Abstract
Systemic lupus erythematosus (SLE) is a multisystem, autoimmune disease that predominantly affects women. Previous findings that duplicated Toll-like receptor 7 (Tlr7) promotes lupus-like disease in male BXSB mice prompted us to evaluate TLR7 in human SLE. By using a candidate gene approach, we identified and replicated association of a TLR7 3′UTR SNP, rs3853839 (G/C), with SLE in 9,274 Eastern Asians (Pcombined = 6.5 × 10−10), with a stronger effect in male than female subjects [odds ratio, male vs. female = 2.33 (95% CI = 1.64–3.30) vs. 1.24 (95% CI = 1.14–1.34); P = 4.1 × 10−4]. G-allele carriers had increased TLR7 transcripts and more pronounced IFN signature than C-allele carriers; heterozygotes had 2.7-fold higher transcripts of G-allele than C-allele. These data established a functional polymorphism in type I IFN pathway gene TLR7 predisposing to SLE, especially in Chinese and Japanese male subjects.
Keywords: functional polymorphism, disease susceptibility, autoimmunity, type I interferon
Systemic lupus erythematosus [SLE; Online Mendelian Inheritance in Man (OMIM) no. 152700] is a multisystem, autoimmune disease with strong genetic and environmental components (1). SLE predominantly affects women, with a female-to-male ratio of approximately 9:1. Male patients with SLE, although rare, tend to have more severe disease and poorer outcome (2), suggesting potential sex dimorphism in the disease development. Although the sex effect has often been attributed to sex hormones, the fact that XXY male subjects have approximately a 14-fold higher risk of developing SLE than 46 XY men indicates that X-linked genes may be risk factors for human SLE (3).
Located at Xp22.2, Toll-like receptor 7 (TLR7; OMIM no. 300365) and its functionally related gene TLR8 (OMIM no. 300366) encode proteins that play critical roles in pathogen recognition and activation of innate immunity (4). They recognize endogenous RNA-containing autoantigens and induce the expression of type I IFN, a pivotal cytokine in the pathogenesis of SLE (5). In lupus-prone BXSB mice, the translocation of a segmental duplication of X chromosome to Y chromosome creates the Y-linked autoimmune accelerator (Yaa) locus, which was associated with autoreactive B cell responses to RNA-related antigens and exacerbation of glomerulonephritis in male mice (6). Although translocated X chromosome segment in Yaa may contain as many as 16 genes, the major gene for causation of the autoimmune phenotypes was identified to be TLR7 (7), making it a potential susceptibility gene for SLE. By using a candidate gene approach, we report herein that a functional polymorphism in 3′UTR of TLR7 is associated with SLE in Chinese and Japanese populations, with a stronger effect in male than female subjects.
Results
Discovery and Replication of the Association of a TLR7 3′UTR SNP with SLE in Eastern Asian Population.
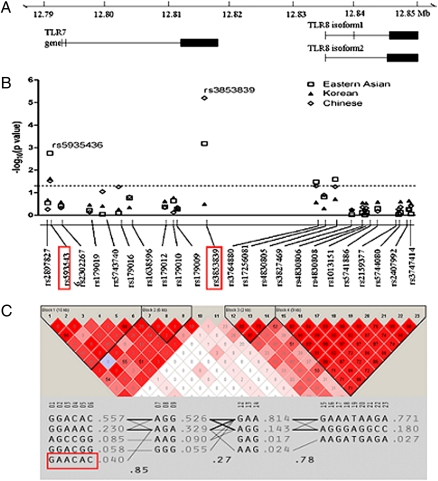
We genotyped 27 SNPs from the TLR7–TLR8 region (12 in TLR7 and 15 in TLR8) in 1,434 SLE cases and 1,591 control subjects of Eastern Asian ancestry using the Beadstation Infinium II platform (Illumina). Eleven SNPs in TLR7 and 12 SNPs in TLR8 that showed a minor allele frequency greater than 0.01 were included in association analysis (Fig. 1B and Table S1). We observed evidence of association with SLE in 2 TLR7 SNPs (rs5935436 and rs3853839) and 2 TLR8 SNPs (rs3764880 and rs4830805; Fig. 1B). Only rs5935436 and rs3853839 in TLR7 remained significant after Bonferroni correction (P = 0.041 and P = 0.016, respectively). The strongest association signal was detected at rs3853839 among Chinese subjects [cases vs. controls, 563 vs. 522; P = 6.3 × 10−6; odds ratio (OR) = 1.67 (95% CI = 1.33–2.08)], but not in Korean subjects [P = 0.32; OR = 0.92 (95% CI = 0.79–1.08); Fig. 1B], suggesting potential genetic heterogeneity of SLE between the two populations.
Fig. 1.
A functional SNP rs3853839 in 3′UTR of TLR7 is associated with SLE in a Chinese population. (A) TLR7 and TLR8 gene structure. (B) Eleven TLR7 SNPs and 12 TLR8 SNPs were genotyped in 1,434 SLE cases and 1,591 healthy controls of Eastern Asian descent. The two SNPs (rs5935436, rs3853839) that showed significant association after Bonferroni correction are highlighted. (C) Two haplotype blocks were constructed based on the strength of LD in each gene region. The R2 values of each SNP pair are depicted. The protective haplotype GAACAC is highlighted.
We next performed a haplotype-based association test using Haploview 4.03 software. The “GAACAC” haplotype formed by rs2897827, rs5935436, rs2302267, rs179019, rs5743740, and rs179016 had a frequency of 3.1% in SLE cases and 4.8% in controls (P = 0.0017; Fig. 1C). Given that only this TLR7 protective haplotype carries the minor allele of rs5935436, we genotyped rs5935436 in replication studies to represent the SLE-associated haplotype. Rs3853839 was in low linkage disequilibrium (LD) with other SNPs in the region. To minimize missing any other common polymorphisms, we sequenced 5′ promoter region (2 kb upstream) as well as three exons (including 3′UTR) of TLR7 in 48 Chinese female patients with SLE, which revealed no additional polymorphism and raised the possibility that rs3853839 might be causal.
To verify the association of rs3853839 and rs5935436 with SLE, we then conducted a replication study in two independent case-control Chinese and Japanese panels. Whereas rs5935436 was not replicated in these studies (P = 0.97 and P = 0.25, respectively), rs3853839 showed a consistent association with SLE in both replication panels [Chinese, 2,340 vs. 2,436; P = 9.0 × 10−4; OR = 1.21 (95% CI = 1.08–1.35); and Japanese, 560 vs. 913; P = 0.007; OR = 1.28 (95% CI = 1.07–1.53); Table 1]. A combined analysis of discovery and replication panels showed compelling evidence supporting that G allele of TLR7 SNP rs3853839 conferred risk for SLE [Pcombined = 6.5 × 10−10, ORcombined = 1.27 (95% CI = 1.17–1.36)]. Furthermore, G allele showed an additive effect in female subjects, as the OR for female homozygous GG versus CC was 1.45 (95% CI = 1.17–1.79) and OR for heterozygous GC versus CC was 1.15 (95% CI = 0.93–1.43), which reinforced a role of rs3853839 in developing SLE. Stratification by clinical subsets including age of disease onset, presence of mucocutaneous manifestations, lupus nephritis, dsDNA, or anti-RNA binding protein antibodies (presence of one or more autoantibodies to Ro/SSA, La/SSB, RNP, and Sm) (8) revealed a weak association of G allele with the presence of anti-RNA binding protein [P = 0.049, OR = 1.16 (95% CI = 1.00–1.35); missing data, 45%], suggesting that TLR7 binding of RNA containing immune complexes might play a role in the initiation and perpetuation of autoimmunity.
Table 1.
Association of rs3853839 with SLE in Eastern Asian populations
| G risk allele frequency | |||||
| Ethnicity and panel | Case/control | Case | Control | P value | OR (95% CI) |
| Eastern Asian*: Discovery | |||||
| Male | 126/229 | 0.83 | 0.73 | 0.038 | 1.79 (1.03–3.13) |
| Female | 1,308/1,362 | 0.79 | 0.76 | 3.00E-03 | 1.22 (1.06–1.39) |
| All | 1,434/1,591 | 0.80 | 0.76 | 6.70E-04 | 1.24 (1.1–1.41) |
| Chinese: Replication 1 | |||||
| Male | 196/931 | 0.92 | 0.81 | 2.31E-04 | 2.73 (1.57–4.74) |
| Female | 2,144/1,505 | 0.83 | 0.80 | 4.24E-03 | 1.19 (1.06–1.34) |
| All | 2,340/2,436 | 0.83 | 0.80 | 9.02E-04 | 1.21 (1.08–1.35) |
| Japanese: Replication 2 | |||||
| Male | 36/390 | 0.89 | 0.69 | 0.014 | 3.51 (1.22–10.2) |
| Female | 524/523 | 0.75 | 0.71 | 0.037 | 1.23 (1.01–1.49) |
| All | 560/913 | 0.75 | 0.70 | 7.00E-03 | 1.28 (1.07–1.53) |
| Eastern Asian*: Combined | |||||
| Male | 358/1,550 | 0.89 | 0.77 | 1.33E-06 | 2.33 (1.64–3.30) |
| Female | 3,976/3,390 | 0.80 | 0.77 | 1.19E-07 | 1.24 (1.14–1.34) |
| All | 4,334/4,940 | 0.81 | 0.77 | 6.50E-10 | 1.27 (1.17–1.36) |
*Includes subjects of Chinese, Korean, or Japanese descent.
Significant Male Effect of rs3853839 in the Risk of Developing SLE.
Of interest, we observed a stronger association of rs3853839 with male SLE. In the discovery panel, G allele showed higher OR in male patients compared with female patients [OR = 1.79 (95% CI = 1.03–3.13) vs. 1.22 (95% CI = 1.06–1.39); Table 1], especially in Chinese subjects [OR = 5.56 (95% CI = 1.85–16.7) vs. 1.54 (95% CI = 1.22–1.96)]. This notable male effect was verified in both replication panels: Chinese male patients with SLE had a significantly higher frequency of G allele than controls [92% vs. 81%; OR = 2.73 (95% CI = 1.57–4.74)]; in contrast, female cases only showed a modest increase of G allele versus controls [83% vs. 80%; OR = 1.19 (95% CI = 1.06–1.35)]. A similar finding was also observed in the Japanese subject replication panel. In the combined analysis, G allele occurred in 89% of male cases (n = 358), but only 77% of male controls (n = 1,550), showing a strong association with SLE (P = 1.33 × 10−6) and significantly higher OR in male versus female subjects [2.33 (95% CI = 1.64–3.30) vs. 1.24 (95% CI = 1.14–1.34); P = 4.1 × 10−4]. The significant male sex effect of rs3853839 in SLE was confirmed by a higher G allele frequency in male cases than female cases (P = 1.1 × 10−4; OR = 1.90 (95% CI = 1.36–2.64)] and by a higher OR in male subjects than the OR calculated by using only female homozygotes [GG vs. CC, OR = 1.45 (95% CI = 1.17–1.79); P = 0.02].
TLR7 SNP rs385839 Confers Elevated Expression of TLR7 in Vivo.
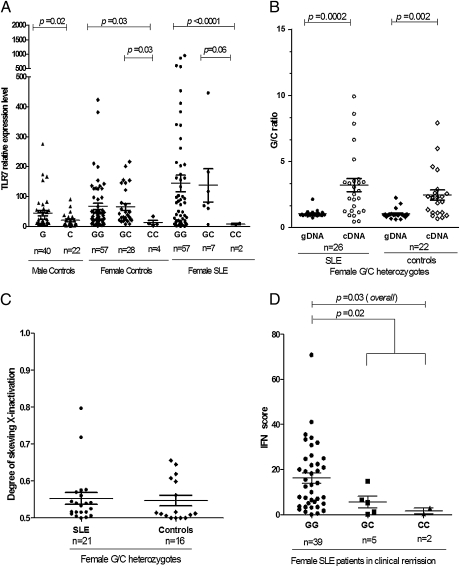
The consistent association of rs3853839 with SLE led us to investigate the potential functional consequences of the G risk allele. We first examined whether the 3′UTR polymorphism affected the expression level of TLR7 transcripts. We isolated peripheral blood mononuclear cells (PBMCs) from selected Chinese individuals carrying various genotypes and measured the mRNA expression levels of TLR7 by using real-time PCR. As shown in Fig. 2A, male controls carrying G allele had significantly higher TLR7 transcripts than those carrying C allele (P = 0.02). Similarly, female cases and controls homozygous for G allele showed notably higher TLR7 mRNA expression than those carrying homozygous C allele (P = 7.4 × 10−6 and P = 0.03, respectively). No sex differences in TLR7 mRNA expression levels were observed between normal controls carrying the same allelic genotype of rs3853839 (P = 0.07 or P = 0.55 for GG females vs. G males or CC females vs. C males, respectively; Fig. 2A). The sex comparison was not replicated in patients with SLE because of insufficient sample size of blood collected from male patients with SLE.
Fig. 2.
TLR7 SNP rs3853839 plays a functional role in TLR7 regulation. (A) Correlation of the TLR7 transcript levels in PBMCs from Chinese individuals with rs3853839 genotypes. G represents the major allele and C represents the minor allele of rs3853839. Each symbol represents an individual, and horizontal lines indicate mean values. (B) Allelic specific transcriptional expression of TLR7 in Chinese female subjects heterozygous for rs3853839 (G/C genotype) by pyrosequencing. TLR7 transcripts containing risk G allele were significantly elevated in PBMCs compared with those containing C allele. (C) XCI analysis in Chinese female individuals was performed by measuring the degree of methylated CAG repeats in AR gene and revealed no significant skewing XCI among SLE cases (n = 21) and normal controls (n = 16). (D) Comparison of IFN signature in PBMCs from clinically inactive female patients with SLE carrying various genotypes using IFN score. IFN score was calculated by combination of mRNA expression levels of four IFN-regulated genes including LY6E, MX1, IFIT1, and IFIT3 as described in Materials and Methods. Patients with SLE carrying GG genotype showed a more pronounced IFN signature in PBMCs compared with those carrying GC and CC genotypes.
To exclude potential bias between individuals, we next assessed the allelic specific transcriptional expression of TLR7 in Chinese female G/C heterozygotes by pyrosequencing. We extracted genomic DNA (gDNA) and cDNA from PBMCs of heterozygous SLE patients and controls and determined the exact ratios of G/C in gDNA and cDNA for each individual. As shown in Fig. 2B, the mean G/C ratio in gDNA of all individuals was 1.05, which was consistent with the theoretical value. However, SLE and control female heterozygotes showed a 2.7-fold higher G/C ratio in TLR7 transcripts than that in gDNA (P = 2 × 10−4 and P = 2 × 10−3, respectively; Poverall = 1.2 × 10−6), meaning that there was significantly higher level of G allele–containing TLR7 transcripts than C allele–containing transcripts in PBMCs, even though there was same amount of gDNA that contains either allele.
To investigate whether the disparity observed between transcript and gDNA was a result of imbalanced X chromosome inactivation (XCI) in female subjects, we analyzed the XCI pattern in these individuals by measuring the degree of methylated CAG alleles in androgen receptor (AR) gene as previously described (9). Fig. 2C showed no significant skewed XCI in either SLE or controls (P = 0.8), suggesting that skewed XCI or somatic selection might not account for the current finding. Taken together, these data suggested rs3853839 polymorphism could play a functional role in the regulation of TLR7 transcript levels in vivo.
To investigate whether allelic differences of rs3853839 corresponded to TLR7 protein levels, we isolated proteins from PBMCs of healthy Chinese male subjects carrying the G or C allele for Western blot analysis. Fig. S1 showed that five of the six male G allele carriers had higher expression levels of TLR7 protein than the two C allele carriers after normalization of the amount of protein loading using β-actin. Although the result was not statistically significant (P = 0.21, probably because of the very small sample size), the finding of higher TLR7 protein expression of the G risk allele carriers was consistent with what we observed at mRNA levels of TLR7.
We next investigated that whether the functional SNP rs3853839 might influence the IFN reaction downstream of TLR7. A prominent feature of patients with SLE is an increased expression of type I IFN–regulated genes (i.e., IFN signature) in PBMCs compared with unaffected controls (10, 11). We measured expression levels of four type I IFN–regulated genes, including lymphocyte antigen 6 complex, locus E (LY6E), myxovirus resistance 1 (MX1), IFN-inducible protein with tetratricopeptide repeats 1 (IFIT1), and IFIT3 by real-time PCR in PBMCs from clinically inactive female SLE patients carrying various rs3853839 genotypes and calculated an IFN score for each individual. As shown in Fig. 2D, female SLE patients carrying GG genotype showed a more pronounced IFN signature in PBMCs compared with those carrying GC and CC genotypes, indicating rs3853839 might modulate expression levels of TLR7, affecting IFN response in vivo.
Discussion
There is compelling evidence supporting the contribution of TLR7 to the development of autoimmunity. Transgenic mice with a twofold overexpression of TLR7 would have increased production of RNA-related autoantibodies and develop spontaneous autoimmunity (7). Whereas a modest increase in TLR7 gene dosage promoted autoreactive lymphocytes with RNA specificities and myeloid cell proliferation, a substantial increase in TLR7 expression caused fatal acute inflammatory pathologic process and profound dendritic cell dysregulation. In contrast, TLR7-deficient mice had ameliorated lupus disease, decreased lymphocyte activation, and decreased serum IgG (8). In addition, inhibitors of TLR7 reduced a number of lupus-associated phenotypes both in the MRL and NZB/W lupus-prone strains (12). Although a genomic duplication of Tlr7 was associated with an increased severity of lupus-like disease in a murine model, the similar finding was not translated to humans with SLE. Using quantitative real-time PCR, a copy number variation (CNV) study in multiple ethnic groups showed that the CN of TLR7 varied in patients with SLE and normal controls, but it was not significantly increased among patients with SLE compared with controls (13). The lack of difference in TLR7 CNV between human SLE and controls led us to explore functional SNPs in the current study.
Using a case-control design, we fine-mapped the TLR7–TLR8 region and identified and replicated the association of the TLR7 3′UTR SNP rs3853839 with SLE. The significant elevation of TLR7 transcripts in PBMCs from individuals carrying G risk allele, along with the notable higher level of G allele-containing TLR7 transcripts in heterozygous participants, supported a functional role rs3853839 played in the regulation of TLR7. The allelic specific expression analysis by pyrosequencing as we presented here is an accurate and valid approach that represents the biologically functional consequence of a certain SNP in vivo; without subjecting to various confounding factors frequently associated with in vitro assays, such as inappropriate reporter gene construct or cell line chosen or lack of suitable condition or agonists applied (14). Further bioinformatic analysis based on current databases did not provide a clue on how the SNP regulates mRNA expression: rs3853839 is not located in the binding site of any transcriptional factors or miRNAs; neither is it in the AU-rich sequence that is usually involved in mRNA decay. The exact mechanism is therefore yet to be elucidated.
It is perplexing that the X-linked TLR7 association does not explain the female dominance of SLE. The observed stronger male effect of rs385839 supports the notion that unique genetic predispositions might exist for male lupus. Given that both Tlr7 duplication in Yaa mice and TLR7 SNP rs3853839 in humans were preferably associated with male lupus, there might be potential epistasis between TLR7 and male sex background. This is supported by previous observation of substantially lower IFN response in male versus female subjects upon TLR7 agonist stimulation (15, 16). The sex difference in TLR7-mediated response has been used to account for the finding of higher viral loads in men than women in early HIV infection (16) and might explain, at least in part, the increased severity of several infectious diseases in male subjects (17). The sex-specific effect demonstrated by TLR7 SNP therefore might have implications that go beyond SLE. It would be interesting to investigate the contribution of TLR7 polymorphism to other diseases with the TLR7 pathway involved, especially in those with sex-specific incidence and manifestations.
To help exclude the possibility that the low LD of rs3853839 with neighboring SNPs is caused by CNV in this region, we used three independent methods including SYBR Green quantitative real-time PCR assay, PmeI pulsed-field gel electrophoresis (PFGE) and TaqI genomic Southern blot analyses to detect the existence of CNV at TLR7-8 locus. Samples from 223 patients with SLE and 139 normal controls were tested by real-time PCR assay (Fig. S2 and SI Materials and Methods), and TLR7–TLR8 CNVs were detected only in patients with SLE, at a frequency of less than 2% (one male patient had a two-copy variant of both TLR7 and TLR8, one female patient had a one-copy variant of TLR7, and another female patient had a one-copy variant of TLR8; Fig. S3). The PmeI-PFGE and TaqI genomic Southern blot analyses performed among 250 subjects, including 203 female and 47 male subjects, with a total of 453 X chromosomes did not show physical evidence of structural variations at TLR7–TLR8 locus (frequency less than 0.2%; Fig. S4 and SI Materials and Methods). Our results were consistent with two recent studies that used customized CGH platforms, which have reported no evidence of greater than a 5% frequency of TLR7–TLR8 CNVs greater than approximately 500 bp in 19 HapMap trios of individuals of European descent from Utah, 20 HapMap trios from Yoruba individuals in Nigeria, 10 HapMap trios of Han Chinese individuals from Beijing, 10 HapMap trios of Japanese individuals from Tokyo, and 10 Korean female individuals, with an estimated statistical power of 95% (18, 19). Additionally, no structural variants at TLR7–TLR8 locus have been described in the current Database of Genomic Variants (http://projects.tcag.ca/variation; RCh37, February 2009). Taken together, it is unlikely that the low LD between rs3853839 and neighboring SNPs is attributed to CNVs of this locus.
Currently, four European and two Asian genome-wide association studies (GWASs) in SLE have been published and more than 20 risk loci have been identified and replicated, which do not include the SLE-associated TLR7 SNP described here (20–25). There are various factors that may influence the association detection in a GWAS, including sample size, study design, cohort demographics (ethnicity and sex composition, for example), analytical strategies, and in the case of rs3853839, SNP selection. Rs3853839, located in a region with poor LD structure, has not been included in commercial predesigned genotyping arrays, which could explain why the previous GWAS failed to capture this risk SNP. To estimate power of these GWAS to identify TLR7 SNP association, we separately examined the male and female samples, set the level of significance (P value) for the subsequent replication study in each GWAS, and assumed the associated allele G with an odds ratio of 1.79 in male subjects and 1.22 in female subjects (OR was identified in our discovery panel study). The power estimate for male samples is less than 1% in Asian GWASs and ranges between 2% and 7% in European GWASs, whereas for female samples it ranges between 1% and 3.4% in Asian GWASs and 3% and 24% in European GWASs. Thus, these studies have inadequate power to identify this TLR7 association in their primary analyses. Our study was focused on the X chromosome; therefore, our correction for multiple testing was less stringent than the others that were conducted genome-wide, providing us greater power. In addition, fairly large sample size of male subjects with SLE and male controls (358 and 1,550, respectively, currently the largest male lupus study to our knowledge) facilitated our ability to detect the stronger effect of rs3853839 in male subjects. However, our male sample size is still not large enough to be comparable to female SLE genetic studies, and large-scale independent studies are needed to verify the effect of TLR7 in men.
In summary, we identified a functional polymorphism in rs3853839 that may confer elevated expression of TLR7 and predisposes to the development of SLE, especially in Chinese and Japanese male individuals. These data provide compelling evidence supporting a causal role for the type I IFN pathway genes in human autoimmunity and highlight the importance to explore sex-specific genetic contribution in a sex-biased disease such as SLE.
Materials and Methods
Clinical Samples.
In the discovery panel, we genotyped 27 SNPs spanning the TLR7 and TLR8 gene region in 1,434 SLE cases and 1,591 controls of Eastern Asian ancestry. These samples were obtained from multiple centers in the United States and Eastern Asia, of which 563 cases and 522 controls were Chinese, 845 cases and 1,022 controls were Korean, and 26 cases and 47 controls were Japanese. The Chinese replication panel included 2,340 independent Chinese SLE cases and 2,436 controls, collected by collaborators in Shanghai (case vs. control: 687 vs. 849), Hong Kong (881 vs. 840), and Taiwan (772 vs. 747). The Japanese replication panel included 560 SLE cases and 913 controls that were recruited from two centers in Japan. All SLE cases fulfilled the 1997 revised American College of Rheumatology criteria for the classification of SLE (26). Informed consent was obtained from all subjects. The study was approved by the human subject institutional review boards or ethical committees at each of the participating locations.
SNP Selection and Genotyping.
In the discovery panel, DNA from each subject was genotyped using custom-designed Beadstation Infinium II platform (Illumina). Genotyping was performed at the Lupus Genetics Studies Unit of the Oklahoma Medical Research Foundation. Twelve SNPs within TLR7 and 15 SNPs within TLR8 that either tagged major haplotypes in the HapMap database (January 2006) or were potentially functional were selected for genotyping. The average call rate for all samples was 98.3%. In the replication panels, rs3853839 and rs5935436 were genotyped by predesigned TaqMan SNP genotyping assay (Applied Biosystems) according to the manufacturer's instructions.
DNA Sequencing.
We sequenced 5′ promoter region (up to 2 kb upstream) as well as three exons (including 3′UTR in exon 3) of TLR7 in 48 Chinese female subjects. Briefly, consecutive overlapping amplicons were amplified from gDNA extracted from peripheral blood leukocytes. Primer pairs used are shown in Table S2.
Allelic Expression of TLR7 in PBMCs.
The mRNA expression level of TLR7 was measured in PBMCs from Chinese individuals carrying different genotypes of rs3853839. Briefly, total RNA were extracted from PBMCs using TRIzol (Invitrogen). RNA samples were then treated with DNase to eliminate gDNA contamination before being reverse-transcribed into cDNA. SYBR Green real-time PCR was used to measure the relative expression of TLR7. RPLP13A was used as internal control. We performed allelic-specific transcription quantification assay to measure the allelic expression of rs3853839 by using pyrosequencing as previously described (14). The allelic ratio for each cDNA and gDNA from each individual was calculated by using PSQMA software (version 2.1; Biotage) and compared by paired Student t test.
IFN Signature in Patients with SLE.
Expression levels of four type I IFN–regulated genes including LY6E, MX1, IFIT1, and IFIT3 were measured by real-time PCR in PBMCs from female patients with SLE carrying various rs3853839 genotypes and normal controls. RPL13A was used as an internal control. The patients with SLE recruited in this part of study were in clinical remission defined by SLE Disease Activity Index 2000 (27) lower than 4 and prednisone dosage less than 15 mg/d. IFN scores were calculated as described in previous studies (10, 11).
XCI Analysis in Female Participants.
We performed XCI analysis in female GC heterozygotes by measuring the degree of methylated CAG repeats in AR gene as previously described (9). Briefly, methylation-sensitive restriction enzyme HpaII was used to digest unmethylated (active X chromosome) DNA, which is thereby unable to amplify during the following PCR. Postdigestion PCR products therefore represent methylated (inactive X chromosome) DNA sequence. PCR products were sized using 5% denaturing polyacrylamide gel electrophoresis in an ABI Prism 3730 DNA automated sequencer, and analyzed by using Genescan Analysis 3.1 software (Applied Biosystems) as described previously (28).
Western Blot Analysis.
Proteins were extracted from PBMCs of Chinese male controls carrying rs3853839 G and C allele (n = 6 and n = 2, respectively) to conduct Western blot analysis (SI Materials and Methods).
CNV Discovery Experiment.
CNV discovery experiments are described in SI Materials and Methods. Study cohorts for CNV using real-time PCR assay and Southern blot analyses are shown in Tables S3 and S4, respectively.
Statistical Analysis.
For single SNP analysis, we used gPLINK 1.062 software (http://pngu.mgh.harvard.edu/∼purcell/plink/gplink.shtml) to evaluate the significance of differences in allelic frequencies of each SNP in the case-control samples. Bonferroni correction was used for multiple testing corrections in the discovery panel. For haplotype-based analysis, we used Haploview version 4.03 software (http://www.broad.mit.edu/mpg/haploview/index.php) to calculate pairwise LD indices R2 and D′, define LD blocks, infer haplotype frequencies, and analyze the significance of associations. We used the Q statistic (weighted sum of squares) to test the heterogeneity of odds ratio between male and female subsets. We used the Mann-Whitney test to compare the mRNA expression level of TLR7 in PBMCs from individuals carrying different genotypes. A paired Student t test was used to compare the G/C ratio between DNA and cDNA. A P value lower than 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank all subjects for participation in this study, and H. Wu and Y. Trejo-Lopez for their help in DNA preparation and organization. This work was supported by the Lupus Research Institute, National Institutes of Health Grants AR043814 (to B.P.T.), AR054459 (to C.Y.Y.), and AR053483 (to J.M.G.); National Natural Science Foundation of China Grant 30971632; Program of the Shanghai Commission of Science and Technology Grants 06JC14050, 07ZR14130, and 08JC1414700; Program of the Shanghai Subject Chief Scientist Grant 07XD14021 (to N.S.); Korea Healthcare Technology R&D Project (Ministry for Health, Welfare and Family Affairs, Republic of Korea) Grants A010252 and A080588 (to S.C.B.); Grant-in-Aid for Scientific Research (B; 19390271) from the Japan Society for the Promotion of Science; Grant-in-Aid for Young Scientists (B; 21790935) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to A.K. and N.T.); and a Veterans Affairs Merit Review Grant (K.M.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001337107/-/DCSupplemental.
References
- 1.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 2.Yacoub Wasef SZ. Gender differences in systemic lupus erythematosus. Gend Med. 2004;1:12–17. doi: 10.1016/s1550-8579(04)80006-8. [DOI] [PubMed] [Google Scholar]
- 3.Scofield RH, et al. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 5.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 7.Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X, et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 11.Fu Q, et al. Association of elevated transcript levels of interferon-inducible chemokines with disease activity and organ damage in systemic lupus erythematosus patients. Arthritis Res Ther. 2008;10:R112. doi: 10.1186/ar2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrat FJ, Meeker T, Chan JH, Guiducci C, Coffman RL. Treatment of lupus-prone mice with a dual inhibitor of TLR7 and TLR9 leads to reduction of autoantibody production and amelioration of disease symptoms. Eur J Immunol. 2007;37:3582–3586. doi: 10.1002/eji.200737815. [DOI] [PubMed] [Google Scholar]
- 13.Kelley J, Johnson MR, Alarcón GS, Kimberly RP, Edberg JC. Variation in the relative copy number of the TLR7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56:3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
- 14.Sun A, Ge J, Siffert W, Frey UH. Quantification of allele-specific G-protein beta3 subunit mRNA transcripts in different human cells and tissues by Pyrosequencing. Eur J Hum Genet. 2005;13:361–369. doi: 10.1038/sj.ejhg.5201334. [DOI] [PubMed] [Google Scholar]
- 15.Berghöfer B, et al. TLR7 ligands induce higher IFN-alpha production in females. J Immunol. 2006;177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 16.Meier A, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fish EN. The X-files in immunity: Sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad DF, et al. Wellcome Trust Case Control Consortium. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 22.Harley JB, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 24.Kozyrev SV, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 25.Yang W, et al. Asian Lupus Genetics Consortium. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 27.Gladman DD, Ibañez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29:288–291. [PubMed] [Google Scholar]
- 28.Lappalainen S, Utriainen P, Kuulasmaa T, Voutilainen R, Jääskeläinen J. Androgen receptor gene CAG repeat polymorphism and X-chromosome inactivation in children with premature adrenarche. J Clin Endocrinol Metab. 2008;93:1304–1309. doi: 10.1210/jc.2007-2707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.