell division, we kept track of the number of cells in a colony, whether each cell was senescent or not, and the telomere lengths of all chromosome ends.
Before running a simulation, a set of initial conditions was chosen as follows: all telomere lengths were initially 3-kg bases (kb), the rate of telomere loss was 100 bases per telomere per cell division, and the critical telomere length was 500 bases. For any set of chosen conditions, the simulation was run 64 times corresponding to 64 cells growing into colonies. The T-SCE rate was set to four values between 0.2 and 0.8 T-SCE per chromosome per cell division. The results were averaged and graphed, and are displayed in Fig. 4 as the senescent cell fraction (fraction of a colony composed of senescent cells) vs. time expressed in units of cell divisions. For comparison, with a T-SCE rate of zero, all cells in a colony would senesce simultaneously when the initial shortest telomere dropped below the critical length, in this case after 26 cell divisions. All cells in our simulated colonies eventually senesced, and in no case did we observe prolonged colony growth. In fact, for each nonzero T-SCE rate, colony growth ceased well short of the point in which colonies with no T-SCE would stop expanding. Thus, contrary to our original hypothesis (16) and in agreement with our subsequent study (23), T-SCE in and of themselves are not able to overcome telomere-driven colony senescence. The model allowed us to investigate further the role of telomere number on colony growth, something that would be difficult to do experimentally. In cells with few telomeres, prolonged colony growth is indeed possible, and in the special (and biologically irrelevant) case of cells with one telomere per cell, an infinite proliferative potential could be achieved above a critical T-SCE rate (23, 39). However in a model of human cells, to prolong colony growth through unequal T-SCE, all 92 telomeres of a cell would need to remain above the critical limit in every division cycle. In contrast, early cell senescence requires no more than one telomere to shorten below the critical limit. Thus, premature replicative senescence would be the probabilistically favored outcome of unequal T-SCE.
Fig. 4.
Senescence in silico. (A) T-SCE accelerate colony senescence. Computer simulations were performed in which colonies grew from single cells. The four curves correspond to T-SCE levels covering a range from 0.2 to 0.8 T-SCE per chromosome per cell division. As T-SCE rates rise, so does the fraction of the colony composed of senescent cells at any cell division. (B) T-SCE constrain colony growth. The four curves generated through computer simulation demonstrate that T-SCE can have a strong negative impact on colony size. With increasing T-SCE rates colonies increase in size more slowly, and the average colony size attained when all cells have senesced decreases almost exponentially.
With any nonzero T-SCE rate, senescent cells accumulate gradually over many cell divisions until a point is reached where the colony possesses only senescent cells. Comparing the curves in Fig. 4 leads to important insight: T-SCE are remarkably effective at accelerating cellular replicative senescence. The implication is that WS and BS cells, with their high T-SCE rates, will have a significantly reduced capacity for colony growth. Therefore, the model predicts WRN cells will enter senescence with longer average telomere length than normal cells. This prediction is supported by experimental data (41). The smoothness of the coherent time evolution as we vary T-SCE levels stems from the large number of cells that were used in the simulations. Fluctuations in the Monte Carlo results (SEM) are expected to be on the order of the inverse square root of the product of the number of cells and the number of telomeres in each cell, a quantity that is negligible. Further studies will quantify more precisely the influence of biological factors that determine the efficacy of the ATM/p53 response to critically short telomeres, as they may be crucial for understanding the susceptibility of WS patients to cancer.
Just how profoundly colony growth may be constrained by T-SCE is illustrated in Fig. 4, where the total numbers of cells in an expanding colony are plotted vs. the number of cell divisions for several T-SCE rates. If there were no T-SCE, a colony would grow to 67 million cells in 26 cell divisions. In contrast, over the 4-fold range of T-SCE rates in our simulation, the average maximum colony size is reduced from more than one order of magnitude to nearly three orders of magnitude.
Discussion
Study of accelerated aging phenotypes affords insight into normal human aging processes, as well as the age-related increase in cancer incidence. Because of the intimate relationship between aging and cancer, monitoring the dynamic nature of telomere function can provide valuable biomarkers of underlying determinants of both cellular aging and carcinogenesis. We interrogated telomeric recombination in the premature aging syndromes WS and BS and in the recently identified progeroid-like syndrome XFE associated with ERCC1-XPF deficiency (34). Theory and experiment were combined to relate the obtained recombination rates to the proliferative capacity of human cells.
We previously observed elevated levels of T-SCE in a murine model of combined Wrn and Terc knockout (42), in which the context of a telomerase-negative background was required to reproduce a classic human WS phenotype (19). Here, these studies were extended in both murine gene knockout and normal human fibroblast gene-expression knockdown models. WRN suppressed recombination specifically in telomeric DNA, but BLM played a more global role in suppressing SCE recombination. BLM and WRN likely function to restrain T-SCE by resolving stalled replication forks, which occur frequently within telomeric DNA; individually, this restraint is incomplete, as either protein alone does not fully compensate for loss of the other. We speculate that either there is an insufficient amount of BLM at telomeres, or perhaps more likely, that the two RecQ helicases act on different DNA lesions/structures (e.g., cross-links vs. G-quadruplexes). It is also possible that strand-specific mechanisms deal with DNA lesions dependent on whether they reside on the strand replicated by leading- or lagging-strand synthesis (22, 43).
The separate roles WRN and BLM play in resolving stalled replication forks may depend on the nature and position of the initiating lesion. If the replication fork arrests because of a lesion or structure occurring in the G-rich telomeric strand (such as a G-quadraplex structure), a 3′ end would be generated, a substrate suitable for initiating RAD51-dependent strand invasion. BLM is known to associate with RAD51 (44), so may function in assisting strand-specific strand invasion and resolution of the resulting Holliday junction.
In contrast, a replication-fork stalling lesion or structure occurring on the C-rich telomeric strand would generate a 5′ end, an unsuitable substrate for initiation of strand invasion. Therefore, other mechanisms must be enlisted for resolution, such as formation of what has been termed a reverse chicken foot, in which newly synthesized DNA can collapse and use itself as a template for replication. After which, it can reverse, bypassing the lesion or prohibitive structure, and replication can proceed (28). It has been proposed that the exonuclease domain on WRN, not found in any other RecQ helicase, may assist in this processing (45). An important aspect of this model is that replication of the strands can be uncoupled—that is, one strand's replication can proceed if the other is stalled—allowing both strand invasion and reverse chicken-foot structure formation to occur.
Examination of cells from the only known human patient presenting with accelerated aging associated with ERCC1-XPF deficiency revealed no influence on recombination in either telomeric or genomic DNA (i.e., T- and G-SCE frequencies were not affected in this background). Either the residual level of ERCC1-XPF activity in the individual was sufficient to circumvent an elevated SCE phenotype, or ERCC1-XPF is not a suppressor of T-SCE and may in fact be required for their formation. Regardless, the premature aging associated with this case is not likely to be mediated by telomere hyper recombination. Instead, premature aging might have been caused by nontelomere-related mechanisms of replicative senescence, or by depletion of telomere reserves brought on by an elevated rate of cell turnover to compensate for frequent cell death.
A WRN cell culture can be thought of as a collection of expanding colonies, each burdened by a type of telomere instability that drives the culture as a whole toward accelerated senescence. Computer simulations suggest that higher T-SCE rates lead to smaller colony sizes after the same number of divisions. Because senescent cells in a colony are incapable of cell division, nonsenescent WS cells have to divide more times to repopulate a colony in comparison with cells without the WRN mutation, which almost certainly contributes to the senescence of WS cell cultures after fewer in vitro passages.
How might elevated T-SCE contribute to cancer risk and premature aging? An in vivo study of the tissues of baboons found the proportion of senescent cells increased with age rising to about 20% in the oldest animals (46). This study suggests that a senescent cell fraction of 0.2 may sufficiently disrupt organ function to cause the effects of advanced old age. Our in silico study implies this level of replicative senescence would be reached at an earlier chronological age when the T-SCE rate is high. Colony growth impairment during the growth spurt of the teen years may also contribute to the small stature of adult WS patients. A causal connection to cancer is likely to be more complex. Compared with normal individuals of the same age, high T-SCE rates cause WS and BS patients to challenge the ATM/p53 guardian barrier with a larger number of cells having short telomeres. On the other hand, accelerated replicative senescence is likely to terminate many premalignant clones before they acquire a full suite of oncogenic mutations, thereby suppressing the rate of tumor formation. With very few parameters, our model can be directly related to experiments with cultured cells. The model, however, is limited in its application to the age-related pathology of tissues and organisms because it lacks important mechanisms, such as genomic instability, reactivation of telomerase, and bypass of checkpoints en route to malignant transformation. In future studies, we hope to incorporate these mechanisms to gain a better appreciation for the interrelationship between cancer and aging, and their mutual dependence on telomere fitness.
In the quest to identify the cause of human aging, much attention has focused on replicative senescence driven by telomere loss. Our studies strongly implicate T-SCE as a second potent driver of replicative senescence. To accurately predict a cell's replicative fate, T-SCE evaluation should be included in studies attempting to relate genetics or lifestyle factors to telomere dynamics and aging.
Materials and Methods
Cell Lines.
Human WS and BS lymphoblasts were obtained from Coriell (WS-AG04103 and BS-GM16375, respectively). Both were grown in RPMI 1640 (HyClone) supplemented with 15% FBS (Sigma-Aldrich) and 1% pen/strep (HyClone). Low passage normal human dermal fibroblasts (Cascade Biologics #C-004–5C) were maintained in α-MEM medium (HyClone) supplemented with 15% FBS (Sigma Aldrich) and 1% pen-strep (HyClone). Ercc1−/− primary MEFs were generated as per ref. 34. Primary human fibroblasts from ERCC1 patient 165TOR were obtained and maintained as previously described (37). All cell lines were incubated at 37 °C in an atmosphere of 95% air and 5% carbon dioxide.
G-SCE Analysis.
Fluorescence-plus-Giemsa was performed according to protocol of Perry and Wolff with some modification (47). Cells were trypsinized and subcultured into medium containing 1 × 10−5M of the thymidine analog BrdU (Sigma-Aldrich) for two cell cycles. Colcemid (0.1 mg/mL; Invitrogen) was added during the final 3 to 4 h to accumulate mitotic figures. Slides were air dried and stained with Hoechst 33258 (0.50 mg/mL; Sigma-Aldrich) for 15 min, exposed to 365 nm UV light (Stratalinker 2400) for 25 min, then stained in 2% Giemsa (Fisher Scientific). Harlequin (differential) staining was visualized with light microscopy and SCE scored as a “color switch” between sister chromatids.
T-SCE Analysis.
CO-FISH was performed as previously described (48) and in SI Materials and Methods. Briefly, cells were subcultured into medium containing BrdU (1 × 10−5M; Sigma-Aldrich) for one cell cycle. Slides were stained with Hoechst 33258 (0.50 μg/mL; Sigma-Aldrich) for 15 min and exposed to 365 nm UV light (Stratalinker 2400) for 25 min. BrdU-incorporated strands were digested with Exonuclease III (3 U/μL; Promega) for 10 min. A Cy-3 conjugated (TTAGGG)3 PNA telomere probe (0.2 μg/mL; Applied Biosystems) was hybridized at 37 °C for 1.5 h. Chromosomes were counterstained with DAPI (Vectashield, Vector Laboratories). Preparations were examined and images captured. T-SCE was scored as a CO-FISH telomere signal split between the two chromatids of a metaphase chromosome.
Statistical Analysis.
A minimum of 50 metaphases per sample was scored, unless 50 scorable metaphases were not available. Error bars were calculated using SEM, and SCE frequencies were calculated on a per chromosome basis.
Small Interfering RNA Transfection.
The siRNA sequences (20 nM) (SI Materials and Methods) were incubated for 30 min in HiPerfect transfection reagent (Qiagen) and Opti-MEM (Invitrogen) at a 2.5:1,000 ratio (2.5 μL transfection reagent/1,000 μL of Opti-MEM), and then added to exponentially growing cells in culture media without FBS or antibiotics. Cells were incubated for 6 h at 37 °C, then culture media with FBS and pen/strep was re-added. Lystates were collected after 24 to 72 h, dependent on time of maximum knockdown for each protein.
Western Blot Analysis.
See SI Materials and Methods for details. The following primary antibodies and their corresponding ratios were used: WRN (c-19) antibody (1:1,000; Santa Cruz), BLM(RBX) antibody [1:1,000; Chemicon/Upstate (Millipore)], actin (1:250; Santa Cruz). The following secondary antibodies were used for detection: for WRN, donkey anti-goat HRP (1:1,000; Santa Cruz); for BLM, goat anti–rabbit HRP (1:1,000; Santa Cruz); for actin, (1:1,000; Santa Cruz).
Supplementary Material
Acknowledgments
We thank Ms. Christine Battaglia for expert technical assistance and critical reading of the manuscript. This research was supported in part by the National Aeronautics and Space Administration (NNJ04HD83G and NNX08AB65G), National Institute of Environmental Health Sciences Grant ES16114 (to L.J.N.), and Ellison Medical Foundation Grant AG-NS-0303-05 (to L.J.N.). K.B.B. was supported by the National Science Foundation while working at the Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006338107/-/DCSupplemental.
References
- 1.Royle NJ, et al. The role of recombination in telomere length maintenance. Biochem Soc Trans. 2009;37:589–595. doi: 10.1042/BST0370589. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Sullivan RJ, Karlseder J. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Sedelnikova OA, et al. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 6.von Zglinicki T. Role of oxidative stress in telomere length regulation and replicative senescence. Ann N Y Acad Sci. 2000;908:99–110. doi: 10.1111/j.1749-6632.2000.tb06639.x. [DOI] [PubMed] [Google Scholar]
- 7.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 8.Pisano S, Galati A, Cacchione S. Telomeric nucleosomes: Forgotten players at chromosome ends. Cell Mol Life Sci. 2008;65:3553–3563. doi: 10.1007/s00018-008-8307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 10.Greider CW, Blackburn EH. Tracking telomerase. Cell. 2004;116(2 Suppl):S83–S86. doi: 10.1016/s0092-8674(04)00053-4. 1 p following S86. [DOI] [PubMed] [Google Scholar]
- 11.Shore D, Bianchi A. Telomere length regulation: Coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 13.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 14.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 15.Bailey SM, Goodwin EH, Cornforth MN. Strand-specific fluorescence in situ hybridization: The CO-FISH family. Cytogenet Genome Res. 2004;107:14–17. doi: 10.1159/000079565. [DOI] [PubMed] [Google Scholar]
- 16.Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–1143. doi: 10.1038/sj.embor.7400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 19.Laud PR, et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 21.Opresko PL, et al. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 22.Arnoult N, Saintome C, Ourliac-Garnier I, Riou JF, Londoño-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blagoev KB, Goodwin EH. Telomere exchange and asymmetric segregation of chromosomes can account for the unlimited proliferative potential of ALT cell populations. DNA Repair (Amst) 2008;7:199–204. doi: 10.1016/j.dnarep.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005 doi: 10.1093/hmg/ddi266. 14 Spec No. 2: R191–R196. [DOI] [PubMed] [Google Scholar]
- 25.Taylor JH. Sister chromatid exchanges in tritium-labeled chromosomes. Genetics. 1958;43:515–529. doi: 10.1093/genetics/43.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi Y, Schneider EL. In vivo sister-chromatid exchange: A sensitive measure of DNA damage. Mutat Res. 1979;60:329–337. doi: 10.1016/0027-5107(79)90023-x. [DOI] [PubMed] [Google Scholar]
- 27.Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Wilson DM, 3rd, Thompson LH. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007;616:11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Cornforth MN, Eberle RL. Termini of human chromosomes display elevated rates of mitotic recombination. Mutagenesis. 2001;16:85–89. doi: 10.1093/mutage/16.1.85. [DOI] [PubMed] [Google Scholar]
- 30.Rudd MK, et al. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 2007;3:e32. doi: 10.1371/journal.pgen.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickson ID. RecQ helicases: Caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 34.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 35.Niedernhofer LJ, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu XD, et al. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 37.Jaspers NG, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niedernhofer LJ, et al. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antal T, Blagoev KB, Trugman SA, Redner S. Aging and immortality in a cell proliferation model. J Theor Biol. 2007;248:411–417. doi: 10.1016/j.jtbi.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 41.Schulz VP, et al. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 42.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 43.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 44.Lahkim Bennani-Belhaj K, et al. The Bloom syndrome protein limits the lethality associated with RAD51 deficiency. Mol Cancer Res. 2010;8:385–394. doi: 10.1158/1541-7786.MCR-09-0534. [DOI] [PubMed] [Google Scholar]
- 45.Sowd G, Wang H, Pretto D, Chazin WJ, Opresko PL. Replication protein A stimulates the Werner syndrome protein branch migration activity. J Biol Chem. 2009;284:34682–34691. doi: 10.1074/jbc.M109.049031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 47.Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974;251:156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- 48.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.