Abstract
Genetic conflicts between sexes and generations provide a foundation for understanding the functional evolution of sex chromosomes and sexually dimorphic phenotypes. Y chromosomes of Drosophila contain multi-megabase stretches of satellite DNA repeats and a handful of protein-coding genes that are monomorphic within species. Nevertheless, polymorphic variation in heterochromatic Y chromosomes of Drosophila result in genome-wide gene expression variation. Here we show that such naturally occurring Y-linked regulatory variation (YRV) can be detected in somatic tissues and contributes to the epigenetic balance of heterochromatin/euchromatin at three distinct loci showing position-effect variegation (PEV). Moreover, polymorphic Y chromosomes differentially affect the expression of thousands of genes in XXY female genotypes in which Y-linked protein-coding genes are not transcribed. The data show a disproportionate influence of YRV on the variable expression of genes whose protein products localize to the nucleus, have nucleic-acid binding activity, and are involved in transcription, chromosome organization, and chromatin assembly. These include key components such as HP1, Trithorax-like (GAGA factor), Su(var)3–9, Brahma, MCM2, ORC2, and inner centromere protein. Furthermore, mitochondria-related genes, immune response genes, and transposable elements are also disproportionally affected by Y chromosome polymorphism. These functional clusterings may arise as a consequence of the involvement of Y-linked heterochromatin in the origin and resolution of genetic conflicts between males and females. Taken together, our results indicate that Y chromosome heterochromatin serves as a major source of epigenetic variation in natural populations that interacts with chromatin components to modulate the expression of biologically relevant phenotypic variation.
Keywords: evolution, heterochromatin, position-effect variegation, regulatory
The Y chromosome is a prime target for the evolution and resolution of genetic conflicts related to the distortion of sex ratios, the evolution of male-limited traits, and antagonistic parent-of-origin influences on male and female progeny (1). Despite theoretical expectations regarding potentially broader roles for male-limited genetic elements, the functional relevance of polymorphic variation in Y chromosomes has mostly been overlooked in both theoretical and empirical studies, although noteworthy exceptions can be found (1–3). The reasons for this neglect stem from the unusual molecular characteristics of the Y chromosome. First, Y chromosomes show dramatically sparse gene numbers with a limited and specialized functional profile; in Drosophila, 13 protein-coding genes are assigned to the Y chromosome, each of which is expressed only during spermatogenesis (4, 5). Second, Y chromosomes have dramatically lower levels of polymorphic, single-nucleotide genetic variation compared with genes in other chromosomes; indeed, theoretical models and sequencing studies have led to the widespread view that Y-linked, single-copy protein coding genes are monomorphic within species of Drosophila (6). In contrast, the Y chromosome shows a great deal of structural polymorphism, as evidenced by the variation in copy number of repeated sequences (7).
The absence of sequence polymorphisms in Y-linked protein-coding genes has cast a shadow of doubt on phenotypic data showing polymorphic Y-linked effects on fitness (8), temperature sensitivity of spermatogenesis (9), sex ratio distortion (3), and behavior (10). Furthermore, population genetic theory suggests that the conditions for stably maintaining Y-linked polymorphisms might be limited (11). The fundamental conundrum has been to reconcile evidence indicating polymorphic phenotypic effects of Y chromosomes with a lack of protein sequence variation (12). Remarkably, the amount of Y-linked DNA (40 Mb) constitutes >20% of the Drosophila genome; based on the large size of the Y chromosome and gene density in the X chromosome, >5,000 genes might be expected to be Y-linked. That only 13 protein-coding genes are located in the Y chromosome underscores the heterochromatic content of the chromosome in the form of multi-megabase stretches of satellite DNA. Thus, the discovery that Drosophila Y chromosomes have polymorphic effects on gene regulation was unexpected (13). This discovery suggests mechanisms through which heterochromatic variation might promote functional variation with consequences for various downstream phenotypes, including fitness.
Heterochromatin represents a large fraction of eukaryotic genomes and is characterized by a high density of sequence repeats that remain condensed through the cell cycle (6, 14, 15). Furthermore, euchromatic and heterochromatic environments present distinct and sometimes opposing requirements for the expression of protein-coding genes. Euchromatic genes are silenced on insertion into heterochromatin, whereas genes that natively reside within heterochromatin might be repressed on their translocation to euchromatin (6, 16). In Drosophila, manipulating the amount of Y-linked heterochromatin results in variable gene expression, with larger amounts of Y-linked heterochromatin leading to decreased heterochromatization of autosomal markers at the boundary between euchromatin and heterochromatin (6, 17).
Thus, the finding of YRV led to the proposal that the Y chromosome might have evolved to become a heterochromatic 40-Mb regulatory giant whose polymorphic functions between individuals are exerted through its contribution to global chromatin dynamics (13, 18, 19). Here we addressed the hypothesis that naturally occurring Y chromosome lineages are polymorphic for genetic elements that may influence global chromatin dynamics within the nucleus. We show that variation among Y chromosome lineages contributes to the heterochromatin–euchromatin balance in the genome. Furthermore, we identify functionally coherent gene sets that are affected by Y chromosome variation. Importantly, coherent patterns can be observed in Drosophila XXY female genotypes in which Y-linked protein coding genes are not expressed.
Results
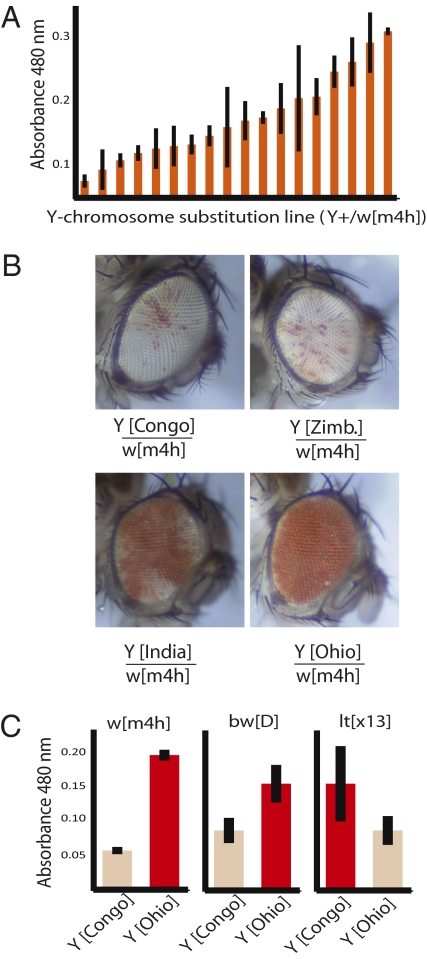
Position-effect variegation (PEV) of mutations affecting the white-eye gene is observed when the gene is moved from its native location to a position near a boundary between heterochromatin and euchromatin (6). PEV in white mutants (e.g., w[m4h], white-mottled 4) is attributed to the spreading of heterochromatin-associated proteins and other modifications into the adjacent euchromatin (6, 20). To address whether Y chromosomes are polymorphic for variation affecting PEV, we generated 16 Y chromosome substitution lines of Drosophila that are identical for all autosomes and X chromosomes and differ only in the origin of the Y chromosome. In white (w[m4h]) mutants, the diagnostic mosaic phenotype arises due to a chromosomal inversion that moves the white gene from its native euchromatic environment to the euchromatic–heterochromatic boundary in the X chromosome (6, 20). Expression of the white gene occurs in cell lineages in which the heterochromatin–euchromatin balance is tipped toward a greater abundance of euchromatin. A lack of white gene expression occurs in cell lineages in which the chromatin balance is tipped toward a greater amount of heterochromatin. We find that Y chromosomes are polymorphic for genetic elements with consequences for w[m4h] PEV; whereas some Y chromosome lineages result in flies with almost completely white eyes, other Y chromosome lineages result in flies with mostly red eyes, with a continuum of effects between these two extremes (Fig. 1 A–C and Table S1). These data show that naturally occurring Y chromosome lineages comprise a rich source of quantitative epigenetic variation that can be detected in somatic tissues and contribute to the balance of heterochromatin and euchromatin around the white locus in w[m4h] mutants.
Fig. 1.
Epigenetic contributions of polymorphic Y chromosomes to global chromatin regulation in somatic tissues. (A) Variation in eye pigmentation in stocks carrying the X chromosome marker w[m4h] and Y chromosomes sampled from diverse localities worldwide. (B) Eye phenotypes showing differential variegated expression for w[m4h] in four Y chromosome backgrounds. These flies are genetically identical except for the origin of the Y chromosome. (C) Differential variegation between Y[Congo] and Y[Ohio] strains for the X-linked marker w[m4h] and second chromosome markers bw[D] and lt[x13].
To address the generality of the effect of naturally occurring Y chromosome lineages on the heterochromatin–euchromatin balance, we investigated two additional loci. One of these loci was bw[D] (brown-eye dominant), in which variegated expression arises due to a dominant insertion of >1 Mb of heterochromatin into the brown locus at its native location in the euchromatin of chromosome 2 (6). Variegated expression of bw[D] is attributed to the recruitment of the brown locus to a heterochromatic compartment of the nucleus. We find that, in agreement with results for white (w[m4h]), Y chromosomes are also polymorphic for their effect on the amount of heterochromatization in the bw[D] locus (Fig. 1C). The requirements for the expression of genes naturally residing within heterochromatin may be opposite for the expression of genes naturally residing within euchromatin (6, 16). Accordingly, the light gene, which is normally located in the pericentric heterochromatin of chromosome 2, shows variegated expression when moved to an euchromatic location in the allele lt[x13] (6). Therefore, we can predict that Y[Ohio] and Y[Congo] might show an opposite effect on the lt[x13] allele relative to what we observed for w[m4h] and bw[D]. Indeed, we find greater expression of the light gene in the Y[Congo] strain than in the Y[Ohio] strain (Fig. 1C). Taken together, these results regarding the effects of epigenetic silencing of variegating alleles of the white, brown, and light genes indicate that polymorphisms within Y-linked heterochromatin affect the balance between heterochromatic and euchromatic compartments within the cell nucleus. Importantly, the effects can be observed in male somatic tissues, in which Y chromosome transcription of single-copy protein-coding genes does not occur. Furthermore, in view of the large magnitude of the effect observed, the data places naturally occurring Y chromosome polymorphisms on a par with several laboratory-generated mutations affecting major chromatin components and chromatin regulators [E(var) and Su(var) genes] (21).
To address the hypothesis that Y-linked polymorphism might differentially affect gene expression in females in the absence of expression from Y-linked protein-coding genes, we generated identical XXY female genotypes that varied only in terms of the origin of the Y chromosome (Fig. S1). The important background information is that XXY genotypes in Drosophila develop into viable females and that Y-linked genes are not transcribed in females (16). Indeed, using several PCR primer sets, we detected no trace of Y-linked protein-coding gene expression (Fig. S2) in XXY female genotypes of Drosophila. We assayed genome-wide gene expression on female flies carrying Y chromosomes with markedly distinct effects on white PEV (Fig. S3). Remarkably, we observed dramatic differences in gene expression when polymorphic Y chromosome lineages were present in the female genotype (Fig. S4). For instance, at P < 0.001, we observed 1,152 genes [false discovery rate (FDR) < 0.05] expressed differentially between XXY[Congo] and XXY[Ohio], with 662 genes (57%) up-regulated in XXY[Congo] and 490 genes (43%) up-regulated in XXY[Ohio]. The data showed a significant positive correlation between the fold change in gene expression in Y[Congo] relative to Y[Ohio] in males and females (ρ = 0.26; P < 0.0001). This recapitulates previous results (13) and identifies the differential expression of genes known to be involved in male fertility. For instance, differentially expressed targets in XXY females include genes coding for protein ejaculatory bulb II, male-specific RNA 84Dc, and fmr1, a gene known to be involved in male courtship (22). These data further suggest a mechanism for YRV that does not require the expression of Y-linked protein-coding genes.
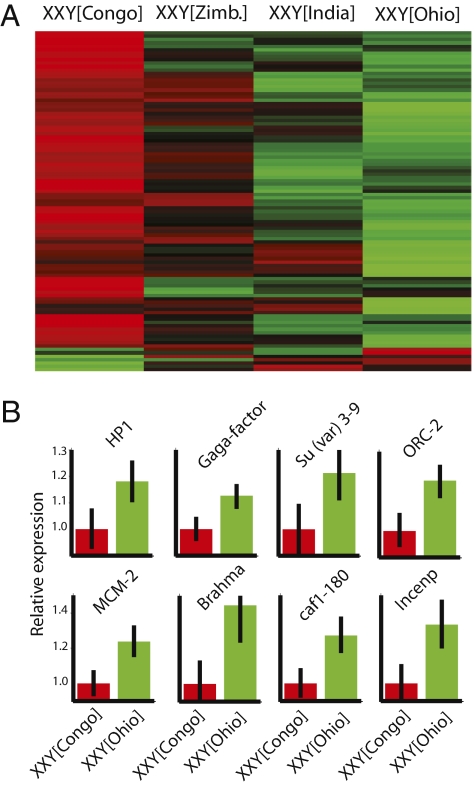
Cytological evidence from two chromatin-associated proteins—the transcriptional activator trithorax-related (GAGA factor), which binds to AAGAG satellites, and the origin of replication complex protein 2 (ORC2), which binds to AT-rich repeats—indicates that they bind to simple sequence repeats in the Y chromosome (23, 24). In view of autoregulatory feedback mechanisms in gene expression (25), we predicted that the expression of these two genes might vary across lines differing in the origin of the Y chromosome. In agreement with our expectations, we find that levels of trithorax-related and ORC2 transcripts differ significantly between XXY[Congo] and XXY[Ohio] (P < 0.001). Thus, we suggest a model in which chromatin regulators might be recruited differentially to polymorphic Y chromosomes, which in turn might affect the steady-state mRNA abundance of these genes. In particular, we tested the hypothesis that other chromatin components and chromatin regulators are responsive to Y chromosome polymorphism and expressed more meagerly in XXY[Congo]. Indeed, we found that >90% of the differentially expressed genes assigned to the Gene Ontology category of chromatin silencing are more meagerly expressed in XXY[Congo] (Fig. 2A and Table S2) (P < 0.01, Fisher's exact test). Furthermore, analyses across several Gene Ontology categories revealed that lower expression of genes in the XXY[Congo] background shows significant enrichment for genes whose products are located in the nucleus (145 genes; P < 10−25, after Bonferroni correction for multiple testing) and have nucleic acid–binding activity (136 genes; P < 10−14). Functionally, the most highly affected genes appear to be involved in the processes of transcription (79 genes; P < 10−7), chromosome organization and biogenesis (29 genes; P < 10−5), DNA packaging (23 genes; P < 10−5), chromatin assembly or disassembly (19 genes; P < 10−5), reproduction (49 genes; P < 10−4), and RNA splicing (23 genes; P < 10−4). The genes identified include not only ORC-2 and GAGA factor, but also such candidates as HP1, Su(var)3–9, MCM2, brahma, centromere identifier, chromatin-assembly factor 1 subunit, caf1-180, and others (Fig. 2B). To further confirm the relevance of differential expression of chromatin components, we used quantitative real-time PCR to assay the expression of key components in Y[Congo] and Y[Ohio] males (Fig. S5). Overall, our analysis revealed a substantial contribution of Y chromosome polymorphisms to the differential expression of chromatin components and chromatin regulators, with several genes previously identified as suppressors of variegation expressed more meagerly in the presence of Y[Congo]. This finding suggests that the enhancer of variegation property of Y[Congo] might be a consequence of the greater availability of Su(var) proteins such as HP1 and Su(var)3–9, among others.
Fig. 2.
Polymorphic Y chromosomes modulate the expression of chromatin components. (A) Heat map of relative expression levels of 101 differentially expressed genes (P < 0.01) belonging to the Gene Ontology category of chromatin silencing. (B) Examples of key chromatin components expressed to a lesser degree in XXY[Congo] (red) relative to XXY[Ohio] (green). Bars denote 95% credible intervals. Relative expression levels are shown, with the lowest expression normalized to 1 (red).
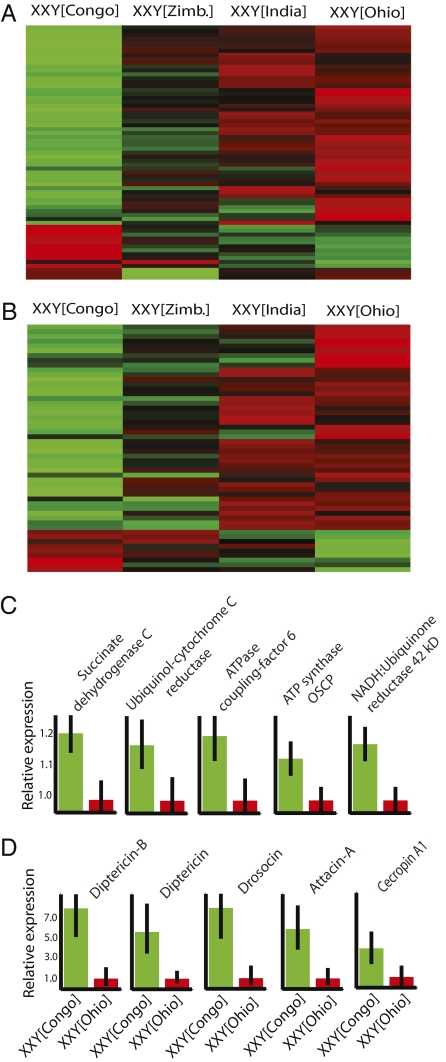
In view of the theoretical expectation that Y chromosomes might mediate the origin and resolution of genetic conflicts (1, 2), we predicted that YRV targets might be enriched for genes with antagonistic fitness effects on males and females. One indicator of this possibility is that genes responding to Y-linked polymorphisms show higher expression in males and lower expression in females compared with genes not affected by YRV (13). Furthermore, in view of potentially strong sibling–sibling competition in the progeny of Drosophila, population genetic theory predicts that harmful interactions between the mitochondria and the Y chromosome might evolve (1). In Drosophila, harmful interactions might be further enhanced through the dramatic mitochondrial remodeling that occurs during spermatogenesis and that leads to the mitochondrial derivatives found in the fruit fly sperm (26). YRV targets are enriched for genes with functional roles related to the mitochondria (13), presumably as a byproduct of genetic interactions between the Y chromosome and mitochondria that occur during spermatogenesis. On the other hand, the effects of the Y chromosome on mitochondrial-related genes might occur independently of spermatogenesis. To test this hypothesis, we searched for genes localized to the mitochondrion and genes involved in electron transport (Table S2) and found significant up-regulation of these genes in XXY[Congo] (Fig. 3 and Table S2) (P < 0.01, Fisher's exact test). Further testing in genes up-regulated in XXY[Congo] found significant enrichment for proteins belonging to the mitochondrial electron transport chain (12 genes; P < 0.05) and with oxidoreductase activity (50 genes; P < 0.01), including both mitochondrial- and nuclear-encoded proteins. These results point to connections between Y chromosome polymorphisms and mitochondrial functions not limited to those occurring during spermatogenesis.
Fig. 3.
Polymorphic Y chromosomes modulate the expression of mitochondria-related and immune-related genes. (A and B) Heat maps of relative expression levels of 65 differentially expressed genes (P < 0.01) belonging to the Gene Ontology category of electron transport (A) and 52 differentially expressed genes (P < 0.01) belonging to the Gene Ontology categories of defense response and immunity (B). (C and D) Examples of mitochondria-related genes (C) and immune-related genes (D) more highly expressed in XXY[Congo] (green) relative to XXY[Ohio] (red). Bars denote 95% credible intervals. Relative expression levels are shown, with the lowest expression normalized to 1 (red).
Evidence suggests that variable immune function might arise as a consequence of genetic conflict (27). To address the role of Y chromosome polymorphism underlying variable immunity, we searched for defense-response and immune-related genes among the targets of YRV. We found that >80% of the differentially expressed genes assigned to the Gene Ontology category of defense response and immunity show greater expression in Y[Congo] relative to Y[Ohio] (P < 0.01) (Fig. 3, Fig. S6, and Table S2). Among genes up-regulated in XXY[Congo], we found 13 targets that show protease inhibitor activity (P = 0.0005). Polymorphic variation in Y chromosomes has been shown to result in the differential expression of transposable elements (13). Variable transposable element activity due to Y chromosome origin can be recapitulated in females; we found an up-regulation of transposable elements in XXY[Congo] relative to XXY[Ohio] (Fig. S7). These patterns were confirmed in contrasts between Y[Congo] and Y[Ohio] males in two different genetic backgrounds (Figs. S6 and S7). This is evidence for a general epigenetic mechanism underlying YRV, which is associated with transposable element activity and differential expression of chromatin components and chromatin regulators.
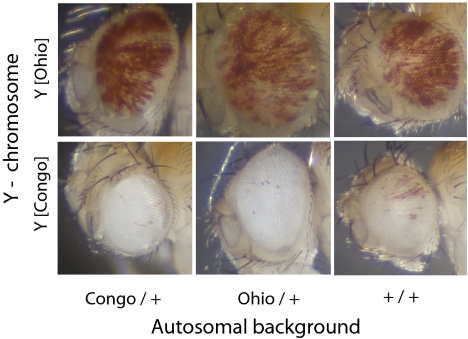
With few exceptions (28), chromatin components are generally conserved and under strong purifying selection within species, which limits the extent of sequence or expression polymorphisms in these genes. On the other hand, the large amounts of heterochromatin found in Y chromosomes might be particularly conducive to harboring polymorphic variation. One reason for this is that the mutation rates of repetitive heterochromatic DNA are unusually high, due to replication slippage and other processes (29, 30); another is that the population genetics of Y chromosomes is unusual in featuring male hemizygosity. Thus, attributes of Y chromosomes might be conducive to the accumulation of heterochromatic variation as well as its expression in males. An important issue is whether the effects of Y chromosomes on gene expression are largely autonomous or whether polymorphic autosomal variation can substantially modify the effects of individual Y chromosomes. To address this issue, we studied the relative contributions of natural variation among Y chromosomes and of dominant modifiers present in the Congo and Ohio genetic backgrounds on the epigenetic variation evidenced by PEV. In these experiments, we swapped the Y chromosomes of Y[Ohio] and Y[Congo] strains while preserving the original genetic backgrounds. We then assessed these Y-swapped strains for their effects on white PEV. Remarkably, we found a silencing effect of the Y[Congo] chromosome in the Ohio genetic background as well as in its original background. Conversely, the Y[Ohio] chromosome (suppressor of variegation leading to a mostly red-eyed phenotype) acted similarly when introgressed into the Congo genetic background as in its original Ohio background (Fig. 4). This result suggests that most of the naturally occurring epigenetic variation in PEV can be attributed to the Y chromosome, with little contribution of dominant, naturally occurring genetic modifiers.
Fig. 4.
The autosomes from Congo and Ohio do not harbor dominant modifiers of Y chromosome-driven PEV. Eye phenotypes showing variegated expression for w[m4h] in Y[Congo] and Y[Ohio] in three backgrounds of autosomes (Congo, Ohio, and laboratory). Autosomes from Congo or Ohio are heterozygous, with a common set of autosomes inherited from a laboratory strain (+) carrying the X-linked w[m4h] marker.
Discussion
Here we have shown that naturally occurring YRV arising from highly heterochromatic Drosophila Y chromosomes can be detected in male somatic tissues and contributes to the epigenetic balance of heterochromatin versus euchromatin at three distinct loci showing PEV. These results point to the Y chromosome as a major contributor to naturally occurring epigenetic variation in Drosophila. The effect of polymorphic Y chromosomes on the amount of heterochromatin-driven silencing of the white gene is of similar magnitude as that of loss-of-function mutants of major chromatin components previously uncovered in genetic screens (21). Furthermore, polymorphic Y chromosomes differentially affect the expression of thousands of genes located in the X chromosome and autosomes of XXY females. These results indicate that expression of Y-linked protein coding genes is not required for YRV. Finally, the gene expression variation identified in these experiments is functionally coherent; it affects chromatin components, transposable elements, and gene sets that might be involved in sexual conflict, including immunity- and mitochondria-related genes. Further characterization of Y-linked variation and its effects on PEV and YRV with larger samples that include intrapopulation variation will be important.
One molecular model for interpreting variegated expression is that this expression reflects the availability of heterochromatin-forming proteins in the nucleus. This model is one of local diffusion of chromatin-modifying enzymes from high-affinity binding sites to nearby low-affinity sites (31). For instance, halving the dosage of genes required for heterochromatin formation, such as HP1 or Su(var)3–9, leads to decreased availability of these proteins and, consequently, to a decrease in the amount of heterochromatin within the nucleus (24). The reduced heterochromatin can in turn restore the wild-type eye color in the white (w[m4h]) PEV system (24). Similarly, a molecular model for interpreting the consequences of polymorphic Y chromosome heterochromatin is that variation in autosomal and X-linked gene expression reflects the limited availability of DNA-binding proteins with heterochromatin-forming and transcription-factor activity in the nucleus. Accordingly, polymorphic Y chromosomes might exert their effects on gene regulation by serving as a differential sink for the binding of chromatin regulators or other DNA-binding proteins, which may lead to the titration of these proteins at other genomic locations (13). The feasibility of such satellite-repeat-sink model has been demonstrated in the case of the transcription factor C/EBP alpha, which binds to satellite repeat alpha in mice (32). Moreover, analyses of enhancer-trap mutants whose expression is modulated by the Y chromosome identified a multi-megabase segment in the Y chromosome that acts as a transregulator of a lacZ reporter expression (33). The sequences mediating the effects were functionally redundant and spatially dispersed across bands h1–h10 of the long arm of the Y chromosome, which coincides with the locations of (AAGAG)n and (AAAGAGA)n repeats that serve as motifs for binding by the GAGA transcription factor (33). Finally, other independent evidence for a chromatin-sink model involving the GAGA factor comes from studies revealing mutant phenotypes of flies fed polyamide compounds that bind specifically to satellite repeats of the type (AAGAG)n (34). Remarkably, we found that the expression of GAGA factor is significantly modulated by polymorphic variation in the Y chromosome. Nevertheless, our data point to several other previously unrelated DNA-binding proteins and suggest that other repeats might also serve as chromatin sinks, possibly leading to a complex dynamics of protein availability at autosomal and X-linked sites.
Variable size of the rDNA array, which is present on the short arm of the Y chromosome, could potentially underlie some of the effects reported here. It was recently reported that variable rDNA array size contributes to variation in PEV phenotype when examined in isolation (35). However, when naturally occurring Y chromosome lines with different rDNA array sizes are probed, PEV phenotypes can be opposite of that expected based on their rDNA size alone. Thus, other loci or segments along the Y chromosome must play significant roles as well. We propose that the effects of naturally occurring Y chromosome lineages on gene expression and PEV are exerted through multiple variable loci located along the Y chromosome. Such variability in the content or length of heterochromatic blocks harboring satellite repeats might be extensive; it arises through a complex dynamics involving repeat homogeneization through interchromosomal and intrachromosomal gene conversion, expansion and contraction of repeats through replication slippage, sister chromatid exchange, intrachromatid exchange, and divergence of repeat units through point mutations (29, 30). Models of mutation–selection balance might be sufficient to account for Y-linked polymorphism within species.
Epistatic Y-linked effects on gene expression and antagonistic X–Y interactions resulting from the altered availability of chromatin components and chromatin regulators at limiting concentrations throughout the genome can be expected. Furthermore, the effects of Y chromosomes on the transcriptional output of various tissues might underlie some of the variation reported here (36). Our results raise the possibility that naturally occurring polymorphic variation in tracts of heterochromatin in the Y chromosome and other chromosomes, including those in the human genome, might serve as important determinants of global chromatin dynamics. The data suggest pathways through which altered chromatin associated with Y chromosome lineages showing activity as suppressors or enhancers of variegation might underlie the expression and resolution of genetic conflicts through the up-regulation of transposable elements, mitochondria-related genes, and immune response genes. Natural polymorphic variation in heterochromatin may be an underappreciated modifier of the differential expressivity and penetrance of ecologically important traits as well as genetic risk factors for disease.
Materials and Methods
Drosophila Stocks.
Y chromosomes from 16 different strains were introgressed into the same laboratory stock background (BL4361) that we used previously (13). This stock is expected to contain very little genetic variation, and upon receipt was subjected to more than eight additional generations of brother–sister mating to reinforce homozygosity of the genomic background. The strains used are listed in Table S1. Crosses for each Y-substitution line were carried out as described previously (13) and shown in Fig. S1. Introgression of Y[Congo] into the Y[Ohio] background and of Y[Ohio] into the Y[Congo] background was done by seven generations of backcrossing. Flies were grown under 24-h light-, temperature-, and humidity-controlled incubators. For gene expression analyses, newly emerged flies were collected and aged for 3 d at 25 °C, after which they were flash-frozen in liquid nitrogen and stored at −80 °C. Whenever females were analyzed, they were collected in less than 7 h on eclosion. All of the females used were unmated females.
Essays for Variegated Expression.
In all assays, males were crossed to females from a stock carrying w[m4h] maintained in a background with the Su(var)3–10[2]. Experiments also were replicated with w[m4h] maintained in a background with Su(var)2–4[01]. Variegation of brown was assessed with allele bw[D]. Variegation of light was assessed with allele lt[x13]. Males were collected, aged for 3 d at 25 °C, and stored at −80 °C. The heads of the males were removed with a blade. Sets of five heads were homogenized with 10 μL of acidified ethanol (30% ethanol acidified to pH 2 with HCl). Eye pigmentation was assessed by spectrophotometric analysis at an optical density of 480 nm. Between four and six biological replicates were used per treatment, with two measurements obtained per replicate. Males displaying typical eye pigmentation phenotypes were imaged using the Snycroscopy Auto-Montage system.
Gene Expression Analyses.
Microarrays were ~18,000-feature cDNA arrays spotted with D. melanogaster cDNA PCR products (13). Total RNA was extracted from whole flies using TRIzol (Life Technologies). cDNA synthesis, labeling with fluorescent dyes (Cy3 and Cy5), and hybridization reactions were carried out using 3DNA protocols and reagents (Genisphere). Slides were scanned using an Axon 4000B scanner (Axon Instruments) and GenePix Pro 6.0 software. Stringent quality control criteria were used to ensure the reliability of foreground intensity reads for both Cy3 and Cy5 channels. Foreground fluorescence of dye intensities was normalized by the Loess method in the R Limma library. The significance of variation in gene expression due to the Y chromosome was assessed with linear models in Limma and with the Bayesian Analysis of Gene Expression Levels (BAGEL) (37). FDRs were estimated based on the variation observed when randomized versions of the original dataset were analyzed. Enrichment in Gene Ontology categories was assessed with GeneMerge (38), using a modified Bonferroni correction. The microarray gene expression data reported herein can be obtained at the Gene Expression Omnibus database (GSE9457 and GSE23612). For quantitative PCR analyses, three biological replicates of 10 adult flies each were sampled in each genotype. qPCR analyses were carried out with the Fast Sybr Green Master Mix (Applied Biosystems). cDNA synthesis was done with the QuantiTect Reverse-Transcription Kit (Qiagen). Real-time PCR profiles were obtained with 7900HT Fast Real-Time PCR (Applied Biosystems). Reactions were checked for the presence of dimers and unspecific amplification. qPCR data were analyzed with REST (39).
Supplementary Material
Acknowledgments
We thank Bernardo Carvalho, Bill Rice, and Ron Woodruff for their careful reading and comments on the manuscript and Silvana Paredes (Texas A&M University) and the Bloomington Stock Center for fly stocks. This work was supported by National Institutes of Health Grant GM084236.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE9457 and GSE23612).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010383107/-/DCSupplemental.
References
- 1.Rice WR, Gavrilets S, Friberg U. Sexually antagonistic “zygotic drive” of the sex chromosomes. PLoS Genet. 2008;4:e1000313. doi: 10.1371/journal.pgen.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice WR. Male fitness increases when females are eliminated from gene pool: Implications for the Y chromosome. Proc Natl Acad Sci USA. 1998;95:6217–6221. doi: 10.1073/pnas.95.11.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho AB, Vaz SC, Klaczko LB. Polymorphism for Y-linked suppressors of sex-ratio in two natural populations of Drosophila mediopunctata. Genetics. 1997;146:891–902. doi: 10.1093/genetics/146.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardo Carvalho A, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009;25:270–277. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295–307. doi: 10.1534/genetics.109.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [Google Scholar]
- 7.Lyckegaard EM, Clark AG. Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86:1944–1948. doi: 10.1073/pnas.86.6.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chippindale AK, Rice WR. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:5677–5682. doi: 10.1073/pnas.101456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohmer C, David JR, Moreteau B, Joly D. Heat-induced male sterility in Drosophila melanogaster: Adaptive genetic variations among geographic populations and role of the Y chromosome. J Exp Biol. 2004;207:2735–2743. doi: 10.1242/jeb.01087. [DOI] [PubMed] [Google Scholar]
- 10.Huttunen S, Aspi J. Complex inheritance of male courtship song characters in Drosophila virilis. Behav Genet. 2003;33:17–24. doi: 10.1023/a:1021095331850. [DOI] [PubMed] [Google Scholar]
- 11.Clark AG. Natural selection and Y-linked polymorphism. Genetics. 1987;115:569–577. doi: 10.1093/genetics/115.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zurovcova M, Eanes WF. Lack of nucleotide polymorphism in the Y-linked sperm flagellar dynein gene Dhc-Yh3 of Drosophila melanogaster and D. simulans. Genetics. 1999;153:1709–1715. doi: 10.1093/genetics/153.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 14.Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hearn MG, Hedrick A, Grigliatti TA, Wakimoto BT. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: Relationship between suppression effect and the amount of Y chromosome. Genetics. 1989;122:793–800. doi: 10.1093/genetics/122.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice WR, Friberg U. Genetics: Functionally degenerate—Y not so? Science. 2008;319:42–43. doi: 10.1126/science.1153482. [DOI] [PubMed] [Google Scholar]
- 19.Malone JH, Oliver B. The sex chromosome that refused to die. Bioessays. 2008;30:409–411. doi: 10.1002/bies.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulze SR, Wallrath LL. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Annu Rev Entomol. 2007;52:171–192. doi: 10.1146/annurev.ento.51.110104.151007. [DOI] [PubMed] [Google Scholar]
- 21.Reuter G, Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee P, et al. Substitution of critical isoleucines in the KH domains of Drosophila fragile X protein results in partial loss-of-function phenotypes. Genetics. 2007;175:1241–1250. doi: 10.1534/genetics.106.068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platero JS, Csink AK, Quintanilla A, Henikoff S. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pak DT, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 25.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall; 2007. [Google Scholar]
- 26.Shoup JR. Spermiogenesis in wild type and in a male sterility mutant of Drosophila melanogaster. J Cell Biol. 1967;32:663–675. doi: 10.1083/jcb.32.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensson EI, McAdam AG, Sinervo B. Intralocus sexual conflict over immune defense, gender load, and sex-specific signaling in a natural lizard population. Evolution. 2009;63:3124–3135. doi: 10.1111/j.1558-5646.2009.00782.x. [DOI] [PubMed] [Google Scholar]
- 28.Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plohl M, Luchetti A, Mestrović N, Mantovani B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Palomeque T, Lorite P. Satellite DNA in insects: A review. Heredity. 2008;100:564–573. doi: 10.1038/hdy.2008.24. [DOI] [PubMed] [Google Scholar]
- 31.Monod C, Aulner N, Cuvier O, Käs E. Modification of position-effect variegation by competition for binding to Drosophila satellites. EMBO Rep. 2002;3:747–752. doi: 10.1093/embo-reports/kvf155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Wu B, Szary J, Kofoed EM, Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J Biol Chem. 2007;282:20868–20876. doi: 10.1074/jbc.M702547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, Timakov B, Stankiewicz RL, Turgut IY. A trans-activator on the Drosophila Y chromosome regulates gene expression in the male germ line. Genetica. 2000;109:141–150. doi: 10.1023/a:1026504721067. [DOI] [PubMed] [Google Scholar]
- 34.Janssen S, Cuvier O, Müller M, Laemmli UK. Specific gain- and loss-of-function phenotypes induced by satellite-specific DNA-binding drugs fed to Drosophila melanogaster. Mol Cell. 2000;6:1013–1024. doi: 10.1016/s1097-2765(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 35.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart AD, Pischedda A, Rice WR. Resolving intralocus sexual conflict: Genetic mechanisms and time frame. J Hered. 2010;101(Suppl 1):S94–S99. doi: 10.1093/jhered/esq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biology. 2002;3:research0071.1-0071.16. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo-Davis CI, Hartl DL. GeneMerge: Post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.