Abstract
Abscisic acid (ABA) is a hormone that controls seed dormancy and germination as well as the overall plant response to important environmental stresses such as drought. Recent studies have demonstrated that the ABA-bound receptor binds to and inhibits a class of protein phosphatases. To identify more broadly the phosphoproteins affected by this hormone in vivo, we used 14N/15N metabolic labeling to perform a quantitative untargeted mass spectrometric analysis of the Arabidopsis thaliana phosphoproteome following ABA treatment. We found that 50 different phosphopeptides had their phosphorylation state significantly altered by ABA over a treatment period lasting 5–30 min. Among these changes were increases in phosphorylation of subfamily 2 SNF1-related kinases and ABA-responsive basic leucine zipper transcription factors implicated in ABA signaling by previous in vitro studies. Furthermore, four members of the aquaporin family showed decreased phosphorylation at a carboxy-terminal serine which is predicted to cause closure of the water-transporting aquaporin gate, consistent with ABA's role in ameliorating the effect of drought. Finally, more than 20 proteins not previously known to be involved with ABA were found to have significantly altered phosphorylation levels. Many of these changes are phosphorylation decreases, indicating that an expanded model of ABA signaling, beyond simple phosphatase inhibition, may be necessary. This quantitative proteomics dataset provides a more comprehensive, albeit incomplete, view both of the protein targets whose biochemical activities are likely to be controlled by ABA and of the nature of the emerging phosphorylation and dephosphorylation cascades triggered by this hormone.
Keywords: mass spectrometry, proteomics, quantitation, metabolic labeling, Arabidopsis thaliana
Abscisic acid (ABA) is a phytohormone that initiates the water removal and overall dormancy program that plant embryos undergo during seed formation and germination and also mediates the cold and drought stress responses that occur in vegetative tissues (1, 2). The receptor for this hormone recently has been identified as a family of small soluble proteins known as PYR/PYLs that are encoded by 14 genes in the Arabidopsis genome (3, 4). Conformational studies using crystallography and NMR, as well as in vitro biochemical experiments, indicate that upon ABA binding these proteins undergo a conformational change that results in an increased affinity for protein phosphatase 2C (PP2C), and this interaction results in inhibition of phosphatase activity (5–8). These results are consistent with genetic data using stable transgenic lines containing knockout and overexpression mutations and with transient expression studies of the receptors and phosphatase (9, 10).
The resulting model predicts that downstream effects of ABA are mediated by early changes in the phosphoproteome induced by changes in PP2C phosphatase activity, which then can activate downstream sucrose non-fermenting 1-related subfamily 2 (SnRK2) kinases (11). To date, the only measurements of ABA-induced changes in phosphorylation have been performed in vitro with selected proteins (12). To obtain a more comprehensive picture of the changes induced by ABA, we performed a quantitative measurement of in vivo changes in the phosphoproteome following treatment with ABA.
Results and Discussion
Two types of metabolic heavy-isotope labeling strategies were used for the precise quantitation of phosphopeptide levels in control Arabidopsis plants compared with those that had been treated with ABA. In the first strategy, the reciprocal-labeling method, a control (untreated) Arabidopsis population was grown in natural-abundance medium (14N), and the experimental Arabidopsis population (ABA treated for 30 min) was grown in medium that was enriched with a 15N heavy isotope. The plants grown in the 15N-enriched medium exhibit greater than 98% isotope incorporation into total protein, as previously shown by our group (13). To control for any isotope effects, we concurrently performed a second experiment in which the isotopic label was reversed (Fig. S1A). Therefore, any changes in phosphorylation levels caused by the addition of ABA show a reciprocal ratio when the isotope label is exchanged between experimental sets (14). This strategy is an effective quantitation platform with each reciprocal set providing a biological replicate and has been used in several laboratories. However, for a time-course of treatment with ABA, a second metabolic-labeling strategy was used that was more amenable to analysis of a larger number of samples. In the second labeling procedure, which we refer to as the “universal standard method,” experimental Arabidopsis plants were grown in natural-abundance medium (14N) and then were left untreated (control) or were treated with ABA for 5, 15, or 30 min. A second Arabidopsis plant population was grown in 15N-enriched medium, (>98%) and similarly treated for use as an internal standard. Plant tissue from the experimental and internal standard populations then were combined at a 1:1 ratio providing (i) an internal standard for each peptide in the control and ABA-treated experimental samples and (ii) a control for any isotope effects (Fig. S1B). After growth and treatment, total protein was isolated from the tissue, digested with protease, enriched in phosphopeptides by chromatography over TiO2 particles, and analyzed by tandem mass spectrometry using an LTQ-Orbitrap with either collision-induced dissociation (CID) or electron transfer dissociation (ETD) fragmentation and triplicate technical replicates per sample. In each experiment the natural-abundance (light) and isotope-labeled (heavy) forms of each phosphopeptide produced distinct isotope envelopes, and the peptide intensities were compared through their extracted ion chromatograms. Utilization of both metabolic-labeling strategies with 15N-enriched growth medium permitted the identification of global phosphopeptide changes in the ABA-treated plants with high confidence.
In total, 17,201 phosphopeptides were identified yielding 4,675 unique phosphopeptide sequences (representing 2,185 different phosphoproteins). For all phosphopeptides, abundance ratios were quantified using the Census software package (15). Of these phosphopeptides, 50 showed a statistically significant difference (P < 0.05) between the ABA-treated and control samples, with 25 showing an 8–493% increase in phosphorylation, and 25 showing a 10–284% decrease (Table 1 and Table S1).
Table 1.
Partial list of phosphopeptides significantly altered in response to ABA treatment (50 μM for 5, 15, or 30 min)
| Average phosphorylation change with ABA | |||||
| ATG accession | Protein | Phosphopeptide | 5 min | 15 min | 30 min |
| Aquaporins | |||||
| AT3G53420, AT2G37170 | Plasma membrane intrinsic protein 2-A/B (PIP2-A/B) | K.SLGS*FRS*AANV.- | N/a | N/a | Decrease (76%) CID/ETD |
| AT4G35100 | Plasma membrane intrinsic protein 3 (PIP3) | K.ALGS*FRS*NATN.- | Decrease (26%) | Decrease (16%) | Decrease (59%) CID/ETD |
| AT2G16850 | Plasma membrane intrinsic protein 2–8 (PIP2-8) | K.ALAS*FRS*NPTN.- | Decrease (36%) | No change | Decrease (19%) ETD |
| AT5G60660 | Plasma membrane intrinsic protein 2–4 (PIP2-4) | K.ALGSFGSFGS*FR.S | No change | No change | Decrease (43%) CID/ETD |
| SnRK2 kinases | |||||
| AT4G33950 | SnRK2.6/OST1 kinase | K.S*TVGTPAYIAPEVLLK.K | N/a | N/a | Increase (358%) ETD |
| AT3G50500 | SnRK2.2 kinase | K.STVGTPAYIAPEILLRQEYDGK.L | N/a | N/a | Increase (493%) ETD |
| AT5G66880 | SnRK2.3 kinase | K.S*TVGTPAYIAPEVLLRQEYDGK.I | N/a | N/a | Increase (124%) ETD |
| AT1G10940 | SnRK2.4 kinase | K.STVGTPAYIAPEVLSRREYDGK.M | N/a | N/a | Increase (239%) ETD |
| Transcription factors | |||||
| AT1G45249, AT1G49720, AT4G34000, AT3G19290, AT3G56850 | ABA-responsive elements-binding factor 2 (ABF2) | R.QGS*LTLPR.T | N/a | N/a | Increase (106%) |
| AT3G56850 | ABA-responsive element binding protein 3 (AREB3) | R.QSS*LYSLTLDEVQNHLGSSGK.A | N/a | N/a | Increase (112%) |
| AT2G21230 | bZIP family transcription factor | R.SIS*GEDTSDWSNLVK.K | Increase (135%) | N/a | N/a |
| AT2G21230 | bZIP family transcription factor | R.SISGEDTSDWSNLVK.K | Increase (214%) | Increase (108%) | N/a |
| AT1G15750 | Topless transcription repressor (TPL) | R.APS*PVNNPLLGGIPK.A | Decrease (18%) | No change | Decrease (69%) |
| Calcium-related | |||||
| AT2G32450 | Calcium-binding EF hand family protein | R.DNNVPVSYS*GNGIPTK.S | No change | No change | Decrease (30%) |
| AT1G20760 | Calcium-binding EF hand family protein | K.S*QDGDYFGSGDDFGGNTAR.A | No change | Decrease (113%) | Decrease (49%) |
| AT1G09210 | Calreticulin-2 (CRT2) | K.DAPAES*DAEDEPEDDEGGDDSDSESKAEETK.S | N/a | Decrease (10%) | Decrease (33%) |
| AT4G35310; AT2G17290 | Calcium-dependent protein kinase 5/6 (CPK5/6) | K.NSLNIS*MR.D | Decrease (72%) | Decrease (22%) | N/a |
| AT1G19870 | IQ-domain 32 (IQD32) | R.KVS*NPSFIAAQSK.F | N/a | N/a | Decrease (66%) |
| AT1G19870 | IQ-domain 32 (IQD32) | K.GTETEEDDLIGTELQGPSAADAAK.I | Decrease (92%) | No change | N/a |
| Drought or ABA responsive | |||||
| AT1G76180 | Early response to dehydration 14 (ERD14) | K.VHIS*EPEPEVK.H | N/a | N/a | Decrease (78%) |
| AT5G40390 | Seed imbibition 1-like (SIP1) | K.SDS*GINGVDFTEK.F | N/a | N/a | Decrease (117%) CID/ETD |
| AT2G25070 | Protein phosphatase 2C (PP2C) | K.KPNPSETEPEDSKPEPSEDEPSSSS.- | Decrease (59%) | Decrease (14%) | Decrease (23%) |
Confirmed sites of phosphorylation are shown in bold followed by (*) as verified by manual MS2 inspection. Ambiguous sites of phosphorylation are denoted by italics. Phosphorylation changes are presented as the average percent change of the treated sample relative to control with three technical replicates each, with significance (P < 0.05) as determined by two-tailed t test. Unless otherwise denoted, identifications were made by CID fragmentation. N/a, not applicable.
Increased Phosphorylation of SnRK2 Kinases and ABA-Responsive Transcription Factors.
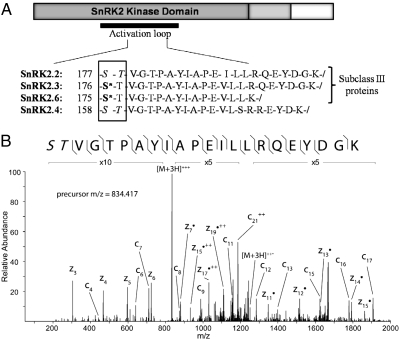
The plant-specific SnRK2 kinase group plays a vital role regulating ABA-induced gene expression. In vitro assays have shown that PP2Cs inhibit SnRK2-mediated phosphorylation of a family of basic leucine zipper (bZIP) transcription factors, ABFs/AREBs, which, when phosphorylated, act as positive regulators of ABA-responsive gene expression (12). It is believed that PP2Cs interact directly with SnRK2s, effectively keeping the kinases in a dephosphorylated state and thus inhibiting their activation (11). Following ABA binding and PP2C inhibition, SnRK2 kinases become phosphorylated at multiple serine/threonine residues in the activation loop and then phosphorylate downstream transcription factors from the bZIP family, as shown previously by in vitro studies (11, 12). Moreover, members of the SnRK2 subclass III family have been shown to be major regulators in ABA signaling (16). In this work, using ETD, we identified all three members of the SnRK2 subclass III family (SnRK2.2, SnRK2.3, and SnRK2.6) following 30-min treatment with ABA. All three members show an increase in phosphorylation in their activation loops following ABA treatment (Fig. 1), confirming the previous in vitro work and extending it to the in vivo system. In addition, the activation loop peptide from a non-subclass III SnRK2 family member, SnRK2.4, was identified as having a similar increase in steady-state phosphorylation. This identification may indicate activation of some non-subclass III SnRK kinases in the presence of ABA, although previously they have not been shown to positively regulate ABA responses.
Fig. 1.
Increased phosphorylation of SnRK2 kinases following ABA treatment. (A) Peptides from the activation loop of the SnRK2 proteins have increased phosphorylation following ABA treatment (30 min). Asterisks indicate validated sites of phosphorylation. When the exact site of phosphorylation could not be determined from the MS2 spectra, all potentially modified residues are shown in italics. (B) MS2 spectra of the SnRK2.2 singly phosphorylated peptide following ABA treatment using ETD fragmentation. Fragment ions detected are shown with the phosphopeptide sequence and annotated in the spectrum. In this case, the phosphorylation site can be isolated only to the first two residues (as noted by italics) because of the lack of distinguishing fragment ions at the N terminus.
Furthermore, 3 of the 25 peptides showing increases in steady-state phosphorylation are members of the bZIP transcription factor family of proteins, which are known to be downstream substrates of the SnRK kinase family. Two, AREB3 and ABF2, are known ABA response element DNA-binding proteins implicated in initiating the long-term changes in gene expression induced by ABA (12, 17), along with a third bZIP family transcription factor (At2g21230). Because of the sequence conservation present in the AREB/ABF protein families, it is possible that both peptides, QSS*LYSLTLDEVQNHLGSSGK and QGS*LTLPR, were derived from the AREB3 (At3g56850) gene product. However, the QGS*LTLPR peptide also may have resulted from the ABF family of proteins (ABF1, 2, 3, and 4), which all contain the conserved sequence region. In fact, it was suggested previously that ABA-dependent phosphorylation of ABF2 at the observed site in this peptide (S86) is important for stress-responsive gene expression (18). Furthermore, the in vitro phosphorylation of this residue, along with others in the ABF2 protein, was shown recently to be necessary for ABA-responsive gene expression (12). Our work shows that an increase in S86 phosphorylation within this peptide is an in vivo response to ABA treatment and additionally points to the previously unrecognized S21 phosphorylation in AREB3 as also participating in the in vivo signaling cascade. Thus, phosphorylation changes previously identified only by in vitro assays have been confirmed following in vivo treatment of Arabidopsis with ABA.
Decreased Phosphorylation of Conserved C-Terminal Serines in Aquaporins.
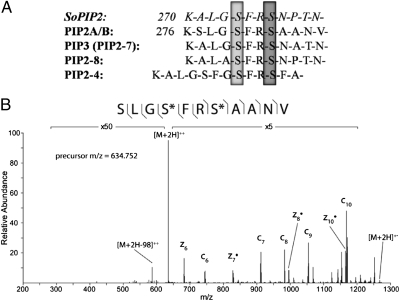
Four members of the water-transporting aquaporin family, plasma membrane intrinsic proteins (PIPs), (19) were found to have a consistently decreased phosphorylation state as compared with control plants (Table 1). Previous in vitro studies have shown that closure of PIP2 in spinach (SoPIP2) is triggered by dephosphorylation of two serine residues, S115 in the cytosolic loop and S274 in the carboxyl-terminal region (20–22); this latter serine is conserved in Arabidopsis homologs. Fig. 2 shows the alignment of the carboxyl-terminal peptide sequences of the SoPIP2 protein with the four Arabidopsis PIP family members found to have significant phosphorylation changes in this study. The decrease in steady-state phosphorylation level in response to ABA in the PIP water transporters is consistent with a decrease in its catalytic activity, as suggested by the closure of the PIP2 channel observed in spinach upon dephosphorylation at this conserved carboxyl-terminal serine residue. Additionally, dephosphorylation detected in this study at the downstream serine residue S283 in Arabidopsis PIP2 was shown previously to occur in response to stressors, such as NaCl (23, 24). Furthermore, the doubly phosphorylated PIP carboxyl-terminal peptides quantified in this study have been identified previously and found to undergo dephosphorylation following osmotic stress (25). These results are consistent with a role of PIP dephosphorylation in preventing rehydration during ABA-regulated seed germination and dormancy and in decreasing water flux in response to drought.
Fig. 2.
Decreased phosphorylation of aquaporins following ABA treatment. (A) Alignment of selected PIP family sequences. Dephosphorylation of S274 (light gray) in SoPIP2 was shown in structural studies to destabilize the “open” conformation of the protein, allowing closure of the water channel (20). This serine is conserved in all PIP family members quantified in this study and was shown to have decreased phosphorylation following ABA treatment. Additionally, the levels of phosphorylation of PIP2A/B in Arabidopsis at S280 (light gray) and S283 (dark gray) have been shown previously to change in response to other stressors (23, 24) and were found to be decreased in this study. (B) MS2 spectrum of PIP2A/B doubly phosphorylated peptide using ETD fragmentation in the control sample. Fragment ions detected are shown with the phosphopeptide sequence and are annotated in the spectrum. Both phosphorylation sites can be specifically assigned (*) by detection of flanking fragment ions around the phosphorylated residues.
Other Identified Phosphorylation Changes.
Several of the 25 peptides that displayed an ABA-induced decrease in phosphorylation are believed to be involved in ABA or other stress responses: At2g25070 (PP2C); At5g40390 (seed imbibition 1-like, SIP1); and At1g76180 (early response to dehydration protein, ERD14), which has been shown previously to be regulated by phosphorylation (26, 27). Additionally, multiple calcium-handling proteins (28), e.g., calcium-binding EF hand proteins (At1g20760/At2g32450), calcium-dependent protein kinase 5/6 (At4g3540), and calreticulin (At1g09210), were found to have decreased phosphorylation levels following ABA treatment. Finally, multiple proteins not previously implicated in ABA action were found to have significantly altered phosphorylation status, including four proteins of unknown function (Table S1). Further exploration of these protein targets may provide insight into previously unknown downstream effectors of the ABA signaling pathway.
Although these findings are meant to guide future research into the downstream mechanisms of ABA action, they do not represent an exhaustive survey of all possible phosphorylation changes. The nature of an untargeted phosphoproteomics survey using a data-dependent acquisition method on a mass spectrometer is inherently biased toward the identification of higher-abundance peptides in the sample. Additionally, the variability arising from instrument ion sampling and slight differences in individual sample handling and phosphopeptide enrichment typically result in imperfect overlap between biological and technical replicates. However, we believe we have achieved an acceptable level of dynamic range, because we are able to identify high-abundance proteins, such as calreticulin, as well as low-abundance transcription factors, such as the ABFs. Furthermore, many of our reported phosphopeptide changes were identified in more than one biological sample, at more than one time point, or by multiple fragmentation methods (Table 1 and Table S1).
Comparison of the ABA Response with That Induced by Calyculin, a Known Phosphatase Inhibitor.
An unexpected observation in these experiments was that, in almost one half of the proteins with ABA-induced changes in phosphorylation, the changes were in the direction opposite that expected from phosphatase inhibition or kinase stimulation, the initial responses currently thought to be induced by the ABA-bound receptor. To provide a dataset with which to compare the ABA-induced changes, we performed parallel experiments with 15N and natural-abundance reciprocally labeled Arabidopsis tissue treated or untreated with 100 nM calyculin-A, a known pharmacological inhibitor of the PP1 and PP2A classes of protein phosphatases (29). Of 996 unique phosphopeptides sequenced and quantified, only 19 peptides showed changes after 30 min of treatment. In contrast to ABA, all calyculin-A–induced changes showed an increase in phosphorylation, consistent with broad inhibition of phosphatase activity (Table S2). Interestingly, although there was little similarity in the list of proteins affected by both ABA and calyculin, four of the ABA targets described in Table 1 and Table S1, PEN3, ADL3, an unknown protein (At2g32240), and FAC1, also are significantly affected by calyculin-A. Three of these affected peptides (PEN3, ADL3, and an unknown protein) show increases in their phosphorylation state consistent with phosphatase inhibition after both treatments. In contrast, the FAC1 peptide shows a decrease in phosphorylation when treated with ABA, suggesting a mechanism of phosphorylation regulation other than phosphatase inhibition. Thus, it seems there is little overlap between the phosphatases and protein kinases affected following the treatment of Arabidopsis tissue with these two compounds. This lack of overlap in the two datasets is consistent with dephosphorylation of individual members of the Arabidopsis phosphoproteome being mediated by these different protein phosphatases.
Conclusion
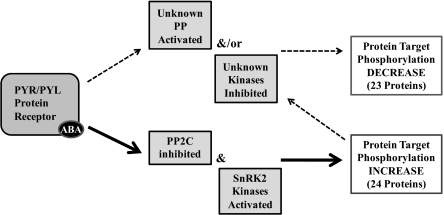
This study has revealed the identity of a number of potential early protein targets of the phosphorylation/dephosphorylation machinery involved in the ABA response in vivo. In particular, the dephosphorylation of conserved carboxyl-terminal serine residues in multiple aquaporin proteins suggests ABA may act to reduce water flux as early as 5 min after ABA treatment. In comparison with the results obtained with calyculin-A, a known protein phosphatase inhibitor, the observations with ABA suggest that a model involving only phosphatase inhibition (or kinase stimulation) is inadequate to describe receptor-mediated, short-term responses. Interaction with and inhibition of protein phosphatases may not be the only means of direct interaction of the ABA receptor family with regulatory proteins (Fig. 3), and other currently unknown ABA signaling pathways that result in decreased phosphorylation of downstream targets must be involved. Possibilities include (i) direct stimulation of a phosphatase or inhibition of a kinase by the ABA–receptor complex or (ii) indirect (downstream) stimulation of a phosphatase or kinase induced by any of the targets of the primary phosphatase inhibition. Regardless of the mechanism, it is clear that the current models solely invoking phosphatase inhibition by the ABA receptor are potentially insufficient and may not fully explain the decreased level of phosphorylation of many proteins observed following ABA treatment.
Fig. 3.
Mechanistic model for ABA-induced changes in protein phosphorylation. Increased protein target phosphorylation is known to occur with activation of SnRK2 kinases as well as from PP2C inhibition (11). Observed decreases in protein target phosphorylation could occur by the hypothetical activation of a phosphatase (PP) resulting from interaction with the ABA receptor complex, or by inhibition of kinases. Both phosphatase activation and kinase inhibition also may occur indirectly, downstream from the primary known effect of phosphatase inhibition.
Materials and Methods
Plant Growth and Sample Preparation.
Wild-type Arabidopsis thaliana Columbia seeds (Lehle Seeds) were grown in medium containing natural-abundance ammonium and potassium nitrate or 15N-enriched ammonium nitrate and potassium nitrate (>98% 15N) (Isotec) as the sole nitrogen source, as previously reported (30). Two methods were used for 15N metabolic labeling: a universal standard method for ABA treatment for 5 min, 15 min, and 30 min and a reciprocal-labeling method for ABA treatment for 30 min.
Reciprocal Metabolic Labeling.
At day 12 of growth, one container of 15N-labeled plants and one container of natural-abundance plants were treated with (+)-cis,trans-ABA (50 uM) for 30 min or with calyculin-A (100 nM) (Biomol International) for 30 min by replacing the medium in the growth box with medium containing the respective additives. Concurrently, one 15N-labeled plant and one natural -abundance plant were left untreated by replacing the growth medium with control medium. Following ABA or calyculin-A treatment, all plants were frozen individually in liquid nitrogen and ground to a fine powder using a mortar and pestle. To create experimental group one, an untreated natural-abundance plant was combined at a 1:1 ratio by weight (frozen, powdered plant tissue) with an ABA-treated 15N-labeled plant, and another natural-abundance plant was combined at a 1:1 ratio with a calyculin-A–treated 15N-labeled plant. Similarly, experimental group two was created by combining an untreated 15N-labeled plant at a 1:1 ratio by weight with an ABA-treated natural-abundance plant. Likewise, an untreated 15N-labeled plant was combined at a 1:1 ratio by weight with a calyculin-A–treated natural-abundance plant. Both ABA- and calyculin-A–treated experimental groups then underwent homogenization separately (4 g total plant tissue) in 40 mL grinding buffer (13) supplemented with phosphatase inhibitors using a sonicator (1-cm probe, 5 × 30 s, 50% duty cycle) while kept on ice. The resulting supernatant was filtered through four layers of Miracloth (Calbiochem) and underwent a soft spin (5,000 × g, 15 min) to remove unbroken cells.
Metabolic Labeling Using a Universal Internal Standard.
At day 12 of growth, one 15N-labeled plant and one natural-abundance plant were treated with cis,trans-ABA (50 uM) for 5, 15, or 30 min by replacing the medium in the growth box with medium containing the respective additives. Concurrently, one container of 15N-labeled plants and one container of natural-abundance plants were left untreated by replacing the growth medium with control medium. Following control or ABA treatment, plant material was individually frozen in liquid nitrogen and ground to a fine powder using a mortar and pestle. All tissues from plants gown in 15N medium were combined and used as a pooled universal internal standard reference. To provide an internal standard for all proteins, each experimental natural-abundance plant was combined at a 1:1 ratio by weight (frozen, powdered plant tissue) with the pooled 15N-labeled universal standard sample. Experimental samples treated with ABA for 5, 15, or 30 min plus their 15N universal standard then underwent homogenization (4 g total plant tissue) in 40 mL grinding buffer (13) supplemented with phosphatase inhibitors using a sonicator (1-cm probe, 5 × 30 s, 50% duty cycle) while kept on ice. The resulting supernatant was filtered through four layers of Miracloth and underwent a soft spin (5,000 × g, 15 min) to remove unbroken cells.
Protein Extraction, Digestion, and Phosphopeptide Enrichment.
Total proteins were isolated from the supernatant using a methanol/chloroform/water extraction as previously described (31). Precipitated proteins were solubilized in 8 M urea containing phosphatase inhibitor mixture (1× PhosStop; Roche). Samples were diluted to 1 M urea, and a BCA protein assay was performed to determine protein concentration in all experimental samples. Samples then were diluted to a total protein concentration of 1 mg/mL. Disulfide bonds were reduced using DTT (5 mM, 30 min), alkylated using iodoacetamide (15 mM, 30 min), and protein digested overnight at 37 °C using trypsin (1:100 ratio of trypsin to protein). Resulting peptide samples were acidified (0.1% TFA) and desalted using a solid-phase extraction procedure (Sep-Pak tC18; Waters) according to manufacturer's instructions. From each experimental sample for ABA and calyculin-A treatments, 5 mg of digested protein was individually subjected to phosphopeptide enrichment using TiO2 particles (4 mg, 5 μm; GL Sciences Inc.) as previously described (32).
Mass Spectrometry and Data Analysis.
Enriched phosphopeptides were solubilized in 0.25% TFA and separated on a reversed-phase analytical column (Magic-C18, 200Å, 3 μm; Michrom Bioresources Inc) placed in-line with a LTQ-Orbitrap mass spectrometer (ThermoFisher) and eluted using a 4-h acetonitrile mobile phase gradient. Each experimental sample was run with triplicate technical replicates. Survey scans (MS1) over the m/z range 300–2,000 were collected in the Orbitrap analyzer at 100,000 resolving power. Tandem mass spectra (MS2) were collected using data-dependent acquisition in the LTQ-ion trap. Data from MS2 spectra were searched using Mascot version 2.2 (Matrix Science) against the Arabidopsis Information Resource (TAIR) Arabidopsis protein database (version 9, released on June 19, 2009) to which was appended a decoy database comprising the same TAIR proteins but with their sequences reversed. For each HPLC/MS analysis, search results were filtered to maintain a 1% peptide false-discovery rate using software developed in house to establish thresholds of mass error and Mascot ion score. Quantitative ratio measurements were made using the Census software package (15) by extracting ion chromatograms within a 30-ppm mass window around the individual isotopologues of the 14N and 15N isotopic envelopes. The areas under the chromatographic peaks generated by the extracted ion chromatograms then were used to calculate the ratio for each peptide identified from each analysis. For the universal standard experiment (5 min, 15 min, and 30 min), a ratio of two ratios was calculated by dividing the ratio of experimental to 15N universal standard and the ratio of control to 15N universal standard (Fig. S1B). Average percent change was calculated for each quantified phosphopeptide at each time point, and statistical significance was determined by performing a two-tailed t test assuming unequal variance when possible (P < 0.05). All peptide sequences and phosphorylation sites reported here were confirmed by inspection of the raw data to verify the peptide sequence assignment and phosphorylation site assignment. When fragment ions were insufficient to determine the phosphorylated residue unambiguously, the peptides are reported with the possible phosphorylated residues indicated. Furthermore, the MS1 spectra were examined to determine that the isotopic envelopes contained no interferences. To extend the results obtained with collision-induced dissociation (CID) fragmentation from the reciprocally labeled 30-min ABA treatment, additional aliquots of the tissues from this time point were mixed and processed as described above. Proteins extracted from these tissues were digested with Lys-C, enriched for phosphopeptides using TiO2, and analyzed on the LTQ-Orbitrap using electron transfer dissociation (ETD) for ion activation. HPLC/MS conditions were the same as those described for trypsin digestion and CID activation, with the inclusion of 100 ms ETD reaction time, precursor charge state-dependent reaction time optimization, and supplemental activation for doubly-charged precursors.
Supplementary Material
Acknowledgments
We gratefully acknowledge the funding of this research through National Science Foundation Grant MCB-0929395.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007879107/-/DCSupplemental.
References
- 1.Christmann A, et al. Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 2006;8:314–325. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- 2.Rodríguez-Gacio MdelC, Matilla-Vázquez MA, Matilla AJ. Seed dormancy and ABA signaling: The breakthrough goes on. Plant Signal Behav. 2009;4:1035–1049. doi: 10.4161/psb.4.11.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 5.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 6.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 7.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida R, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 11.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol Cell Proteomics. 2007;6:860–881. doi: 10.1074/mcp.M600347-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Huttlin EL, et al. Discovery and validation of colonic tumor-associated proteins via metabolic labeling and stable isotopic dilution. Proc Natl Acad Sci USA. 2009;106:17235–17240. doi: 10.1073/pnas.0909282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uno Y, et al. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang JY, Kim DG, Kim YO, Kim JS, Kang H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol. 2004;54:713–725. doi: 10.1023/B:PLAN.0000040900.61345.a6. [DOI] [PubMed] [Google Scholar]
- 20.Törnroth-Horsefield S, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 21.Johansson I, et al. Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell. 1998;10:451–459. doi: 10.1105/tpc.10.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson I, Larsson C, Ek B, Kjellbom P. The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell. 1996;8:1181–1191. doi: 10.1105/tpc.8.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prak S, et al. Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics. 2008;7:1019–1030. doi: 10.1074/mcp.M700566-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Hsu JL, et al. Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed Arabidopsis thaliana. Proteome Sci. 2009;7:42–58. doi: 10.1186/1477-5956-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics. 2007;6:1711–1726. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol. 2001;45:263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- 27.Alsheikh MK, Heyen BJ, Randall SK. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J Biol Chem. 2003;278:40882–40889. doi: 10.1074/jbc.M307151200. [DOI] [PubMed] [Google Scholar]
- 28.MacRobbie EA. Signal transduction and ion channels in guard cells. Philos Trans R Soc Lond B Biol Sci. 1998;353:1475–1488. doi: 10.1098/rstb.1998.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara H, et al. Calyculin A and okadaic acid: Inhibitors of protein phosphatase activity. Biochem Biophys Res Commun. 1989;159:871–877. doi: 10.1016/0006-291x(89)92189-x. [DOI] [PubMed] [Google Scholar]
- 30.Nelson CJ, Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics. 2007;7:1279–1292. doi: 10.1002/pmic.200600832. [DOI] [PubMed] [Google Scholar]
- 31.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 32.Sugiyama N, et al. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.