Abstract
The development of biomolecular devices that interface with biological systems to reveal new insights and produce novel functions is one of the defining goals of synthetic biology. Our lab previously described a synthetic, riboregulator system that affords for modular, tunable, and tight control of gene expression in vivo. Here we highlight several experimental advantages unique to this RNA-based system, including physiologically relevant protein production, component modularity, leakage minimization, rapid response time, tunable gene expression, and independent regulation of multiple genes. We demonstrate this utility in four sets of in vivo experiments with various microbial systems. Specifically, we show that the synthetic riboregulator is well suited for GFP fusion protein tracking in wild-type cells, tight regulation of toxic protein expression, and sensitive perturbation of stress response networks. We also show that the system can be used for logic-based computing of multiple, orthogonal inputs, resulting in the development of a programmable kill switch for bacteria. This work establishes a broad, easy-to-use synthetic biology platform for microbiology experiments and biotechnology applications.
Keywords: gene expression, kill switch, RNA engineering, synthetic biology, systems biology
Synthetic biology seeks to reprogram organisms to alter natural biological behavior or perform novel functions, using engineered gene circuits, pathways, and whole genomes (1–8). Engineered gene circuits, in this regard, are typically constructed by assembling well-characterized biological components into unique architectures to achieve these unnatural capabilities, which are commonly used in biotechnology applications. A need also exists for synthetic biomolecular devices that interface with endogenous systems to track, probe, and influence, rather than reprogram, cellular physiology. Synthetic gene expression platforms that could accomplish this task would be well suited for use in a wide variety of phenotypic and genetic analyses aimed at expanding our knowledge of natural biomolecular components and networks, as well as enhancing our understanding of network dynamics under an array of conditions.
In this work, we showcase an engineered riboregulator system that controls gene expression posttranscriptionally through highly specific RNA–RNA interactions (9). RNA molecules are useful components in synthetic biomolecular devices (10, 11) due to their large sequence space, the number of unique structures that can be adopted, the predictability of structure stability, and the range of functions enabled by these structures. Together, these features make RNA molecules ideal for interfacing with biological systems given that they can be used to control gene expression, sense biomolecules, and serve as foundational devices that can perform complex cellular behavior. As such, the engineered, RNA-based device space is remarkably diverse. Other successful examples include independent transcription–translation networks based on orthogonal mRNA–ribosome pairs (12), riboswitches in which RNA aptamers directly control translation (13–15), riboswitches composed of interacting RNA aptamers and mRNA destabilizing ribozymes (16, 17), and RNA-based transcription factors that activate gene expression (18, 19).
In our riboregulator system (Fig. 1), distinct promoters independently regulate the transcription of two RNA species—a cis-repressed mRNA (crRNA) and a noncoding, trans-activating RNA (taRNA). Initially, target gene translation of the crRNA is blocked by a stem loop, which spontaneously forms in its 5′-untranslated region (UTR), sequesters the ribosome binding site (RBS), and prevents ribosome docking; stem loop formation is mediated by a short sequence (cis-repressive sequence, ≈25 nt) located within the mRNA 5′-UTR that binds the RBS. The taRNA contains a sequence (≈26 nt) that is complementary to the cis-repressive sequence and is capable of activating translation. When both RNA species are expressed, the taRNA targets the crRNA and out-competes the cis-repressive element, leading to stem loop unwinding, availability of the RBS for ribosome binding, and translation of the target protein. Recently, this system was applied as the basis for a genetic circuit that can count user-defined inputs (20).
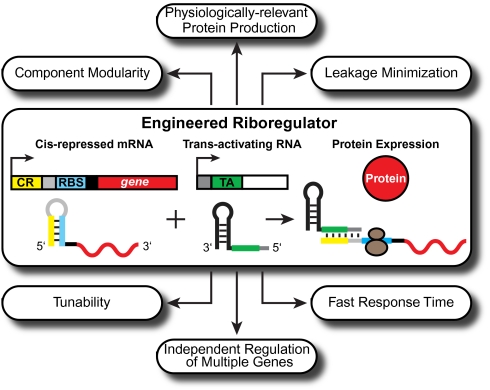
Fig. 1.
The riboregulator and its features. Our riboregulator system regulates gene expression posttranscriptionally through the specific interaction between the crRNA and taRNA (9). The cis-repressed mRNA (crRNA) is composed of the cis-repressive sequence (CR, yellow), the RBS (blue), a stem loop (light gray), and the target gene (red). The trans-activating RNA (black) contains the trans-activating sequence (TA, green) and the stem loop binding sequence (dark gray). To initiate translation, the taRNA binds the crRNA and grants the ribosome (brown circles) access to the RBS.
The riboregulator system possesses a unique set of collective features (Fig. 1), which makes it an ideal synthetic biology platform for interfacing with and exploring different microbial systems. These features include component modularity, physiologically relevant protein production, leakage minimization, fast response time, tunability, and the potential to be scaled up to independently regulate multiple genes simultaneously. Here we apply the riboregulator system in a series of in vivo experiments aimed at demonstrating these advantageous features. We tracked GFP fusion protein expression from environmentally sensitive promoters in wild-type cells, highlighting the system’s physiologically relevant protein production and component modularity. We also analyzed the systems-level biological response to expression of the CcdB toxin, illustrating the platform’s low leakage. Additionally, we sensitively perturbed the response dynamics of a core biological response network following DNA damage, showcasing the tunable gene expression and fast response time one can achieve with the riboregulator. Finally, we computed orthogonal inputs that affect inner membrane permeability and outer membrane integrity, respectively, to combinatorially lyse cells, demonstrating how interoperable riboregulators can be used to independently regulate multiple genes inside a cell and create a programmable kill switch for bacteria. Together, these studies show how the riboregulator system facilitates microbiology experiments that would otherwise prove difficult to execute using commonly used methods and present an exciting synthetic biology tool for inducing rapid bacterial cell death.
Results and Discussion
Physiologically Relevant Protein Production and Component Modularity.
The riboregulator platform is well suited for physiologically relevant expression of a target gene, an important factor when interfacing an artificial system with cellular networks (9). Meaningful protein localization studies require tight control over fluorescent fusion protein expression at physiologically relevant levels where common technical issues (e.g., inclusion body formation and obscuring membrane localization with excess cytoplasmic fluorescence) are avoided. To demonstrate the effectiveness of our synthetic riboregulator system in this capacity, we used it to track the expression of TonB, a protein that bioenergetically facilitates iron import and exhibits a unique localization pattern (21). The N terminus of TonB localizes within the cytoplasmic membrane, whereas its C terminus shuttles back and forth in the periplasm from the inner to the outer membrane (22). Because GFP does not normally fluoresce in the periplasm (23), an N-terminal GFP-TonB fusion was constructed; the fusion design was based on a previous study and included a short helical linker to enhance expression (Fig. 2A) (22, 24). The anhydrotetracycline (aTc)-inducible PLtetO-1 promoter (25) regulated crRNA and taRNA expression for the first series of experiments described below.
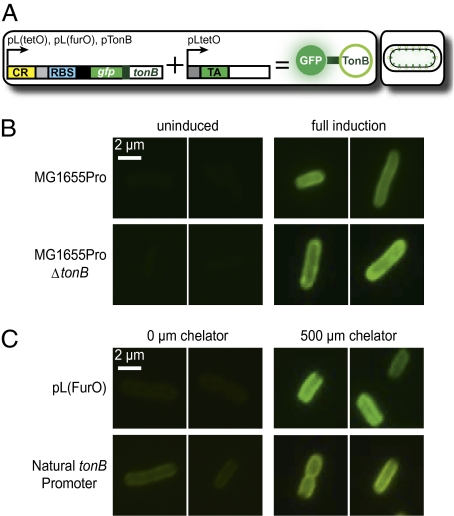
Fig. 2.
Fluorescent imaging of GFP-TonB. All images are representative fluorescent micrographs taken at 90 min postinduction. (A) Riboregulator system. (B) Micrographs of GFP-TonB fluorescence in MG1655Pro and MG1655ProΔtonB cells. (C) Micrographs of GFP-TonB expression from PLfurO and pTonB in MG1655Pro. Additional micrographs are presented in Fig. S1.
We consistently found that micrographs of fully induced MG1655ProΔtonB Escherichia coli cultures clearly showed proper localized fluorescence at the inner membrane by 1.5 h after induction (Fig. 2B). Conversely, we routinely observed dim fluorescence when MG1655ProΔtonB cultures were uninduced. Often, the native, untagged version of the target gene is knocked out in successful protein localization studies. To determine the potential influence of native TonB expression on the localization pattern of riboregulated GFP-TonB, we also monitored fluorescence in MG1655Pro E. coli. Upon induction, we found that there was no significant difference in the degree of localization or GFP-TonB fluorescence when the genomic copy of tonB was present or absent (Fig. 2B). These findings imply that intracellular localization dynamics of a riboregulated gene are not affected by the native expression of that gene, thereby highlighting the physiological relevance of the synthetic system. Moreover, these results suggest that riboregulated GFP fusion construct expression does not exert epigenetic effects on the natural system being explored.
Another important aspect of the riboregulator design is component modularity, which extends to the target gene and promoter elements. The modularity feature allows for the creation of basic sensors that use both synthetic and natural promoters to report on environmental conditions. Perhaps more importantly, this feature readily provides for the integration of a riboregulated gene of interest into the framework of natural response networks. We exploited the modularity of the riboregulation system by incorporating environmentally sensitive promoters—namely synthetic PLfurO (26) and natural pTonB (27)—as regulators of crRNA transcription. The transcriptional regulator Fur, which binds to Fur-operator sites in the presence of iron, represses both promoters (26–28). Derepression of Fur-regulated promoters can be achieved by addition of 2,2′-dipyridyl, an iron chelator that causes iron starvation.
After addition of dipyridyl, both environmentally sensitive riboregulator systems successfully sensed iron starvation, displaying strong fluorescence and proper localization of GFP-TonB (Fig. 2C). By retaining PLtetO-1 as the taRNA promoter, we controlled the sensitivity of the de facto iron sensor through the taRNA concentration. The micrographs in Fig. 2C were obtained following full induction of the taRNA, allowing us to observe the highest degree of fluorescence possible with chelator present. However, full taRNA induction may be responsible for the increased amount of background fluorescence in the no chelator cultures relative to the uninduced cultures in the previous experiment (Fig. 2 B and C). Nonetheless, a significant difference between cells in the presence and the absence of iron chelator was evident, especially in the PLfurO setup.
GFP-TonB localization in the inner membrane was clearly observed without saturation of fluorescence, regardless of native TonB expression. To track a protein of interest at a high resolution, physiologically relevant gene expression is critical and renders knocking out the native gene unnecessary. Along these lines, ectopic expression of a GFP reporter from our platform does not significantly alter the natural behavior of the system under study. Furthermore, exploiting component modularity by using iron-sensitive promoters allows for the integration of our fusion construct into the natural iron homeostasis regulon. This intertwining of a synthetic device with a biological response allows for measurement of the timing and quantification of gene expression. Thus, the riboregulator is an effective tool for precisely measuring regulon dynamics, as well as easily tracking related protein localization. With respect to biotechnology and biomonitoring applications, the designed component modularity dictates that the number of biosensor setups that can be constructed with a riboregulator is constrained only by the supply of identified, environmentally sensitive promoters.
Leakage Minimization.
In the riboregulator system, cis-repression significantly suppresses target gene translation (9), resulting in very low leakage. This suppression permits successful cloning and controlled expression of toxic proteins with high sensitivity at the mRNA level. The bactericidal effect of such proteins has rendered thorough in vivo, systems-level characterization of the pathways and networks leading to toxic peptide-induced cell death prohibitive, even in wild-type backgrounds. This is especially true of strains with impaired defense or recovery systems, where genetic analyses of the comparatively more vulnerable, mutant bacterial strains may provide opportunities to find important subnetworks or entirely alternative networks that may otherwise be masked or repressed by primary, global responses.
To illustrate the suitability of our riboregulator platform for the discovery of activated biological networks during cell death achieved by highly toxic proteins, we used it to tightly regulate the expression of CcdB, a potent, DNA-damaging toxin. Once free from CcdA, its cognate antitoxin, CcdB achieves its toxic effects via binding with the GyrA subunit of DNA-bound DNA gyrase (29, 30). This interaction converts DNA gyrase into a protein mediator of cell death and results in DNA damage (31), cellular filamentation (32), and the induction of the SOS DNA damage repair response (33); notably, these effects of CcdB poisoning have been explored in vitro or in unnatural in vivo setups.
Previously, we compared the effects of gyrase poisoning, by fluoroquinolone antibiotics and riboregulated CcdB, on survival and primary stress response activation in wild-type MG1655 E. coli (26). The ability of the toxin to induce DNA damage was apparent in the strong up-regulation of SOS-related genes and the related decrease in viability (26). In the present study, we transformed the CcdB riboregulator plasmid (Fig. 3A) into highly sensitive MG1063 E. coli to determine if our experimental platform was suitable for exacting systems biology-based determination of alternative sensing mechanisms, secondary response pathways, and subnetwork activation. Derived from the same parental strain as MG1655, MG1063 cells encode the mutant recA56 allele, which has been shown in vivo to be deficient in the RecA functions of recombination, induction of LexA cleavage and the SOS response, and SOS mutagenesis (34).
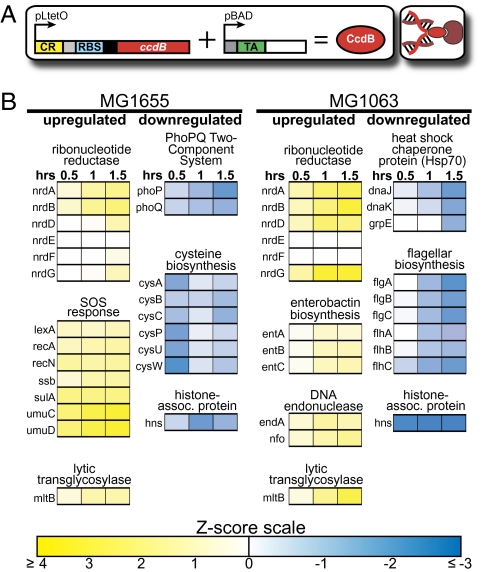
Fig. 3.
Transcriptome response to CcdB expression. (A) Riboregulator system. (B) Portion of the time-course gene expression response of CcdB-expressing MG1655 and MG1063 cells. Yellow and blue represent up-regulation and down-regulation, respectively. See SI Text for details concerning our analysis methods and Dataset S1 for the full microarray results.
Because MG1655 and MG1063 both lack the transcriptional repressor, TetR, CcdB crRNA was constitutively transcribed from the PLtetO-1 promoter (25) in our setup. As evidenced by the successful transformation of only those cells receiving the CcdB riboregulator plasmid (Fig. S2), the translational repression afforded by our system efficiently prevented undesired CcdB expression, thereby demonstrating the effectiveness of the designed cis-repressive secondary structure and allowing for assays that explored the deadly phenotypic effect of CcdB in both strains (Figs. S2 and S3). In addition to these assays, microarray analysis was performed to determine the transcriptome response of MG1063 cells to fully induced CcdB expression and to compare these expression changes to the wild-type MG1655 response. Significantly changing genes (z-score) were determined on a genewise basis, and microarray data were enriched by transcription factor (35, 36) and biochemical pathway classifications (37, 38). By using this approach, we could focus on the regulators and biological function of those genes in our datasets to identify components of the physiological cascade that lead to CcdB-induced cell death in different bacterial strains and determine variations between their stress responses.
Systems-level transcriptome profiling of MG1063 cells uncovered a unique set of coordinated gene expression changes following CcdB poisoning in the absence of RecA functionality and an intact SOS DNA damage repair response (Fig. 3B). As anticipated, we did not observe appreciable expression of SOS-related genes in MG1063 cells, but we did find significant and noncanonical up-regulation of both aerobic (nrdAB) and anaerobic (nrdDG) ribonucleoside reductases (39). In contrast, MG1655 cells exhibited up-regulation of only the aerobic ribonucleoside reductase operon along with SOS activation, suggesting that MG1063 cells sense both the need to synthesize nucleotides for (at least) DNA replication and a lack of their utilization, while seeking out alternative mechanisms to deal with this problem. Additionally, several interesting transcriptional changes with bioenergetic implications were also observed in MG1063 cells, namely up-regulation of fatty acid biosynthesis (Dataset S1), enterobactin iron-siderophore synthesis and DNA endonuclease (class I, endA; class IV, nfo) expression and down-regulation of DnaJ/DnaK/GrpE heat shock chaperone system expression and flagellum biosynthesis. Finally, among the unique transcriptional changes observed in wild-type MG1655 cells were significant down-regulation of the PhoPQ two-component Mg2+ sensor and cysteine biosynthesis-related gene expression.
Leakage minimization and a tight off state make our riboregulator system ideal for use in detailed in vivo analyses (e.g., transcriptome profiling and determination of time- or cycle-dependent effects) of, in principle, any protein, regardless of its toxicity or the genetic background of the host cell. Here, riboregulation of CcdB permitted the identification of the core pathways and primary responses following gyrase poisoning in wild-type cells and, perhaps more importantly, the discovery of potentially critical secondary biochemical effects in a strain where the primary response to DNA damage is genetically impaired. Further research is needed to determine whether these secondary networks (or potential subnetworks) are simply backup responses or are supporting responses masked by the SOS network.
Tunable Gene Expression and Fast Response Time.
The riboregulator can be used to achieve precise levels of gene expression in a tunable, reliable fashion, much like a rheostat. Tunable gene expression is critical if one is aiming to perturb a system with high precision and obtain desired concentrations of proteins within a target network. Accordingly, the ability to vary the concentration of such proteins may result in graded genetic and phenotypic responses. Thus, our system facilitates more sensitive analyses of a target network than typical experiments that rely upon overexpression or gene knockouts alone.
We used the riboregulator to tunably regulate the mutant repressor, LexA3 (Ind−), to demonstrate the system’s ability to sensitively and rapidly interrogate endogenous genetic networks. Under homeostatic conditions, the transcription factor LexA represses the genes of the SOS response regulon, including itself (40). Upon recognition of DNA damage, LexA is cleaved through autocatalysis promoted by an activated form of RecA, a multifunctional regulator (41). LexA3 (Ind−) is a single amino acid mutant of LexA that is resistant to autocatalytic cleavage and therefore is a potent suppressor of the SOS response (42). In our study, LexA3 expression was riboregulated in MG1655Pro E. coli (Fig. 4A), and norfloxacin, a fluoroquinolone antibiotic, was used to generate DNA damage, thereby inducing the SOS response. MG1655Pro cells encode tetR and lacIq (providing increased LacR), affording for particularly tight repression of the aTc-inducible crRNA promoter, PLtetO-1, and the IPTG-inducible taRNA promoter, PLlacO-1, respectively (25).
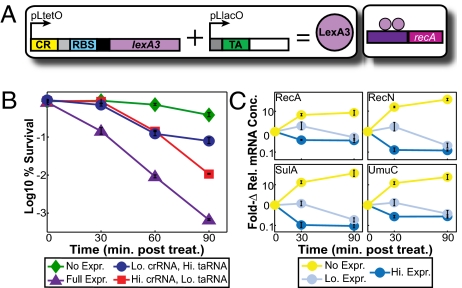
Fig. 4.
Effect of LexA3 expression on the SOS response. All samples, except for untreated controls, were treated with norfloxacin at various expression levels of LexA3. Graphs depict the triplicate mean ± SEM. (A) Riboregulator system. (B) Log % survival of cells. (C) Fold changes in relative mRNA concentrations for selected SOS genes. See Dataset S2 for the full qPCR data set.
We first determined the phenotypic response to riboregulated repressor expression by measuring the survival of norfloxacin-treated cultures. Full induction of the LexA3 repressor during drug treatment dramatically decreased survival by almost 3-log at 90 min posttreatment, compared with <1-log in cultures in which repressor expression was not induced (Fig. 4B). This decrease in viability was apparent at 30 min posttreatment, illustrating the fast response time of riboregulation. Importantly, we observed a graded phenotypic response when we tuned the concentrations of aTc or IPTG, thus lowering the levels of crRNA or taRNA, respectively.
To characterize the ability of our system to alter the response dynamics of a specific biological network, we performed quantitative PCR (qPCR) on four LexA-regulated genes that are highly expressed during SOS response induction: recA, recN (DNA repair recombination protein), sulA (cell division inhibitor), and umuC (important for SOS mutagenesis and repair) (40, 43). Norfloxacin was added to cultures to induce the SOS response, and the relative mRNA concentrations for these four genes were measured at various LexA3 expression levels. As expected, when repressor expression was not induced, mRNA transcript levels for each network gene studied increased dramatically, up to 37-fold by 90 min posttreatment in the case of sulA (Fig. 4C). Full induction of the riboregulator not only prevented this jump in relative mRNA levels, but also reduced the number of network mRNA transcripts between 2.5- and 9.5-fold relative to untreated cultures. Therefore, our qPCR data were consistent with the observed decrease in cell viability. Again, the rapid response time of our system was demonstrated by the knockdown of transcript levels by 30 min posttreatment.
We also explored the effect of decreased LexA3 expression on SOS gene expression by tuning the taRNA concentration. For each studied gene, we established a reliable graded response of relative mRNA levels at 30 min posttreatment (Fig. 4C). Interestingly, we found that regardless of repressor expression levels (i.e., high or low), relative mRNA concentrations converged to a minimum level by 90 min posttreatment. Low repressor expression (high crRNA–low taRNA) led to a temporal delay in the drop of mRNA transcript levels (Fig. 4C), and this delay was manifested in the almost 1-log increase in survival of the corresponding sample at 30 min posttreatment (Fig. 4B). Thus, tinkering with response dynamics directly impacted the survival phenotype. These findings indicate that the riboregulator system can be used to test the robustness of a network to perturbations in the timing or magnitude of component gene expression, synthetically simulating network behaviors potentially elicited by volatile, nonlaboratory conditions.
Although all four genes investigated exhibited a similar response to repressor expression after norfloxacin treatment, the riboregulator system preserved the natural differences in gene expression among these regulon members and, presumably, within the network (Fig. S4). For example, recA mRNA was significantly more abundant than umuC mRNA, regardless of the LexA3 expression level (40). Thus, riboregulated repressor altered response dynamics while preserving the natural architecture of the network.
Tunability facilitates detailed study of networks and can be used to analyze threshold responses and identify complementary networks. Furthermore, tuning within a physiologically relevant range of expression is needed to avoid attaining only simple network responses, e.g., step functions. Riboregulation of a repressor rapidly and precisely influenced our model network during a stimulus to achieve graded transcription rates of network genes and graded levels of a phenotypic response. The repressor-mediated network manipulation approach can be extended to tunably repress any gene by modifying its promoter to include binding sites for a repressor. Using this type of setup, individual nodes of a response could be dampened, therefore probing a natural circuit by “shorting” particular connections.
Independent Regulation of Multiple Genes.
Different variants of the riboregulator platform can be constructed by exploiting the sequence specificity of RNA interactions (9). Minimal reciprocal changes in the cis-repressive and trans-activating sequences give rise to orthogonal riboregulator systems, whereby induction of a specific taRNA will affect only its cognate crRNA. From a synthetic biology viewpoint, the orthogonal riboregulators can be designed to integrate multiple inputs and function as logic gates. In principle, it is feasible to use these riboregulators to create a fast, powerful, programmable kill switch for microbes.
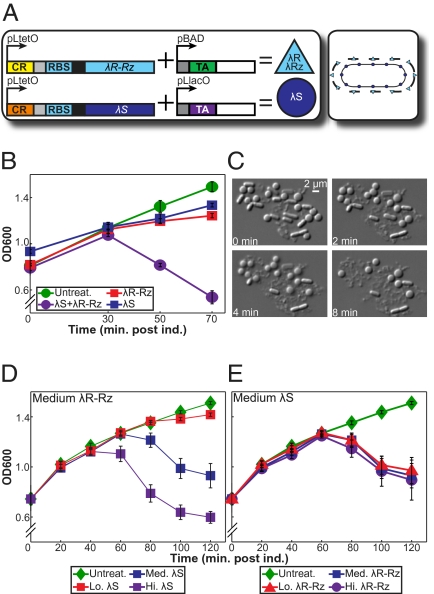
To demonstrate the potential of using orthogonal riboregulators to rapidly kill bacterial cells through AND gate logic, we used the λ-phage lysis genes: λR, λRZ, and λS (44). λR, an endolysin, and λRZ, an endopeptidase, degrade peptidoglycan (PG) but cannot access the PG layer without the holin λS permeabilizing the inner membrane (45). Previous studies have shown that cell lysis occurs only when both λR and λS are present at significant concentrations (46). In addition, past work has revealed that λR expression alone has no effect on cells (47), and λS expression alone results in a loss of cell viability but no cell lysis (46). Although not required for lysis in most synthetic setups, λRZ overlaps λR in the λ-phage genome and is necessary for cell lysis in λ-phage infected cells (46, 48). Therefore, we cloned λR-RZ and λS under the control of two different riboregulator variants; these two independent systems were subcloned into one plasmid to ensure that both variants were present at the same copy number (Fig. 5A). This riboregulated AND gate requires aTc, IPTG, and arabinose to flip the kill switch and lyse E. coli.
Fig. 5.
Independent regulation of λ-phage lysis proteins in a bacterial kill switch. Graphs depict the triplicate mean ± SEM. (A) Riboregulator system. (B) Effect of λR-RZ and λS expression on OD600. Addition of glucose was necessary to further reduce PBAD leakage and prevent lysis (see SI Text and Fig. S5 for details). (C) Images illustrating cell lysis were obtained from a movie file (Movie S1) that was recorded at 45 min postinduction. (D) Variable λS expression at constant, medium λR-RZ expression. See Fig. S6 for full dataset. (E) Variable λR-RZ expression at constant, medium λS expression.
We measured optical density (OD600) to monitor the potency of the kill switch. Full activation of λR-RZ and λS resulted in a sharp decrease in OD600 starting at ≈30 min postinduction (Fig. 5B); the occurrence of lysis was confirmed by microscopy (Fig. 5C). The cell wall was weakened by endolysin activity, and the rod shape of E. coli devolved to a spheroid shape and eventually collapsed into a pile of debris. When expressing only λR-RZ or λS, the steep decline in OD600 was not observed. In these latter experiments, we observed a small decrease in growth rate and no lysis (Fig. 5B). Because aTc-inducible PLtetO-1 (25) regulates both λR-RZ and λS transcription, these results show that the specificity between the taRNA and cognate crRNA, plus the inhibition of translation from the cis-repressive element, is sufficient to achieve independent regulation of the two genes and keep the uninduced kill switch tightly off.
The independent control afforded by orthogonal riboregulation is also particularly useful for studying a biological response stimulated by a combination of factors; e.g., separately regulating the stimuli enables insight to be gained into the robustness of the response as well as the relationship between the individual inputs. We exploited this feature to determine the effect on lysis of separately tuning the concentration of λR-RZ and λS. Decreasing the λS concentration at a constant level of λR-RZ induction resulted in significant reductions in the magnitude of cell lysis as measured by OD600 (Fig. 5D). Comparatively, the λS concentration was held constant and a similar range of λR-RZ expression was established. As observed in Fig. 5E, there was no significant difference in OD600 between high and low induction of λR-RZ. These results suggest that the holin concentration is the most important determinant in the degree of lysis observed.
The highly specific interactions between complementary crRNA and taRNA sequences enable the construction of simple logic circuits, such as AND, OR, NAND, and NOR gates. In this study, an AND gate functions as a programmable kill switch. By computing three inputs, the kill switch exploits leakage minimization to keep the cells alive until the precise induction conditions are met, at which point the switch rapidly induces widespread lysis. Due to the system’s component modularity, the kill switch can be programmed to use any inducer, any change in environmental conditions, or any combination of the two with the appropriate promoters. Going forward, the flexibility and effectiveness of our cell suicide platform make it an exciting tool for biotechnology applications.
Conclusions
In this work, we demonstrate and harness unique features of the riboregulator, an RNA-based, synthetic gene expression system. The advantages of riboregulation over other gene expression systems include expression of physiologically relevant amounts of a target gene, modularity of both the crRNA and the taRNA promoters, tight off state that minimizes leakage, fast response times, tunable regulation of gene expression, and the ability to combine orthogonal riboregulators to independently control the expression of multiple genes. We successfully leveraged these features to interface our platform with microbial physiology in a variety of manners, demonstrating that the platform is appropriate for a number of advanced microbiology experiments and developing a potent, programmable kill switch for bacteria.
The specific RNA–RNA interactions used by the riboregulation platform during trans-activation of gene expression provide a powerful means for the exploration of bacterial gene networks and response pathways. Perhaps more importantly, as demonstrated in our CcdB toxin and cell lysis experiments, cis-repression of mRNA translation provides tight enough regulation for performing exacting microbiological tasks, despite the absence of transcriptional control. On the basis of designed modularity, however, the ability to incorporate any promoter into the system opens up countless additional possibilities for unique experimental setups and biotechnology applications. Expanding the currently limited list of well-characterized natural and engineered promoters is needed to improve and extend the already considerable utility of the platform. As such, the significant present capacity of the synthetic riboregulator device to successfully integrate into microbial physiology, and to track, tune, and terminate biological systems, can be enhanced only by additional layers of transcriptional control.
Materials and Methods
Plasmid Construction, Strains, and Reagents.
Basic molecular biology techniques were implemented as previously described (49). On the basis of the published design (9), we constructed various riboregulator systems (Table S1), each containing the ColE1 high-copy replication origin and kanamycin resistance gene. See SI Text for details on plasmid construction. For experimental purposes, we used four related E. coli K-12 derivative strains, MG1655 (F−, λ−; ATCC no. 47076) (50), MG1063 (F+, λ−, recA56, thi; Yale Coli Genetic Stock Center no. 6199) (51), MG1655Pro (F−, λ−, Spr, lacR, tetR) (25), and MG1655ProΔtonB. In all experiments, we used a combination of the following inducers: anhydrotetracycline (aTc; Sigma), arabinose (Acros), IPTG (Fisher), and 2,2′-dipyridyl (Sigma).
Microscopy.
To track GFP-TonB, we used a Nikon Eclipse Ti microscope with a 100× objective, outfitted with a CoolSnap HQ2 CCD camera (Photometrics), operated with NIS-Elements Advanced Research 3.0 software. The Nikon Intensilight C-HGFIE provided fluorescent light.
RNA Microarrays.
To measure the transcriptome response to CcdB expression, we used Affymetrix E. coli Antisense Genome arrays and an Affymetrix GeneChip Scanner 3000. See SI Text for details on RNA extraction, cDNA preparation, GeneChip array preparation, array data analysis, and systems biology analyses.
Growth Analyses.
To calculate the survival of norfloxacin-treated cultures containing the LexA3 riboregulator, we performed a serial dilution and calculated CFU/mL and survival with the following formulas:
See SI Text for details on the cell viability assay.
To monitor lysis in cultures containing the λR-RZ and λS riboregulators, we measured OD600 with a SPECTRAFluor Plus (Tecan). To record the movie confirming the occurrence of lysis, we used a Nikon Eclipse 80i microscope with a 40× objective, outfitted with a CoolSnap HQ CCD camera (Roper Scientific), operated with NIS-Elements Advanced Research 3.0 software.
Network Manipulation Real-Time PCR.
We performed quantitative PCR using the Roche LightCycler 480. We estimated the relative RNA concentrations for recA, recN, sulA, and umuC using relative quantification and using lplT, a lysophospholipid transporter, and rrsH, 16S ribosomal RNA, as reference genes. We also analyzed a no cDNA template control and an RNA only (no reverse transcriptase during cDNA preparation) control. See SI Text for details on RNA extraction, cDNA preparation, qPCR protocol, and qPCR data analysis.
Supplementary Material
Acknowledgments
We thank Ahmad Khalil for microscopy assistance and advice on data presentation. We thank Sheetal Modi for qPCR assistance and Raffi Afeyan, Henry Lee, Kevin Litcofsky, and Jonathan Winkler for helpful discussions. We are grateful to Mary Berlyn of the Yale Genetic Stock Center for help in selecting the appropriate E. coli strains used in the CcdB analysis. This work was supported by the National Institutes of Health through the National Institutes of Health Director’s Pioneer Award Program, Grant DP1 OD003644, by the National Science Foundation, and by the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: D.J.D., F.J.I, C.R.C., and J.J.C. have a patent for the riboregulator system pending.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009747107/-/DCSupplemental.
References
- 1.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: New engineering rules for an emerging discipline. Mol Syst Biol. 2006;2:2006–0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 5.Khalil AS, Collins JJ. Synthetic biology: Applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherji S, van Oudenaarden A. Synthetic biology: Understanding biological design from synthetic circuits. Nat Rev Genet. 2009;10:859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tigges M, Fussenegger M. Recent advances in mammalian synthetic biology-design of synthetic transgene control networks. Curr Opin Biotechnol. 2009;20:449–460. doi: 10.1016/j.copbio.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 10.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 11.Davidson EA, Ellington AD. Synthetic RNA circuits. Nat Chem Biol. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- 12.An W, Chin JW. Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci USA. 2009;106:8477–8482. doi: 10.1073/pnas.0900267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 15.Topp S, Gallivan JP. Riboswitches in unexpected places—a synthetic riboswitch in a protein coding region. RNA. 2008;14:2498–2503. doi: 10.1261/rna.1269008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yen L, et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 17.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha S, Ansari AZ, Jarrell KA, Ptashne M, Jarrell KA. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 2003;31:1565–1570. doi: 10.1093/nar/gkg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 22.Kaserer WA, et al. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol. 2008;190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 25.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postle K. Aerobic regulation of the Escherichia coli tonB gene by changes in iron availability and the fur locus. J Bacteriol. 1990;172:2287–2293. doi: 10.1128/jb.172.5.2287-2293.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 29.Tam JE, Kline BC. Control of the ccd operon in plasmid F. J Bacteriol. 1989;171:2353–2360. doi: 10.1128/jb.171.5.2353-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki S, Takiguchi S, Miki T, Horiuchi T. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. Antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J Biol Chem. 1992;267:12244–12251. [PubMed] [Google Scholar]
- 31.Couturier M, Bahassi el-M, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 32.Jaffé A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer S, Bailone A, Devoret R. SOS induction by thermosensitive replication mutants of miniF plasmid. Mol Gen Genet. 1985;198:456–464. doi: 10.1007/BF00332939. [DOI] [PubMed] [Google Scholar]
- 34.Lauder SD, Kowalczykowski SC. Negative co-dominant inhibition of recA protein function. Biochemical properties of the recA1, recA13 and recA56 proteins and the effect of recA56 protein on the activities of the wild-type recA protein function in vitro. J Mol Biol. 1993;234:72–86. doi: 10.1006/jmbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- 35.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salgado H, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34(Database issue):D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camon E, et al. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32(Database issue):D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Ed. Washington, DC: American Society for Microbiology; 2006. [Google Scholar]
- 41.Little JW. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 42.Markham BE, Little JW, Mount DW. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981;9:4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang IN, Smith DL, Young R. Holins: The protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 46.Garrett JM, Young R. Lethal action of bacteriophage lambda S gene. J Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrett J, et al. Cell lysis by induction of cloned lambda lysis genes. Mol Gen Genet. 1981;182:326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- 48.Young R, Way J, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: A new gene affecting cell lysis. J Mol Biol. 1979;132:307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 50.Bachmann B. In: Derivations and Genotypes of Some Mutant Derivatives of Escherichia coli K-12. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd Ed. Neidhardt F, et al., editors. Washington, DC: American Society for Microbiology; 1996. pp. 2460–2488. [Google Scholar]
- 51.Guyer MS. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978;126:347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.