Abstract
The cytoplasmic Myc protein (c-Myc) regulates various human genes and is dysregulated in many human cancers. Phosphorylation mediates the protein activation of c-Myc and is essential for the function of this transcription factor in normal cell behavior and tumor growth. To date, however, the targeting of Myc as a therapeutic approach for cancer treatment has been achieved primarily at the nonprotein level. We have developed a molecular imaging sensor for noninvasive imaging of c-Myc activity in living subjects using a split Firefly luciferase (FL) complementation strategy to detect and quantify the phosphorylation-mediated interaction between glycogen synthase kinase 3β (GSK3β) and c-Myc. This sensor system consists of two fusion proteins, GSK 35–433-CFL and NFL-c-Myc, in which specific fragments of GSK3β and c-Myc are fused with C-terminal and N-terminal fragments of the split FL, respectively. The sensor detects phosphorylation-specific GSK3β–c-Myc interaction, the imaging signal of which correlates with the steady-state and temporal regulation of c-Myc phosphorylation in cell culture. The sensor also detects inhibition of c-Myc activity via differential pathways, allowing noninvasive monitoring of c-Myc–targeted drug efficacy in intact cells and living mice. Notably, this drug inhibition is detected before changes in tumor size are apparent in mouse xenograft and liver tumor models. This reporter system not only provides an innovative way to investigate the role of functional c-Myc in normal and cancer-related biological processes, but also facilitates c-Myc–targeted drug development by providing a rapid quantitative approach to assessing cancer response to therapy in living subjects.
Keywords: c-Myc phosphorylation, drug development, GSK3β, split reporter complementation, monitoring of drug efficacy
The myc gene encodes transcription factors (N-Myc, c-Myc, and L-Myc) that regulate up to 15% of all vertebrate genes, which are essential to almost every aspect of cell behavior, including cell growth and proliferation, cell cycle progression, differentiation, and apoptosis (1). The c-Myc protein in particular coordinates the integration of extracellular and intracellular signals as the central hub for cellular cues (2). In light of these functions, it is not surprising that expression of c-Myc is tightly regulated in normal cells. Normally, cells exhibit low steady-state levels of c-Myc expression when in a nonproliferative state. In the presence of stimulatory signals, such as developmental cues or mitogens, c-Myc is phosphorylated at Ser-62 (S62) through Ras-induced ERK pathway activation (3), which temporarily activates and stabilizes the protein. On removal of the stimuli, phosphorylated S62 is recognized by glycogen synthase kinase-3β (GSK3β), which further phosphorylates Thr-58 (T58) and leads to ubiquitination and rapid degradation by proteasome (4). The phosphorylation-mediated temporary c-Myc activation is essential for many cellular processes, including entry into cell cycle phases, biogenesis of ribosomes, response to oxidative stress, and induction of apoptosis (5).
The tight control of c-Myc activity is defective at multiple levels in almost all human cancers, where the protein is constitutively activated and stabilized. This also makes c-Myc an attractive candidate for targeted cancer therapy (6). Current strategies are aimed mainly at down-regulating c-Myc by inhibiting gene expression, such as using antisense oligonucleotides and RNAi to compete for binding to the c-Myc promoter, its coding region, or downstream target genes (7–9). Although these approaches can inhibit tumor growth and promote apoptosis to certain extent, the main disadvantages are the instability of the short oligonucleotides used and the difficulty of in vivo delivery (6). Some attempts to repress c-Myc at the protein level (e.g., the use of small molecules to disrupt c-Myc interaction with other factors) have shown promise in cell culture (10, 11). To date, approaches to regulating phosphorylation-mediated c-Myc activity, which is essential for sustaining the growth of many tumors (5), have been limited. ERK kinase inhibitors PD98059 and U0126 decrease the c-Myc phosphorylation level in vitro (12), but there has been no study of their effect on tumor growth. Atorvastatin (AT), a member of the statin family, was unexpectedly found to reduce phosphorylation of c-Myc by inhibiting 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-coA) reductase and in turn preventing c-Myc–induced lymphomagenesis (13), although the exact molecular mechanism remains unclear. The unavailability of methods to noninvasively monitor c-Myc activity has hindered further understanding of Myc cancer biology and contributed to delays in c-Myc–targeted drug development (14).
Multimodality molecular imaging has emerged as a key spectrum of technologies to advance our understanding of disease mechanisms and accelerate drug discovery and development (15). In particular, reporter gene imaging strategies based on protein-assisted complementation of split luciferases are emerging as powerful tools for detecting and quantifying induced protein interactions and functional protein modifications in vivo, such as ubiquitination and phosphorylation (16–19). To noninvasively monitor and image phosphorylation-mediated c-Myc activation, we developed a new split Firefly luciferase (FL)-based sensor system, in which the complementation of the split FL is induced by phosphorylation-mediated interaction between GSK3β and c-Myc. The complemented FL activity resulting from this interaction is specific to c-Myc phosphorylation and correlated with the steady-state and temporal regulation of c-Myc phosphorylation in cell culture. The sensor system also allows monitoring of c-Myc–targeted drug efficacy in intact cells and living mice. This new imaging sensor may provide insight into the role of functional c-Myc in cancer biology and help accelerate the discovery and development of new, more specific anti–c-Myc drugs.
Results
Sensor System for Imaging of c-Myc Phosphorylation.
Because c-Myc activation is mediated by S62 phosphorylation, we designed a sensor system to image this phosphorylation by detecting the phosphorylation-induced interaction between GSK3β and c-Myc. We used protein-assisted split luciferase complementation systems developed by our laboratories and another group for imaging protein–protein interactions in living subjects (19–22). Through fusion of an activation motif of c-Myc and a phospho recognition domain of GSK3β with the two inactive split luciferase fragments (Fig. 1B), S62 phosphorylation is expected to result in reporter complementation by inducing the interaction between the c-Myc activation motif and the GSK3β recognition domain. Further T58 phosphorylation will result in greater approximation of the split fragments and complementation of the reporter and restoration of bioluminescence (Fig. 1A). The endogenous GSK3β and c-Myc proteins also may interact with the fused c-Myc activation motif and the GSK3β recognition domain, respectively; however, these competitive bindings do not result in reporter complementation.
Fig. 1.
Validation of our sensor system for imaging of c-Myc activation. (A) Schematic strategy of imaging c-Myc activation via detection of S62 phosphorylation-mediated GSK3β–c-Myc interaction. GSK3β and c-Myc fragments are fused with the C-terminal and N-terminal part of the inactive split FLs, respectively. On growth signals stimulation (e.g., EGF-mediated Ras activation), c-Myc is phosphorylated at S62, which induces recognition by the priming phosphate site of GSK3β (25). Further phosphorylation by the active site of GSK3β at T58 brings split luciferases into closer proximity and restores bioluminescence reporter activity. (B) Diagram of different split luciferase fusion constructs. Different truncations of GSK3β and the c-Myc activation motif (51–69 aa) are fused with the C-terminal and N-terminal fragment of split luciferases, respectively, as indicated. Numbers indicate the location of the amino acid in the sequence of full-length GSK3β. The two phosphorylation sites, S62 and T58, are in bold type. (C) Bioluminescence imaging (BLI) of single and paired fusion constructs. NFL-SH2-SH2 is constructed by replacing the c-Myc motif with two SH2 domains of PI3K. **P < 0.01 by the unpaired t test.
The selected c-Myc activation motif included the 51–69 aa in the Myc homology box I (MBI) of c-Myc (SI Experimental Procedures), which is part of the Myc transactivation domain that mediates c-Myc activation (23) but is not sufficient for ubiquitin-mediated proteasomal degradation (24). Several truncations of GSK3β [GSK(N7), GSK(35–350), and GSK(35–433)] were constructed (Fig. 1B and SI Experimental Procedures) based on the crystal structure and functional domains of GSK3β (25) for determination of the optimal sensitivity and specificity to c-Myc phosphorylation. Two types of split luciferase fragments—human codon–optimized split Renilla luciferase (hRL) and split FL—were used for fusion and compared for signal strength (Fig. 1B). The split sites, NhRL229/ChRL230 and NFL398/CFL394, were chosen based on previous studies in our lab (19, 21) for optimal intrinsic complementation efficiency.
To test the dependence of the split luciferase complementation on GSK3β–c-Myc interaction, we introduced the mock DNA (vector alone), a single sensor construct (GSK Full-CFL or NFL-c-Myc), a single sensor construct with a scrambled fusion construct (GSK Full-CFL/NFL-SH2-SH2), and paired sensor constructs (GSK Full-CFL/NFL-c-Myc) into SKBR3 breast cancer cells (Fig. 1C), in which c-Myc is constitutively activated (26). No single sensor construct resulted in split FL complementation, whereas the pairing of GSK Full-CFL and NFL-c-Myc resulted in a >10-fold higher complemented FL activity compared with the pairing of GSK Full-CFL and NFL-SH2-SH2 (Fig. 1C). We also compared the signal strength of paired sensor constructs using split hRL fusion with those using split FL fusion. All paired split hRL fusion constructs resulted in much lower complemented hRL activity than the paired split FL fusion constructs (Fig. 2A), suggesting higher signal yield with the split FL fusion. Meanwhile, among the split FL fusion constructs, the pairing of different GSK3β truncations with NFL-c-Myc resulted in different levels of complemented FL activity, with the highest activity detected with the full-length GSK3β.
Fig. 2.
Optimization of the sensor constructs. (A) Comparison of complemented RL activity and complemented FL activity. Split RL fusion and split FL fusion of different truncations of GSK3β were coexpressed with split RL fusion and split FL fusion of the c-Myc activation motif, respectively, and subjected to BLI. *P < 0.05 by the unpaired t test. (B) BLI of S62 phosphorylation specificity of different truncations of GSK3β. WT, S62A, and S62D mutants of NFL-c-Myc were coexpressed with CFL-fused different truncations of GSK3β as indicated and subjected to BLI. *P < 0.05 by the unpaired t test. NS, no significant difference.
To test the specificity of different GSK3β truncations to c-Myc phosphorylation, we introduced the mutation S62A, which abolishes phosphorylation, and the mutation S62D, which mimics the constitutive phosphorylation of NFL-c-Myc. Different CFL-fused GSK3β truncations were coexpressed with NFL-c-Myc, NFL-c-Myc S62A, or NFL-c-Myc S62D in SKBR3 cells, except GSK N7-CFL, which had very low signal yield (Fig. 2A). Compared with NFL-c-Myc, NFL-c-Myc S62A resulted in significant reduction of complemented FL activity when paired with GSK 35–433-CFL (P < 0.05), but not when paired with GSK 35–350-CFL or GSK Full-CFL, and NFL-c-Myc S62D resulted in recovery of complemented FL activity when paired with GSK 35–433-CFL and GSK 35–350-CFL, but not when paired with GSK Full-CFL (Fig. 2B). These results suggest that only GSK 35–433-CFL is able to confer specificity to S62 phosphorylation of NFL-c-Myc. Thus, the combination of GSK 35–433-CFL and NFL-c-Myc constitutes the optimal sensor over all other combinations of constructs for detecting phosphorylation-mediated interaction between GSK3β and c-Myc.
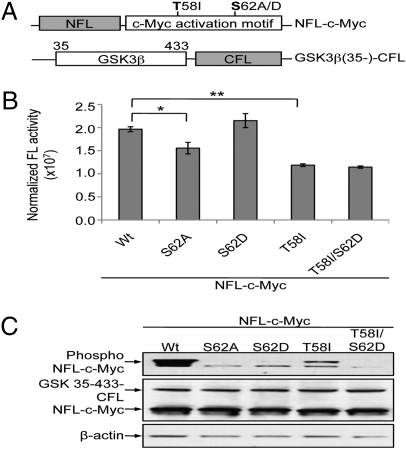
To further characterize the phosphorylation dependence of the complementation between GSK 35–433-CFL and NFL-c-Myc, we introduced a mutation T58I (Fig. 3A), which frequently occurs in lymphomas to prevent c-Myc from T58 phosphorylation by GSK3β and subsequent degradation (27). We also paired T58I with the S62A or S62D mutation to compare the impact on reporter complementation of phosphorylation at T58 and that at S62 (Fig. 3B). Coexpression of GSK 35–433-CFL with the T58I mutation in SKBR3 cells resulted in a greater reduction in complemented FL activity than coexpression with the S62A mutation (Fig. 3B; P < 0.01). The T58I/S62A double mutations resulted in a similar reduction as the T58I single mutation (Fig. S1), whereas the T58I/S62D double mutations did not result in recovery of the complemented FL activity, unlike the S62D single mutation (Fig. 3B). These results suggest that depletion of T58 phosphorylation compromises S62 phosphorylation-induced NFL-c-Myc interaction with GSK 35–433-CFL and further complementation of the split FL. Western blot analysis of the cell lysates revealed the sensor phosphorylation status with a phospho c-Myc antibody against T58 and/or S62 (Fig. 3C). The NFL-c-Myc was highly phosphorylated, whereas the S62A mutation abolished all phosphorylation, and the T58I mutation abolished it partially. The antibody did not recognize the phospho group of Asp, and thus the T58 phosphorylation in the S62D mutant was revealed as a faint band, and the T58I/S62D mutant had no detectable band. Taken together, these results suggest that S62 phosphorylation of NFL-c-Myc is required for T58 phosphorylation, which in turn affects S62 phosphorylation-induced split FL complementation.
Fig. 3.
Characterization of the phosphorylation dependence of the complementation. (A) Diagram of the sensor system. Mutations at T58 and S62 are indicated. (B) BLI of split FL complementation of NFL-c-Myc mutants and GSK 35–433-CFL. WT and mutant NFL-c-Myc as indicated were coexpressed with GSK 35–433-CFL and subjected to BLI. *P < 0.05; **P < 0.01 by the unpaired t test. (D) Western blot analysis of cell lysates shown in C, using phospho T58/S62 c-Myc antibody, FL antibody, and β-actin antibody.
Steady-State Imaging of c-Myc Phosphorylation in Intact Cells.
We transiently transfected the sensor, GSK 35–433-CFL/NFL-c-Myc, into Chinese hamster ovary (CHO), MCF-7, 293T, SKBR3, and HT1080 cells to test its ability in detecting the steady-state level of c-Myc phosphorylation. The complemented FL activity in CHO cells was 2.6-fold higher than that in MCF-7 cells, 17.9-fold lower than that in 293T cells, 54.9-fold lower than that in SKBR3 cells, and 1.1-fold higher than that in HT1080 cells (Fig. 4A). These sensor signals also were correlated with the steady-state differential levels of c-Myc phosphorylation in those cells (R2 = 0.90; Fig. 4C), as revealed by Western blot analysis of the cell lysates (Fig. 4B).
Fig. 4.
Detection of differential c-Myc phosphorylation level in normal cells and cancer cells. (A) The sensor system was transfected in the indicated cell lines and subjected to BLI. Complemented FL activity was normalized to coexpressed RL activity and plotted against drug concentrations on a logarithmic scale. (B) Western blot analysis of the indicated cells using phospho T58/S62 c-Myc, c-Myc, and β-actin antibody. (C) Plot of the correlation coefficient between the c-Myc phosphorylation level and the normalized FL activity of each type of cell (R2 = 0.90).
Dynamic Imaging of c-Myc Activation on Serum Stimulation.
On growth factor stimulation, c-Myc exhibits temporal activation, followed by a rapid decline to basal levels (4). To test the sensor's ability to detect dynamic regulation of c-Myc phosphorylation, we performed a growth stimulation assay. CHO cells were transfected with the sensor and serum-starved for 24 h, and then stimulated with 20% serum for 0 h, 1 h, 3 h, and 5 h. The complemented FL activity increased after serum stimulation, peaked at 3 h, and then declined rapidly to the basal steady-state level by 24 h (Fig. 5A). This dynamic change in the sensor signal also was correlated with temporal regulation of the endogenous c-Myc phosphorylation level (R2 = 0.87; Fig. 5C), as revealed by Western blot analysis (Fig. 5B).
Fig. 5.
Detection of the temporal activation of c-Myc on serum stimulation in cells. (A) CHO cells, transfected with the sensor system, were serum-starved for 24 h, then serum-stimulated for the indicated time and subjected to BLI as described in SI Experimental Procedures. (B) Western blot analysis of CHO cells on serum stimulation for the indicated time using phospho c-Myc, c-Myc protein, and β-actin antibodies. (C) Plot of correlation coefficient between the fold change of c-Myc phosphorylation level and the fold change of FL activity at each time point (R2 = 0.87).
Imaging of c-Myc Phosphorylation Inhibition in Intact Cells.
To test the ability of the sensor to monitor c-Myc phosphorylation levels on drug treatment, we established the SKBR3 and 293T stable cells (SK ST and 293T ST) that constitutively express the sensor. We also constructed SKBR3 and 293T stable cells that constitutively express the full-length FL (SK FST and 293T FST) for the control of direct drug effect on FL activity. The SK ST cells were treated with the MAP kinase inhibitors (PD98059 and U0126) and AT, all of which induced dose-dependent reduction of complemented FL activity after normalization to their effect on the FL activity of SK FST cells. AT induced greater reduction in complemented FL activity with lower drug concentrations than U0126, which in turn induced greater reduction than PD98059. These differential signal inhibitions also were correlated with the decreased endogenous c-Myc phosphorylation level (Fig. 6A and Figs. S2–S4). In 293T ST cells, however, AT induced a dose-dependent increase in complemented FL activity after normalization to its effect on the FL activity of 293T FST cells, which also was correlated with the increase in endogenous c-Myc phosphorylation level (Fig. 6A and Fig. S5). SKBR3 cells transfected with the mutant sensor GSK3β/NFL-c-MycS62A were also subjected to PD98059 treatment, but did not respond to phosphorylation inhibition of c-Myc (Fig. S6).
Fig. 6.
Detection of inhibition of c-Myc phosphorylation in intact cells. (A) SKBR3 and 293T stable cells, constitutively expressing the c-Myc sensor (SK ST and 293T ST) or the full-length FL (SK FST and 293T FST) (Figs. S2–S5) were treated with the indicated drugs at the indicated concentrations and subjected to BLI as described in SI Experimental Procedures. Western blot analyses of SK and 293T cells after drug treatment were performed using phospho c-Myc, c-Myc protein, and β-actin antibodies (Figs. S2–S5). (B) Plot of the correlation coefficients between the fold change of c-Myc phosphorylation level and the fold change of FL activity at different concentrations of indicated drugs. Each R2 value is indicated.
Noninvasive Imaging of c-Myc Phosphorylation in Mice.
To test the in vivo application of the sensor system, we first transiently transfected SKBR3 cells separately with equal DNA molar amounts of the vectors pcDNA3.1(+), GSK 35–433-CFL/NFL-c-Myc or GSK 35–433-CFL/NFL-c-Myc T85I and implanted them s.c. in different locations in each nude mouse (n = 3; Fig. 7A). hRL was also coexpressed for control of transfection efficiency. At 16 h after implantation, the three xenografts of each mouse all showed hRL activity on i.v. injection of its substrate coelenterazine (Clz) (Fig. 7B). However, on i.p. injection of FL substrate D-Luciferin (D-Luc), xenografts carrying pcDNA3.1(+) had very low complemented FL activity, whereas those carrying GSK 35–433-CFL/NFL-c-Myc had 10.6-fold higher activity and those carrying GSK 35–433-CFL/NFL-c-Myc T85I had 4.7-fold higher activity after normalization to their hRL activity (Fig. 7C). These results suggest that the complemented FL activity determined by the phosphorylation status of NFL-c-Myc can be imaged and quantified in a mouse xenograft model. We also transiently expressed the sensor system in the liver of two transgenic mice using hydrodynamic injection. These mice carried a liver-specific transgene of c-Myc under control of the Tet system (13). Both mice had doxycycline discontinued, followed by a 29-day course of treatment with AT in one mouse and with PBS in the other mouse. After treatment, the two mice demonstrated no significant difference in liver tumor size. On the addition of Clz, both mice showed similar hRL activity, whereas on the addition of D-Luc, the mouse treated with AT had a much lower complemented FL activity than that treated with PBS (Fig. 7D). This signal reduction was consistent with the reduced endogenous c-Myc phosphorylation and the histological changes in the liver tissue samples (Fig. 7E).
Fig. 7.
BLI of c-Myc phosphorylation in living mice. (A) Diagram of the implantation location of the SKBR3 cells transiently transfected with combinations of plasmids as indicated. (B) Representative pictures of mice subjected to RL imaging with Clz and to FL imaging with D-Luc. An autoluminescent Clz signal is seen in the liver (Left). (C) Photon output for the complemented FL activity was normalized to that for the RL activity (FL/RL) and plotted as the fold change to the FL/RL of the vector. (D) Eμ-tTA/Tet-O-MYC transgenic mice (n = 2) under AT or PBS treatment were subjected to RL imaging with Clz and FL imaging with D-Luc after hydrodynamic injection as described in SI Experimental Procedures. (E) Western blot analysis of the liver tissue samples using phospho c-Myc, c-Myc, and α-tubulin antibodies and H&E staining of the samples.
In Vivo Monitoring of Drug Inhibition of c-Myc Phosphorylation.
To further demonstrate the ability of the sensor system to monitor drug effects on c-Myc phosphorylation and tumor growth in vivo, we implanted SK ST cells s.c. to monitor phospho c-Myc levels and SK FST cells to control for cell numbers and the direct drug effect on FL activity in nude mice (n = 10; Fig. 8A). The mice underwent gavage with AT (20 μL, 1.25 mM; n = 5) or with PBS (20 μL; n = 5) every other day. The complemented FL activity of SK ST xenografts began to rapidly increase 10 d after implantation in the PBS-treated group, but increased only slightly in the AT-treated group (Fig. 8, Left); yet there was no detectable difference in tumor size between the two groups. The FL activity of the SK FST xenografts increased gradually in both groups; up to about 18 d after implantation, FL activity was higher in the PBS-treated group compared with the AT-treated group (Fig. 8B, Right). Normalization of the complemented FL activity of SK ST to that of SK FST showed a dramatic increase of sensor signal in early tumorigenesis in the PBS group, followed by a decrease due to the increase of tumor cell number. In the AT group, a sustained decrease in the sensor signal was seen (Fig. S6). These results indicate that the sensor detected the immediate c-Myc activity in tumorigenesis before tumor growth and thus noninvasively monitored the inhibitory effect of AT that eventually resulted in reduced tumor growth. The decreased sensor signal in the AT-treated group also was consistent with the decreased c-Myc phosphorylation levels in tumor tissue samples at 23 d after implantation (Fig. 8C).
Fig. 8.
BLI of AT inhibition of c-Myc phosphorylation in living mice. (A) SKBR3 stable cells, constitutively expressing the c-Myc sensor (SK ST) or the full-length FL (SK FST), were implanted s.c. as indicated and treated with AT or PBS as described in SI Experimental Procedures. Representative FL images on the indicated days of treatment are shown. (B) (Left) Photon outputs of SK ST cells with AT or PBS treatment plotted together to show the AT inhibitory effect on the complemented FL activity. (Right) Photon outputs of SK FST cells with AT or PBS treatment plotted together to show the AT inhibitory effect on the full-length FL activity. (C) Western blot analysis of xenograft tissue samples using phospho c-Myc, c-Myc, and α-tubulin antibodies. Representative blots are shown.
Discussion
Because oncogenic activation of the c-Myc gene involves insertional mutagenesis, chromosomal translocation, and gene amplification (6), noninvasive monitoring of c-Myc activity at the transcriptional level using reporter genes is challenging. The phosphorylation-mediated activation of c-Myc protein contributes to the actual function of c-Myc in many normal and cancerous molecular processes. Thus, the fact that S62 phosphorylation is required for the interaction between GSK3β and c-Myc makes imaging the phospho signal and activation of c-Myc by detection of GSK3β–c-Myc interaction an attractive proposition. Consequently, we developed an imaging sensor based on a protein-assisted split luciferase complementation strategy by fusing GSK3β fragments and a c-Myc activation motif with the C-terminal and N-terminal fragments of split FL, respectively. The resulting complemented FL activity was dependent on the interaction between the GSK3β fragment and the c-Myc activation motif (Fig. 1C). We also showed that the 35–433 aa fragment of GSK3β was sufficient to confer specificity to S62 phosphorylation of NFL-c-Myc, but not other truncations (Fig. 2). The deleted 1–34 aa fragment of GSK3β actually contains the autoinhibition site Ser-9, which prevents substrate binding to the phospho priming site (25); thus, the GSK 35–433-CFL fusion protein is not susceptible to the intrinsic inhibition of GSK3β in cancer cells. On a similar note, the 19 aa activation motif contains the phospho sites for c-Myc activation, but is not sufficient for ubiquitin-mediated proteasomal degradation (24). We also compared split RL fusion and split FL fusion constructs and found that the latter resulted in a much higher photon output, which also may have an advantage over the hRL fusion in living subjects due to a more red-shifted wavelength, allowing greater light penetration through the body. Accordingly, GSK 35–433-CFL/NFL-c-Myc comprises a sensor that generates complemented FL activity, measurable through an imaging signal, on S62 phosphorylation of c-Myc.
We further characterized the phosphorylation dependence of the split FL complementation by introducing a T58I mutation, and found that this resulted in a greater reduction of complemented FL activity than the S62A mutation, suggesting that T58 phosphorylation is more efficient in inducing reporter complementation. This also may imply that T58 phosphorylation is required for high-affinity binding of GSK3β to c-Myc, which in turn brings two inactive split FL fragments closer for complementation of FL reporter (Fig. 1A). However, the Western blot analysis indicated that T58 phosphorylation was dependent on S62 phosphorylation (Fig. 3C), which is consistent with the endogenous phosphorylation process of c-Myc on activation. Therefore, by including both phosphorylation sites within its constructs, the developed sensor enables imaging of S62 phosphorylation that induces efficient split FL complementation.
We validated the sensor in intact cells and in living mice, demonstrating that the sensor signal (complemented FL activity) is closely correlated with the endogenous c-Myc phosphorylation level in culture cells and living mice, either at steady-state, on serum stimulation, or on drug treatment. Our results indicate that the sensor system provides a means to noninvasively image and quantify c-Myc phosphorylation and activation with high sensitivity in living subjects. Thus, this sensor might provide insight into previously undetermined roles of phosphorylation in many c-Myc functions, including apoptosis, angiogenesis, and tumorigenesis (5), by noninvasively reporting the phosphorylation status in cells under varying levels of spatial and temporal regulation. Recent stem cell studies have implicated Myc transcription factors in the control of pluripotency and self-renewal properties of tumor stem cells (28). Using the sensor presented herein might help unveil the relevant molecular mechanisms. Future generation of transgenic mice that incorporate the sensor in a tissue-specific manner might help reveal the role of c-Myc activation for in vivo orthotropic tumor development and progression (Fig. 7D). There are various conditional mutant Myc transgenic mice in which overexpression of WT or mutant Myc is reversible under control of the Tet system (13, 29–32). Crossing the sensor transgenic mice to these conditional mutant Myc transgenic mice can allow the noninvasive study of the relationship between Myc gene deregulation and Myc protein activation in the induction and maintenance of tumorigenesis in different tumor models. Because the c-Myc activation motif is also conserved in N-Myc and L-Myc, this sensor also may be used to investigate the role of phosphorylation in the function of these proteins in brain and lung cancer, respectively.
c-Myc is considered an attractive candidate for targeted cancer therapy because of its dysregulation in most human cancers. Recent studies in transgenic mouse models hold further promise for Myc as a valid anticancer target (31). Our sensor system has the potential to accelerate the discovery of c-Myc–targeted molecular therapeutics by providing a rapid, noninvasive readout of c-Myc activity in various in vitro assays as well as high-throughput screening of chemical libraries, with subsequent further validation of their effects in xenograft or transgenic mouse models. Clearly, the advantages of using our sensor system for earlier discovery and validation of candidate drugs (Fig. 8) include significant reductions in the time, cost, and effort involved in c-Myc–targeted drug development. Furthermore, the quantitative readout of c-Myc activity in living subjects on drug treatment could potentially help establish future dosing parameters in preclinical models to guide initial clinical trials and eventual clinical applications.
Although our findings are promising, the present study has some limitations, and more work is needed to improve the c-Myc phosphorylation sensor. Neither the S62A mutation nor the T58I mutation of NFL-c-Myc completely abolished the sensor signal, suggesting that there may be some nonspecific binding between the GSK3β fragments and the c-Myc activation motif, as is also known to occur between endogenous GSK3β and c-Myc (33). By altering the residues surrounding S62 and T58 of NFL-c-Myc, or the residues around the binding sites of GSK3β, it may be possible to diminish the non–phosphorylation-mediated binding between the two fusion proteins. While using sensor stable cells to monitor drug effects, we used full-length FL as a control for direct drug effects on FL activity. Future studies examining drug effects on split FL versus full-length FL may be helpful as well. Moreover, in the mouse xenograft model, it might have been of benefit to control for intrinsic cell numbers by constitutively expressing hRL in the sensor stable cells in one xenograft, rather than using FL stable cells in a separate xenograft on the same mouse. This also could have helped reduce the tumor burden on the mice during prolonged monitoring of drug effects on survival rate. Because of the limited number of mice used, we killed the mice only on the final day of imaging and analyzed the phosphorylation levels in tumor tissue. In future experiments, larger cohorts of mice may be used to obtain tissue samples for examining the correlation between the sensor signal and the actual c-Myc phosphorylation level at several different time points.
In conclusion, we have developed a sensor system for the noninvasive imaging of c-Myc activation in cells and living mice. With a greater appreciation of the ever-expanding role of Myc in all aspects of cell behavior, this sensor will provide new avenues to investigate the mechanisms of Myc-associated normal and neoplastic cell growth and death, as well as to accelerate the development of Myc-targeted cancer therapeutics.
Experimental Procedures
Details on plasmids, selection of fusion fragments, cell culture and Western blot analysis, luciferase activity assays, serum stimulation assays, and drug inhibition assays are provided in SI Experimental Procedures.
BLI of Living Mice.
All animal handling was performed in accordance with Stanford University's Animal Research Committee guidelines. BLI of mouse xenografts was performed with the IVIS Spectrum optical imaging system (Xenogen) as described previously (18), with some modifications. At 4 h after transient transfection with the indicated constructs (Fig. 7A), 5 × 106 SKBR3 cells were implanted s.c. into the indicated locations of nude mice (nu/nu; n = 3). At 16 h after cell implantation, a Clz PBS (0.3 μg/μL; 100 μL) solution was injected i.v. for RL imaging. Then, 8 h later, FL imaging was acquired with D-Luc PBS solution (45 μg/μL; 100 μL) via i.p. injection. Subsequently, 5 × 106 each of SK ST and SK FST cells were implanted s.c. into nude mice (nu/nu; n = 10) as indicated (Fig. 8A). FL imaging was performed 1 day after the implantation and every other day thereafter. After each FL imaging, half of the mice underwent gavage with AT PBS solution (1.25 mM; 20 μL), and the other half did so with PBS only (20 μL). After 23 d of treatment, the mice were killed, and xenograft tissue samples were collected for Western blot analysis of phospho c-Myc, c-Myc, and α-tubulin proteins.
Hydrodynamic Injection.
Eμ-tTA/Tet-O-MYC transgenic mice (13) (n = 2) were removed from doxycycline treatment at 8 wk of age and underwent gavage with AT or PBS as described above. At 29 d after treatment, these two mice underwent imaging studies to characterize the liver phospho c-Myc level after hydrodynamic injection (34) of the c-Myc sensor. In brief, 5 μg of the single vector version of the sensor plasmid was mixed with 1 μg of hRL plasmid (as a control for transfection efficiency) into 2 mL of a 0.9% saline solution and injected within 8 s via the tail vein using a 3-mL syringe fitted with a 27-gauge needle. FL imaging was acquired with D-Luc at 22 h after injection. RL imaging was acquired with Clz at 48 h after injection. The mice were killed after imaging, and liver tissue samples were collected for Western blot analysis of phospho c-Myc, c-Myc, and α-tubulin proteins and for H&E staining (Histo-Tec Laboratory).
Data Analysis.
Curve fitting, correlation coefficients, and Student's t test were obtained using Microsoft Excel. Data are presented as mean ± SD for in cellulo experiments and as mean ± SEM for in vivo experiments. P < 0.05 was used as cutoff point for statistical significance.
Supplementary Material
Acknowledgments
We thank Dr. Rosalie Sears for her helpful advice on the phosphorylation of c-Myc oncoprotein. This work was supported in part by National Cancer Institute Grants R01 CA82214 and ICMIC P50 CA114747 (to S.S.G.). H.F.-M. is supported by National Institutes of Health Training Grant IH R25T CA118681. T.F.M. received partial salary support from the National Institute for Health Research's Cambridge Biomedical Research Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007443107/-/DCSupplemental.
References
- 1.Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Sodir NM, Evan GI. Nursing some sense out of Myc. J Biol. 2009;8:77.1–77.4. doi: 10.1186/jbiol181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 5.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Wang YH, et al. Knockdown of c-Myc expression by RNAi inhibits MCF-7 breast tumor cells growth in vitro and in vivo. Breast Cancer Res. 2005;7:R220–R228. doi: 10.1186/bcr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura S, Maekawa T, Hirakawa K, Murakami A, Abe T. Alterations of c-myc expression by antisense oligodeoxynucleotides enhance the induction of apoptosis in HL-60 cells. Cancer Res. 1995;55:1379–1384. [PubMed] [Google Scholar]
- 9.Kim HG, Miller DM. Inhibition of in vitro transcription by a triplex-forming oligonucleotide targeted to human c-myc P2 promoter. Biochemistry. 1995;34:8165–8171. doi: 10.1021/bi00025a023. [DOI] [PubMed] [Google Scholar]
- 10.Berg T, et al. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc Natl Acad Sci USA. 2006;103:6344–6349. doi: 10.1073/pnas.0601418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shachaf CM, et al. Inhibition of HMGcoA reductase by atorvastatin prevents and reverses MYC-induced lymphomagenesis. Blood. 2007;110:2674–2684. doi: 10.1182/blood-2006-09-048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 15.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 16.Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. Locating a protein–protein interaction in living cells via split Renilla luciferase complementation. Anal Chem. 2003;75:4176–4181. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- 17.Luker GD, Pica CM, Song J, Luker KE, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 18.Chan CT, et al. Molecular imaging of the efficacy of heat shock protein 90 inhibitors in living subjects. Cancer Res. 2008;68:216–226. doi: 10.1158/0008-5472.CAN-07-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulmurugan R, Gambhir SS. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein–protein interactions. Anal Chem. 2007;79:2346–2353. doi: 10.1021/ac062053q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulmurugan R, Gambhir SS, Paulmurugan R, Gambhir SS. Monitoring protein–protein interactions using split synthetic renilla luciferase protein-fragment–assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luker KE, et al. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Muller J, Eilers M. Ubiquitination of Myc: Proteasomal degradation and beyond. Ernst Schering Found Symp Proc. 2008;2008(1):99–113. doi: 10.1007/2789_2008_103. [DOI] [PubMed] [Google Scholar]
- 25.Dajani R, et al. Crystal structure of glycogen synthase kinase 3 beta: Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 26.Robanus-Maandag EC, et al. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC–amplified breast carcinomas. J Pathol. 2003;201:75–82. doi: 10.1002/path.1385. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia K, et al. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 28.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Fest T, Guffei A, Williams G, Silva S, Mai S. Uncoupling of genomic instability and tumorigenesis in a mouse model of Burkitt's lymphoma expressing a conditional box II–deleted Myc protein. Oncogene. 2005;24:2944–2953. doi: 10.1038/sj.onc.1208467. [DOI] [PubMed] [Google Scholar]
- 30.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucek L, et al. Modeling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Riggelen J, et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 2010;24:1281–1294. doi: 10.1101/gad.585710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.