Abstract
Two profilin isoforms (PFN1 and PFN2a) are expressed in the mammalian brain. Although profilins are essential for regulating actin dynamics in general, the specific role of these isoforms in neurons has remained elusive. We show that knockdown of the neuron-specific PFN2a results in a significant reduction in dendrite complexity and spine numbers of hippocampal neurons. Overexpression of PFN1 in PFN2a-deficient neurons prevents the loss of spines but does not restore dendritic complexity. Furthermore, we show that profilins are involved in differentially regulating actin dynamics downstream of the pan-neurotrophin receptor (p75NTR), a receptor engaged in modulating neuronal morphology. Overexpression of PFN2a restores the morphological changes in dendrites caused by p75NTR overexpression, whereas PFN1 restores the normal spine density. Our data assign specific functions to the two PFN isoforms, possibly attributable to different affinities for potent effectors also involved in actin dynamics, and suggest that they are important for the signal-dependent fine-tuning of neuronal architecture.
Keywords: hippocampus, neuronal cytoskeleton, p75 neurotrophin receptor, actin, dendritic complexity
Neuronal plasticity depends on functional changes at synapses and, additionally, on the spatial and temporal modulation of neuronal architecture, which is induced by the transmission of external signals to the cytoskeleton. Among the proteins engaged in the organization of the actin cytoskeleton are profilins (1) that bind to monomeric actin, polyproline-stretch proteins, and membrane-bound phospholipids (reviewed in ref. 2). In the mammalian brain, two different profilin isoforms are found: profilin 1 (PFN1), which is ubiquitously expressed in all eukaryotic cells, and profilin 2a (PFN2a), which is tissue-restricted and shows its highest expression level in the brain (3, 4). The cell- and tissue-specific role of profilins remains poorly understood. In particular, the precise function of neuronal PFN2a is still unclear. Recent evidence points to pre- and postsynaptic functions of both isoforms. Experiments with cultured hippocampal neurons revealed activity-dependent targeting of PFN1 (5) and PFN2a (6) into spines of excitatory neurons. Furthermore, Lamprecht et al. (7) demonstrated a stimulus-dependent accumulation of profilin, without isoform specification, in spines of neurons in the rat amygdala. In addition, NMDA receptor activation was seen to correlate with changes in spine morphology, a process apparently involving PFN2a, RhoA, and the RhoA-specific kinase ROCK (8). In contrast, data derived from a KO mouse indicate that PFN2a acts presynaptically, by controlling vesicle exocytosis and presynaptic excitability (9). The aim of the current study was to unravel the physiological role of PFN2a in regulating dendrite morphology and spine stability of mature pyramidal neurons. We used a loss-of-function approach inducing RNAi-mediated knockdown of PFN2a in hippocampal neurons. Furthermore, we investigated whether PFN2a might be involved in the regulation of actin dynamics downstream of known effectors of neuronal morphology, such as the pan-neurotrophin receptor p75 (p75NTR) in the adult nervous system (10, 11). We found that on knockdown of PFN2a, the number of both dendrites and spines is significantly reduced in hippocampal pyramidal neurons. The defective phenotype is reversed after reintroduction of PFN2a. Concomitant expression of PFN1 rescued the loss of spines but did not restore dendritic complexity. The differential effects of the two isoforms reside in their participation in two different actin signaling pathways. Coexpression experiments with p75NTR and profilins revealed that PFN1 and PFN2a cooperate in preventing p75NTR-dependent morphological alterations. These findings demonstrate that PFN2a indeed plays a crucial role in the maintenance of dendritic structure in mature hippocampal neurons and exerts PFN1-independent as well as redundant functions.
Results
Knockdown of PFN2a Reduces Dendritic Complexity of CA1 Pyramidal Neurons.
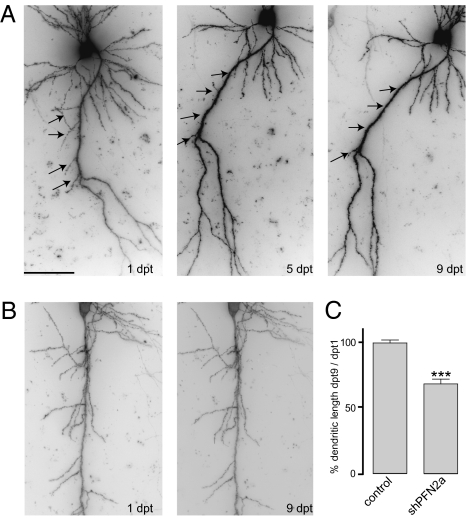
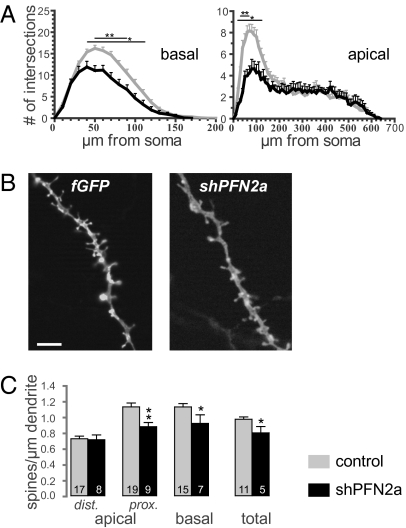
A vector-based RNAi approach was used to unravel the specific function of PFN2a in dendrite morphology and spine stability of hippocampal CA1 neurons. Knockdown of PFN2a by the vector short hairpin PFN2a (shPFN2a) (Fig. S1A) was confirmed in primary hippocampal neurons by immunocytochemistry using a PFN2a-specific antibody (12) and quantified as a reduction in PFN2a protein level of 73.3 ± 2% (SI Text and Fig. S1 C and D). To analyze fully developed principal neurons (13), organotypic hippocampal cultures were transfected at 7 d in vitro and imaged 1, 5, and 9 d posttransfection. Five days after transfection with shPFN2a, CA1 neurons showed a pruning of already existing dendrites that was not observed in control cells transfected with farnesylated GFP (fGFP) only (Fig. 1 A and B, arrows). The changes in dendrite structure remained stable until the last imaging time point. Quantification of changes in dendritic length (approximately the proximal 400 μm of the apical dendrite) revealed a 32% reduction compared with control cells (Fig. 1C). To analyze changes in dendrite structure in detail, we fixed transfected cells 7 d posttransfection and performed a Sholl analysis, plotting the number of dendritic branches in relation to their distance from the neuronal soma. Both apical and basal dendrites of shPFN2a-transfected CA1 neurons displayed a significant reduction of dendritic intersections when compared with control cells (Fig. 2A). Expression vectors against luciferase (shRNA against firefly luciferase, sifluc) or expressing fGFP alone were used as controls. Because no significant differences between the two control conditions were observed (Fig. S2), the results of the two approaches were combined.
Fig. 1.
Pruning of dendrites in shPFN2a-expressing cells. Hippocampal slice cultures were biolytically transfected with shPFN2a (A) or fGFP (B) at 7 DIV, and CA1 neurons were imaged at the indicated time points. Five days posttransfection (dpt), pruning of previously existing apical dendrites was observed in shPFN2a-expressing cells (arrows) that was not detectable in neurons transfected with fGFP. Changes in dendritic structure remained stable up to dpt 9. (C) Dendritic loss was quantified by measuring the first ≈400 μm of the apical dendrite at 1 and 9 dpt. Profilin2a-deficient neurons lost 32% of dendritic length, whereas the length of control cells was unaltered during the imaging period. (***P < 0.001) (Scale bar: A, 50 μm.)
Fig. 2.
Spine density is reduced in shPFN2a-transfected CA1 neurons. (A) Dendritic complexity of basal and apical dendrites was compared between control neurons (n = 25) and shPFN2a-transfected cells (n = 11) using Sholl analysis. (B) High-resolution images of representative basal dendrites of cells in organotypic cultures expressing fGFP or shPFN2a. (C) Spine density of control cells as well as cells expressing shPFN2a. *P < 0.05; **P < 0.005. (Scale bar: 5 μm.)
In a second set of experiments, we used a gain-of-function approach by overexpressing YFP-PFN2a for 48 h driven by a truncated CMV promoter. The overexpression was quantified to approximately 4-fold of the endogenous protein. These cells showed a slight but nonsignificant increase in overall dendritic complexity. A detailed Sholl analysis confirmed this impression for the basal as well as apical dendritic compartment (Fig. S3A).
In summary, our gain- and loss-of-function experiments demonstrate that PFN2a is an important regulator of CA1 pyramidal neuron morphology in the mature hippocampus.
PFN2a Controls Spine Stability in CA1 Pyramidal Neurons.
According to a previous report (6), PFN2a is targeted to dendritic spines in an activity-dependent manner and stabilizes these structures, thus interfering with structural plasticity. Therefore, we analyzed dendritic spine density changes in PFN2a-deficient cells. Detailed spine density counts were performed for the different dendritic compartments—basal dendrites as well as proximal and distal apical dendritic compartments—on CA1 pyramidal neurons fixed 7 d posttransfection. As for dendritic complexity, we found PFN2a to be important for spine maintenance. Specifically, shPFN2a-transfected neurons showed a significantly reduced spine density in the basal and proximal apical dendritic compartments (Fig. 2 B and C, spine P values provided in Table S1). Total spine density, too, was significantly reduced in shPFN2a-expressing CA1 neurons as compared with control cells (Fig. 2C). Neurons overexpressing PFN2a showed no significant alterations in the number of spines. However, a slight reduction in spine number could be observed in the basal dendritic compartment (Fig. S3B), whereas the spine density of the apical compartment and total spine density were unaltered in comparison with control neurons (Fig. S3B). Hence, spine maintenance requires a precisely regulated level of PFN2a.
PFN1 Does Not Compensate for the Reduction in Dendritic Complexity but Restores Spine Density After PFN2a Knockdown.
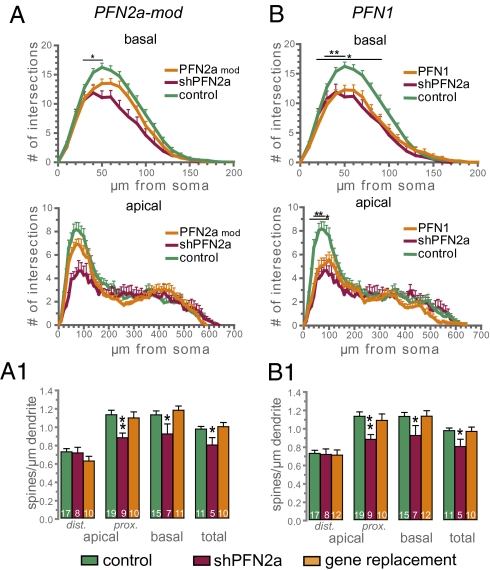
We then analyzed whether the effects observed following knockdown of PFN2a could be reversed by introducing an RNAi-resistant PFN2a mutant (Fig. S1A). CA1 neurons transfected with shPFN2a and the RNAi-resistant PFN2a-mod (shPFN2a-mod) showed an overall normal dendritic morphology (Fig. 3A). Specifically, the Sholl analysis of the apical dendritic compartment yielded data not significantly different from control cells, whereas in the basal dendritic compartment, only the region 30–50 μm from soma was significantly reduced (Fig. 3A). Moreover, the spine density of shPFN2a-mod–transfected cells was almost identical to control spine numbers (Fig. 3A1). These results underline that the vector-transfected cells synthesize PFN2a protein faithfully and that the effects of PFN2a knockdown on neuronal morphology are specific for PFN2a and not an unspecific consequence attributable to the RNAi approach.
Fig. 3.
PFN1 cannot rescue the shPFN2a-dependent reduction in dendritic complexity. Sholl analysis (A) and spine density (A1) of CA1 pyramidal neurons in organotypic slice cultures expressing a polycistronic construct containing shPFN2a and PFN2a-mod, an RNAi-resistant PFN2a mutant (PFN2a mod, n = 17). Sholl analysis (B) and spine density (B1) of CA1 pyramidal neurons in organotypic slice cultures expressing a polycistronic construct containing shPFN2a and PFN1 (PFN1, n = 16). Remarkably, coexpression of PFN1 does not prevent the significant reduction in dendritic complexity induced by the knockdown of PFN2a but it prevents the reduction in spine density. *P < 0.05, **P < 0.005.
To investigate whether the morphological alterations seen in neurons following PFN2a knockdown are specific for this isoform, we replaced endogenous PFN2a with YFP-PFN1 in hippocampal pyramidal neurons (Fig. 3B). The dendritic morphology of CA1 neurons showed an almost identical phenotype to those transfected with shPFN2a alone. In both, the basal and apical compartment dendritic complexity was significantly reduced as compared with cells transfected with control plasmids (Fig. 3B, gene replacement PFN1). However, the loss of spines observed in shPFN2a-transfected cells did not occur in cells overexpressing PFN1, because spine numbers were similar to control levels (Fig. 3B1).
PFN1 and PFN2a Can Compensate for Discrete Aspects of p75NTR-Dependent Morphological Alterations in Primary Hippocampal Neurons.
We then asked whether PFN2a could act as a link between signaling of surface receptors located at the membrane and the actin cytoskeleton. Several receptors are known to be involved in the regulation of neuronal morphology not only during development but in the adult nervous system. Receptors for neurotrophic factors [NGF, BDNF, neurotrophin (NT) 3, and NT4/5] are well-known modulators of neuronal morphology; thus, we chose p75NTR as a candidate molecule. Overexpression of this receptor in primary hippocampal neurons induces a reduction of dendritic complexity in the proximal dendritic compartment (11), quite similar to CA1 cells depleted of PFN2a by shPFN2a transfection. In addition, p75NTR has been shown to affect the activity of the small GTPase RhoA (14, 15), which, in turn, regulates dendritic branching and spine density (reviewed in ref. 16).
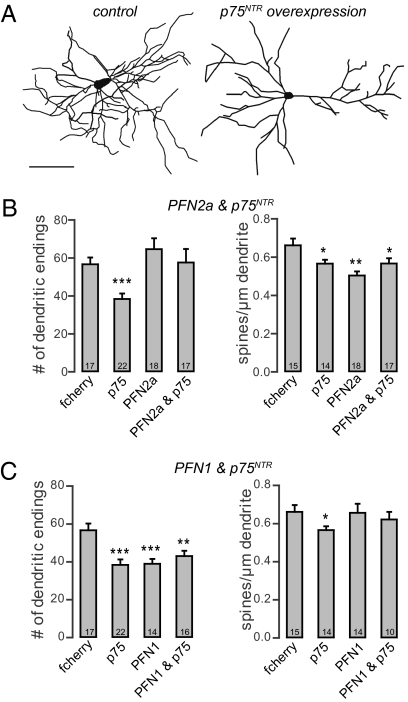
Because it has been reported in p75NTR KO mice (11) that dendritic complexity and spine density in pyramidal hippocampal neurons are increased, we wondered whether there might be a direct connection between the levels of p75NTR and profilins and whether mice lacking p75NTR might compensate for their neuronal defects by up-regulation of profilins. We found that both isoforms are up-regulated in p75NTR KO mice (Fig. S4). We first transfected primary hippocampal neurons with p75NTR. These neurons showed no signs of degeneration as swellings or retraction bulbs but displayed a significant simplification of the dendritic tree as indicated by a reduction in the number of dendritic endings when compared with control cells (Fig. 4 A–C, details provided in Table S2). The overexpression of p75NTR as well as profilin isoforms was verified by immunocytochemistry. We then asked whether the expression of PFN2a would be sufficient to rescue the p75NTR-mediated loss of dendrites. Indeed, when p75NTR and PFN2a were overexpressed in the same cells, the number of dendrites was identical to that of control neurons (Fig. 4B). Furthermore, the expression of PFN2a in dissociated cultures mimicked the increase in dendrite number as observed in organotypic cultures (Fig. 4B). The overall dendritic length was not affected by either treatment. Because the level of p75NTR is also negatively correlated with the number of dendritic spines (11), we then quantified spine numbers of control cells and neurons overexpressing p75NTR, PFN2a, or both proteins. As expected, the number of spines on p75NTR-overexpressing cells was significantly reduced (Fig. 4B). Notably, and in contrast to the dendritic phenotype, p75NTR-dependent spine loss was not rescued by overexpressing PFN2a within the same neurons (Fig. 4B). Moreover, spine numbers in neurons overexpressing PFN2a alone were also significantly reduced (Fig. 4B). Taken together, our results show that in primary hippocampal neurons, PFN2a is able to compensate for the reduction in dendritic complexity induced by p75NTR overexpression, whereas the p75NTR-induced loss of spines is not prevented. To determine whether this reversal of p75NTR-dependent morphological alterations is PFN2a-specific, we expressed PFN1 alone in hippocampal neurons as well as PFN1 together with p75NTR. Analysis of the total dendritic endings revealed that PFN1 could not restore the dendritic simplification induced by p75NTR overexpression (Fig. 4C). Moreover, overexpression of PFN1 alone significantly reduced the number of dendrites when compared with control neurons (Fig. 4C). Surprisingly, we found the number of spines in p75NTR and PFN1 coexpressing neurons to be similar to that in control cells (Fig. 4C), indicating that PFN1 can prevent the p75NTR-dependent loss of dendritic protrusions. The expression of PFN1 alone did not lead to a significant change in spine number (Fig. 4C).
Fig. 4.
PFN2a but not PFN1 can compensate for p75NTR-dependent dendritic loss in primary hippocampal neurons. Neurolucida reconstructions of primary hippocampal neurons (21 DIV) expressing fcherry. (A) Control cell expressing fcherry only and a hippocampal neuron transfected with p75NTR and fcherry. (B) Histograms showing the number of dendritic endings and spine density of neurons expressing only fcherry as a control, p75NTR, PFN2a, or PFN2a and p75NTR. (C) Histograms showing the number of dendritic endings and spine density of cells transfected with fcherry as a control, p75NTR, PFN1, or PFN1 and p75NTR. *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bar: 100 μm.)
To address the question of which molecular mechanisms could account for the differential role of the two profilin isoforms in actin-based neuronal architecture, we looked at their interaction with actin nucleators. We focused on two members of the formin family, both binding partners of profilins via their polyproline stretch-containing FH1 domain (17, 18) and important mediators of actin polymerization (19), and analyzed their binding affinity for the profilin isoforms. Although PFN1 and PFN2a both reacted with FH1 of mDia1 in pull-down assays, coimmunoprecipitation revealed that in brain lysates, only PFN2a forms complexes with mDia2 (Fig. S5).
In summary, we show that both profilin isoforms significantly influence neuronal architecture but have complementary effects on mature hippocampal neurons (working model illustrated in Fig. 5). Whereas overexpression of PFN1 reduces the number of dendrites but leaves spine numbers unaffected, overexpression of PFN2a slightly increases dendritic complexity and leads to a loss of dendritic spines at the same time. Reduction of the PFN2a level by RNAi knockdown severely affects both dendritic complexity and spine numbers. Furthermore, our results suggest discrete roles for both profilin isoforms downstream of p75NTR.
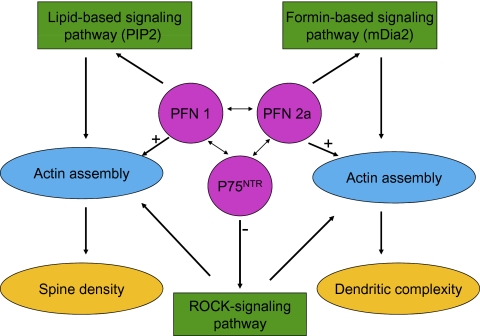
Fig. 5.
Proposed model for the action of both profilin isoforms in different neuronal compartments. Interactions of PFN1, PFN2a, and p75NTR shape dendritic and spine morphology during processes of synaptic plasticity. The three proteins react with different signaling pathways to regulate actin assembly. PFN1 preferentially binds to PIP2 (26), PFN2a binds with the formin mDia2 (this study), and both interactions stimulate actin assembly, but the actin assembly is directed to different neuronal compartments (single arrows). In contrast, p75NTR negatively controls the ROCK pathway (14, 15), which modulates spine density and dendritic complexity. Both profilin isoforms and p75NTR communicate with each other (this study, double arrows), and can thus influence each of the pathways. For reasons of clarity, the known direct interactions between profilins and members of the ROCK signaling cascade (RhoA and ROCK) are not included in this scheme.
Discussion
The relevance of two different isoforms of profilin (PFN1 and PFN2a) in the mammalian brain has been tackled in a number of studies, especially because only PFN1 is essential for cell survival (3). Although PFN2a is expressed predominantly in the brain, PFN1 also shows high expression levels there, indicating that both proteins might have crucial but possibly distinct functions (4). Interestingly, the ratios between the different isoforms vary significantly among different brain regions. The ratio of PFN2a to PFN1 is especially high in areas of the brain that show a high amount of activity-dependent synaptic plasticity, such as the hippocampus and cortex (20), but nowhere in the adult central nervous system is either of them expressed alone.
Recent studies in PFN2a KO mice reported an overall normal brain anatomy and neuronal morphology. Processes of functional plasticity, such as long-term potentiation and long-term depression, as well as learning were normal in these mice. These findings led investigators to suggest a role for PFN2a in controlling vesicle exocytosis (i.e., a presynaptic function) (9). However, compensatory effects cannot be ruled out, as may be concluded from the observation of an initial but transient increase in the number of sprouting neurites from young PFN2a−/− neurons (21).
By using RNAi-mediated acute knockdown of PFN2a in mature pyramidal neurons, we were able to analyze the postsynaptic role of PFN2a without possible compensatory effects. The number of dendrites as well as spines was reduced in PFN2a-deficient pyramidal neurons, suggesting that PFN2a plays a crucial role in the actin-dependent stability of these structures. This is consistent with studies in nonneuronal cells, where profilins have been shown to increase the density of submembranous actin networks, thereby stabilizing these dynamic structures (22, 23). Thus, one might expect functional equivalence of both isoforms in actin-based processes. Such an assumption is supported by findings that PFN2a is the ubiquitously expressed form clearly responsible for general actin-based motility in birds (12). Yet, our data presented here show that functional equivalence between PFN1 and PFN2a is not the case in neurons: The decrease in PFN2a-specific dendritic morphology could not be prevented by replacement with exogenous PFN1, whereas spine density was comparable to control levels. Thus, we see a specific function of PFN2a in stabilizing dendrite architecture and a concerted action of the two profilin isoforms in maintaining dendritic spines (Fig. 5). These results, together with our findings demonstrating a differential response to the activity of a surface receptor that also controls neuronal architecture, indicate that PFN1 and PFN2a differ in their interactions with the actin cytoskeleton. Biochemical studies revealed that PFN1 and PFN2a have comparable affinities for actin (reviewed in ref. 24). Hence, rather than being based on differences in direct profilin-actin interaction, such diverse variability may concern differential affinities for the profilin partners that are also involved in regulating the actin dynamics in neurons; for example, proteins interacting with the polyproline binding site on profilins. Both isoforms can form complexes with a variety of polyproline stretch proteins (reviewed in ref. 2), but they might have differential preferences. Indeed, different affinities for PFN1 and PFN2a have been revealed for the polyproline stretch of SMN, a protein vital for neuronal survival in mice and humans (25), and in biochemical analyses of the complexes present in extracts of murine brain, the polyproline motifs of the actin regulator Mena/Vasp were found to be associated with PNF2a but not with PFN1 (4, 26). Furthermore, there is evidence for differential roles of profilin isoforms in the performance of formins, a family of powerful actin regulators (19). Formins drive the assembly of actin bound to diverse profilin isoforms by significantly different rates (27), and in Schizosaccharomyces pombe, the failure of human PFN1 to compensate for yeast profilin could be assigned to an incompatibility with the respective yeast formin, indicating that there are indeed isoform-specific differences in the interaction of profilins with formins (28). Here, we show that only PFN2a is bound to the formin mDia2 in brain lysates, whereas PFN1 was not detectable in the corresponding immunoprecipitates. On the other hand, for purified bovine profilins, it has already been shown that PFN1 has a higher affinity for the membrane lipid PIP2 than PFN2a (26). Such differences may well be the basis for differential actin filament assembly, as required for either dendritic complexity or spine density.
Both profilin isoforms considered here are also involved in transmitting signals from the neuronal membrane downstream to the actin cytoskeleton. A membrane-bound neurotrophin receptor, p75NTR, modulates the small GTPase RhoA, which binds to mDia1 and can activate the Rho kinase ROCK (14, 15, 17, 18, 29). RhoA and ROCK have been shown to control actin stability during neuritogenesis in a PFN2a-dependent manner (21). On the other hand, PFN1 has been shown to influence neuritogenesis in PC12 cells in a ROCK-dependent manner (30). Future research will provide details about the signaling mechanism and signaling partners involved.
Based on the information available and the data reported here, we support the view that PFN1 and PFN2a are both important for actin-based neuronal plasticity. Their role in regulating actin dynamics and in signaling involves discrete as well as cooperative activities, and their role can be explained by the following working model (Fig. 5): PFN1 preferentially binds to PIP2 (26), whereas PFN2a interacts preferentially with the formin mDia2, as we show in this study (Fig. S5). We propose that both interactions stimulate actin assembly but direct them to different neuronal compartments. In contrast, p75NTR negatively controls the ROCK pathway (14, 15), which modulates spine density and dendritic complexity. Taken together, both profilin isoforms and p75NTR communicate with each other, and can thus influence each of the pathways. Consequently, this provides neurons with a higher degree of flexibility for the fine-tuning of structural plasticity in the response to extracellular signals.
Materials and Methods
Cell Culture.
Organotypic slice cultures of postnatal day 5/6 mice (C57 Bl/6) were prepared as previously described (31). To reduce the number of nonneuronal cells, antimitotic drugs (uridine, cytosine-β-D-arabinofuranoside·hydrochloride and 5-fluoro-2′-deoxyuridine) were applied for 24 h 3 d after preparation. Primary cultures of mouse hippocampal neurons were prepared using mice (C57 Bl/6) at embryonic day 18. Embryos were decapitated, and the brains were kept in ice-cold Gey's balanced salt solution supplemented with glucose. Cells were plated at high density (105) on poly-L-lysine–coated coverslips (13 mm) and kept in Neurobasal medium (Gibco) supplemented with 2% (vol/vol) B27 (Gibco) and 0.5 mM Glutamax (Gibco) at 37 °C, 5% (vol/vol) CO2, and 99% humidity.
Transfection.
To visualize dendritic complexity, the neurons were transfected with constructs carrying farnesylated forms of either enhanced GFP (fGFP) (Clontech–Takara) or mcherry (fcherry) (32) under the control of a CMV promoter. Coexpression of fGFP and YFP-PFN2a or YFP-PFN1 was confirmed by microscopy. Neurons expressing both plasmids were easily identified by high fluorescence intensity in the neuronal soma attributable to fGFP at the membrane and YFP in the cytoplasm. Organotypic cultures were transfected at 7 and 12 days in vitro (DIV), respectively, using the Helios gene gun system (Biorad). Bullet preparation was performed using a ratio of 2:1 (milligrams of gold per microgram of DNA). Slices were transfected by shooting at a pressure of 100 psi through tissue culture inserts with a pore size of 3 μm to prevent gold clumps from damaging the cultures. Primary hippocampal cultures were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. As already observed, the efficiency of cotransfection using Lipofectamine 2000 was nearly 100%. Coexpression was confirmed by immunocytochemistry.
Constructs.
RNAi constructs were based on pRNATU6.3/Hygro (Genscript). For PFN2a-specific knockdown ds oligodeoxynucleotide, GATCCGGATAACCTGATGTGCGATGGCGAACCATCGCACATCAGGTTATCCTTTT was inserted into the vector. GFP-cDNA was exchanged against fGFP-cDNA derived from pEGFP-F (Clontech–Takara). For gene replacement, synthetic cDNAs encoding RNAi-PFN2a-mod or human PFN1 (Geneart) fused to YFP were ligated into the PFN2a-specific shRNA vector. The expression of the YFP-profilin fusion proteins was driven by a truncated CMV promoter (33). To generate recombinant profilin, cDNAs of the profilin mutant were subcloned into pET21 (Novagen). The mouse full-length p75NTR cDNA [GenBank accession no. BC038365 (11)] was inserted into pSP70 vector (Promega).
Immunocytochemistry and Immunoblotting.
Organotypic (14 DIV) as well as primary hippocampal (21 DIV) cultures were fixed over night at 4 °C with 4% (wt/vol) formaldehyde and permeabilized with 0.2% Triton X-100 in phosphate buffer. All primary antibodies were incubated at 4 °C. Rabbit polyclonal anti-human p75NTR (Promega) was used at a dilution of 1:500 (3 d) for organotypic cultures and 1:4,000 (overnight) on dissociated neurons. Monoclonal mouse anti-PFN2a antibody (12) was diluted 1:100 for organotypic cultures (7 d) and 1:200 on dissociated neurons (overnight). Secondary anti-mouse or anti-rabbit antibodies conjugated with Cy2, Cy3, or Cy5 (Jackson ImmunoResearch) were incubated 1:500 in PBS for 2 h at room temperature. Immunoblotting was performed with HeLa cell extracts after SDS/PAGE. BiPro-tagged PFN1 was identified using anti-BiPro antibody as described (34).
Image Acquisition and Analysis.
For live imaging of organotypic hippocampal cultures, the slices were transferred into modified HBSS (35) supplemented with Fungizone (1.25 μg/mL, Gibco), penicillin (10,000 U/mL), and streptomycin (10 mg/mL). Imaging was performed using an upright Olympus Cell^M imaging station controlled by the Cell^M software and equipped with a 20× 0.8-N.A. objective (Olympus). After image acquisition, the cultures were transferred back into the incubator in culture medium containing the same antibiotics as above. Images of fixed neurons were acquired using an Axioplan 2 Microscope (Zeiss) equipped with an Apotome (Zeiss) controlled by Axiovision software (Zeiss). To cover entire CA1 neurons in the organotypic cultures, several z-stacks with a slice interval of 1 μm were acquired using a 20× 0.8-N.A. Plan-APO objective (Zeiss). In dissociated cultures, plain fluorescence images of hippocampal neurons were acquired. For analysis of spine density in organotypic cultures, parts of basal and both proximal and distal apical dendrites were imaged at a higher magnification with a 63× 1.4-N.A. Plan-APO oil immersion objective (Zeiss) and a z-stack thickness of 0.5 μm. In dissociated neurons, spines of secondary as well as tertiary dendrites were imaged in the middle part of the dendritic tree using the same settings as above. Morphological analysis was performed using Neurolucida and Neurolucida explorer software (Microbrightfield). The values obtained for Sholl analysis (36), spine density, or dendrite number and length were exported to Excel (Microsoft) and Graphpad Prism (Graphpad Software, Inc.) for statistical analysis using a paired Student's t test (two-tailed and two-sample unequal variance); significance was set at P < 0.05. For Sholl analysis, data significance was considered only if more than two adjacent points showed P values less than 0.05. All data are shown as the mean + SEM.
Supplementary Material
Acknowledgments
We thank Diane Mundil, Tania Messerschmidt, and Jasmin Will for outstanding technical assistance, Sabine Zessin for experimental help, and Sabine Buchmeier for the production and careful characterization of the isoform-specific profilin antibodies. This work was supported by the Deutsche Forschungsgemeinschaft (Grant FOR471).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004406107/-/DCSupplemental.
References
- 1.Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 2.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 3.Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witke W, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuhoff H, et al. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21:15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- 6.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 7.Lamprecht R, Farb CR, Rodrigues SM, LeDoux JE. Fear conditioning drives profilin into amygdala dendritic spines. Nat Neurosci. 2006;9:481–483. doi: 10.1038/nn1672. [DOI] [PubMed] [Google Scholar]
- 8.Schubert V, Da Silva JS, Dotti CG. Localized recruitment and activation of RhoA underlies dendritic spine morphology in a glutamate receptor-dependent manner. J Cell Biol. 2006;172:453–467. doi: 10.1083/jcb.200506136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pilo Boyl P, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehler S, Gallo G, Veien E, Letourneau PC. p75 neurotrophin receptor signaling regulates growth cone filopodial dynamics through modulating RhoA activity. J Neurosci. 2004;24:4363–4372. doi: 10.1523/JNEUROSCI.0404-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zagrebelsky M, et al. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25:9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murk K, Buchmeier S, Jockusch BM, Rothkegel M. In birds, profilin-2a is ubiquitously expressed and contributes to actin-based motility. J Cell Sci. 2009;122:957–964. doi: 10.1242/jcs.041715. [DOI] [PubMed] [Google Scholar]
- 13.De Simoni A, Griesinger CB, Edwards FA. Development of rat CA1 neurones in acute versus organotypic slices: Role of experience in synaptic morphology and activity. J Physiol. 2003;550:135–147. doi: 10.1113/jphysiol.2003.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita T, Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat Neurosci. 2003;6:461–467. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]
- 16.Koh CG. Rho GTPases and their regulators in neuronal functions and development. Neurosignals. 2006-2007;15:228–237. doi: 10.1159/000101527. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe N, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs A, Rothkegel M, Klar M, Jockusch BM. Characterization of functional domains of mDia1, a link between the small GTPase Rho and the actin cytoskeleton. J Cell Sci. 2001;114:3663–3672. doi: 10.1242/jcs.114.20.3663. [DOI] [PubMed] [Google Scholar]
- 19.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambrechts A, et al. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol Cell Biol. 2000;20:8209–8219. doi: 10.1128/mcb.20.21.8209-8219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Silva JS, et al. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkel T, Theriot JA, Dise KR, Tomaselli GF, Goldschmidt-Clermont PJ. Dynamic actin structures stabilized by profilin. Proc Natl Acad Sci USA. 1994;91:1510–1514. doi: 10.1073/pnas.91.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothkegel M, et al. Plant and animal profilins are functionally equivalent and stabilize microfilaments in living animal cells. J Cell Sci. 1996;109:83–90. doi: 10.1242/jcs.109.1.83. [DOI] [PubMed] [Google Scholar]
- 24.Schlüter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 25.Giesemann T, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 26.Lambrechts A, et al. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezezika OC, et al. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: Insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284:2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol. 2008;28:5196–5208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambrechts A, et al. Profilin-I-ligand interactions influence various aspects of neuronal differentiation. J Cell Sci. 2006;119:1570–1578. doi: 10.1242/jcs.02884. [DOI] [PubMed] [Google Scholar]
- 31.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 32.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 33.Boshart M, et al. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 34.Wittenmayer N, et al. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15:1600–1608. doi: 10.1091/mbc.E03-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang SB, Stein V, Bonhoeffer T, Lohmann C. Endogenous brain-derived neurotrophic factor triggers fast calcium transients at synapses in developing dendrites. J Neurosci. 2007;27:1097–1105. doi: 10.1523/JNEUROSCI.3590-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.