Abstract
Objective
To detect genetic polymorphisms associated with high serum interferon alpha (IFN-α) in juvenile dermatomyositis (JDM) and explore interactions between associated polymorphisms.
Study design
Eighty-five children of European ancestry with definite/probable JDM were studied. Selected genetic polymorphisms which were associated with high IFN-α in twelve untreated patients newly diagnosed with JDM were genotyped in a validation cohort of 73 children with JDM, and analyzed for gene-gene and gene-sex interactions.
Results
Newly diagnosed untreated children with JDM carrying both the osteopontin (OPN) rs28357094G and tumor necrosis factor alpha (TNF-α) -308 A alleles had significantly increased serum IFN-α. These two polymorphisms were genotyped in the validation cohort, and the OPN rs28357094G allele was more common in female subjects with JDM (OR=3.97, p=0.012). This OPN allele was most strongly enriched in female carriers of TNF-α -308A as compared with male carriers of TNF-α -308A (OR>9.0, p=7.2×10−3).
Conclusions
These data support a complex gene-gene-sex interaction between the OPN and TNF-α promoter regions in JDM, defining a high serum IFN-α subgroup within JDM. This suggests pathogenic synergy between the OPN and TNF-α loci in females with JDM, which may underlie some of the increased incidence of this condition in females.
Keywords: Idiopathic Inflammatory Myopathies, Genetics, Systemic Lupus Erythematosus, Osteopontin, Tumor Necrosis Factor Alpha
Juvenile dermatomyositis (JDM) is a severe multisystem autoimmune disease of childhood (1). Although the pathogenesis of JDM is unknown, genetic factors have been associated with disease susceptibility (2, 3) as well as severity and specific manifestations (4). JDM is 2 to 3 times more common in females than in males (1): the reasons for this sex disparity are not understood. Treatments for JDM involve immunosuppressive therapies which are frequently only partially effective and confer a significant risk of side effects (1).
Interferon alpha (IFN-α) is a pleiotropic type I interferon which exerts a number of pro-inflammatory effects, and is involved in viral defense. Dysregulation of the IFN-α system has been observed in a number of autoimmune diseases, including systemic lupus erythematosus (SLE) (5, 6), Sjogren syndrome (7, 8), and JDM (9, 10). In SLE and both juvenile and adult dermatomyositis, increased IFN-α signaling has been associated with more severe disease and increased disease activity (10–12). Microarray studies have shown massive upregulation of IFN-α-induced transcripts in muscle biopsies (13) from children with symptoms of JDM of both long and short untreated disease duration (14), and to a lesser extent, in the isolated peripheral blood mononuclear cells from untreated patients with JDM (9, 15). In addition, serum levels of active IFN-α are elevated in many children with JDM (10).
High serum IFN-α is a heritable risk factor for SLE (16, 17). It has been shown that serum IFN-α levels are influenced by a number of SLE genetic risk factors, including protein tyrosine phosphatase non-receptor type 22 (PTPN22) (18), interferon regulatory factor 5 (IRF5) (19), osteopontin (OPN) (20), and others (21, 22). The PTPN22 genetic association is shared between SLE and JDM (23, 24), suggesting that the genetic background of these two disorders will be similar to some degree. In this study, we genotyped children with JDM at loci linked to increased serum IFN-α in SLE, and explored relationships between genotype at these loci and serum IFN-α in JDM. Interestingly, in the initial analysis we observed a gene-gene association between two genes on different chromosomes (OPN and TNF-α), and this combination defined the high IFN-α subgroup of patients. The OPN locus has been previously associated with a sex-related effect on SLE susceptibility and serum cytokine profiles in SLE (20, 25). To follow up this finding, we explored gene-gene and gene-sex interactions between polymorphisms in the promoter regions of these two cytokine genes in JDM.
Methods
Genomic DNA samples from 85 children with definite/probable JDM by Bohan and Peter criteria (26) were studied, and 12 of these children had a serum sample available from the time of initial diagnosis prior to any treatment. Sixty-two of the subjects were female and 23 were male. All of the children included in the study were of self-reported European-American ancestry. All subjects had genotyping data available for the tumor necrosis factor alpha (TNF-α) -308 promoter polymorphism (rs1800629), which was performed at Children’s Memorial Hospital using standard techniques. Genomic DNA from 112 sex-matched controls of European ancestry was obtained from the University of Chicago Translational Research in the Department of Medicine (TRIDOM) registry. These controls were screened for the absence of autoimmune or inflammatory conditions by medical record review. The study was approved by the institutional review boards at both institutions (University of Chicago IRB #15701B, Children’s Memorial Hospital IRB#10778), and age appropriate informed consent was obtained from all subjects.
The reporter cell assay for IFN-α has been described in detail elsewhere (16, 27). Reporter cells were used to measure the ability of patient sera to cause IFN-induced gene expression. The reporter cells (WISH cells, ATCC #CCL-25) were cultured with 50% patient sera for 6 hours, and then lysed. cDNA was made from total cellular mRNA. cDNA was then quantified using real-time PCR using an Applied Biosystems 7900HT PCR machine with the SYBR Green fluorophore system. Forward and reverse primers for the genes MX1, PKR, and IFIT1, which are known to be highly and specifically induced by IFN-α, were used in the reaction (16). GAPDH was amplified in the same samples to control for background gene expression. The amount of PCR product of the IFN-α-induced gene was normalized to the amount of product for the housekeeping gene GAPDH in the same sample. The relative expression of each of the three tested IFN-induced genes was calculated as a fold increase compared with its expression in WISH cells cultured with media alone. Results from the IFN-α assay were standardized to a healthy multi-ancestral reference population as previously described, and a serum IFN-α activity score was calculated based upon the mean and SD of the reference population (16).
Genotyping
Twelve untreated children with JDM were genotyped at single nucleotide polymorphisms (SNPs) in OPN (rs11730582, rs28357094, rs6532040, and rs9138), IRF5 (rs2004640, rs3807306, rs10488631, and rs2280714), and PTPN22 (rs2476601). 73 additional children with JDM were genotyped at the OPN rs28357094 SNP. Each of these SNPs conformed to Hardy-Weinberg equilibrium (p≥0.01 for all markers). The IRF5 rs2004640, rs3807306, rs10488631, and rs2280714 SNPs were chosen as they designate a risk haplotype for SLE in European ancestry (28). The OPN SNPs were chosen based upon our previous work examining IFN-α in the context of OPN genotype in SLE (20). Genotyping was performed using ABI Taqman Assays-by-Design primers and probes on an ABI 7900HT PCR machine.
Statistical Analysis and Power Estimate
Fisher exact test was used to analyze categorical genotype data, and odds ratios were calculated using standard methods. The non-parametric Mann-Whitney U test was used to compare quantitative IFN-α data between two genotype subgroups. P-values shown are uncorrected for multiple comparisons. Spearman rho was used to detect correlation between alleles. Logistic regression models were used to examine gene-gene interaction between OPN rs28357094 and TNF-α -308 alleles. In this analysis, each SNP was included as a predictor variable, and a multiplicative interaction term was also included to detect evidence for a multiplicative (non-additive) interaction between these loci on different chromosomes. In case-control genetic analysis, we estimate that given our sample size we would have 80% power to detect an allele with an odds ratio of 2.20 or greater with an alpha of 0.01 in an additive model [CaTS Genetic Power Calculator by Skol et al (29)]. In a multiplicative model, we would be powered to detect down to an odds ratio of 1.94 with similar measurements.
Results
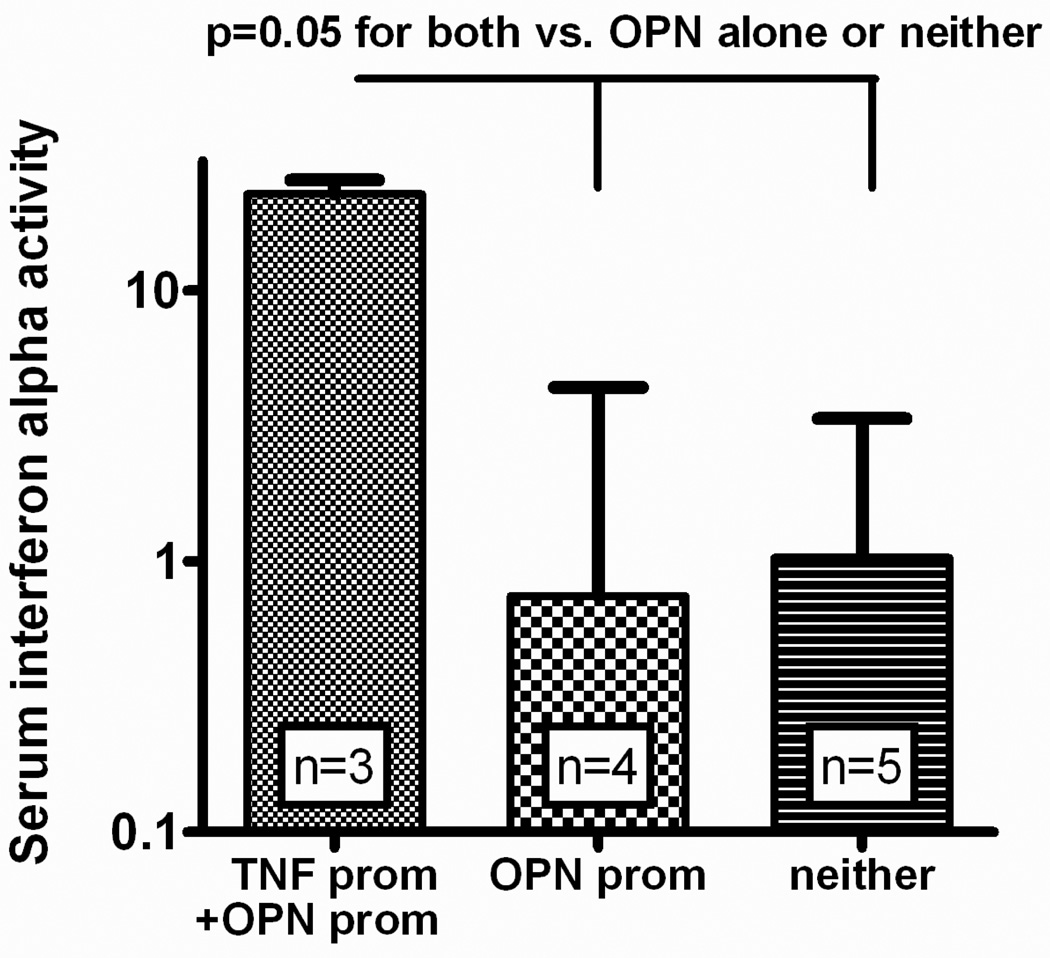
We examined a number of genetic polymorphisms in untreated patients with JDM to detect genotype-IFN-α relationships. The TNF-α -308 A allele was significantly associated with increased serum IFN-α. When the other SNP genotypes were examined, it was apparent that the OPN rs28357094 G allele was also associated with increased serum IFN-α, and carriers of both the TNF-α -308 and OPN rs28357094 alleles were a high IFN-α subgroup (Figure). SNPs in IRF5 and PTPN22 did not show any significant relationship with serum IFN-α in this cohort (data not shown). Given the strong influence of these two promoter polymorphisms on serum IFN-α, we tested for an association between these two genotypes which are found on different chromosomes and would be expected to assort independently (OPN is on chromosome 4, TNF-α is on chromosome 6). Surprisingly, the OPN rs28357094 G and TNF-α -308 A alleles were found together more often that would be expected by chance in this small data set (Spearman rho = 0.59, p=0.04).
Figure.
Serum IFN-α activity in patients with JDM stratified by presence of the TNF-α -308 A promoter polymorphism and the minor allele of the OPN promoter SNP rs28357094. TNF prom = presence of the TNF-α -308 A allele, OPN prom = presence of the OPN rs28357094 G allele, neither = subjects who lacked both the TNF -308 A allele and the OPN rs28357094 G allele. None of the subjects who carried the A allele of TNF-α -308 lacked the OPN rs28357094 G allele, so the three columns represent all patients studied. Bars represent the median, and error bars show the interquartile range. P value calculated using the Mann-Whitney U test, comparing the “TNF prom + OPN prom” column to the other two columns combined.
We next attempted to validate this association in a larger European ancestry cohort composed of 73 subjects in total. We did not see a significant overall correlation between the OPN rs28357094 G allele and the TNF-α -308 A allele in this replication set (Spearman rho = 0.04, p = 0.75). In the 62 female subjects, there was a non-significant trend toward correlation between the rs28357094 G allele and the TNF-α -308 A allele (Spearman rho = 0.16, p = 0.22), and in the 23 male subjects the opposite trend was observed (Spearman rho = −0.33, p = 0.12). When we examined allele frequencies in the joint cohort of 85 children with JDM, the OPN rs28357094 G allele was significantly more common in females than in males (OR = 3.97 (1.3–11.9), p = 0.012) (Table I). This was not observed with the TNF-α -308 A allele (OR = 1.35 for females vs. males, p = 0.62).
Table 1.
Allele frequencies in male vs. female JDM patients for the TNF-α -308 and OPN rs28357094 polymorphisms
| TNF-α -308 | OPN rs28357094 | |
|---|---|---|
|
Female (n=62) |
0.250 A | 0.274 G |
|
Male (n=23) |
0.196 A | 0.087 G |
|
Odds ratio: female vs male |
1.35 (0.5–3.7) |
3.97 (1.3–11.9) |
|
p value: female vs male |
0.62 | 0.012 |
n= number of subjects, frequency is indicated as a decimal, so 0.250 = 25.0% frequency.
P value calculated using two-sided Fisher’s exact test.
We next tested whether the association between OPN rs28357094 G and TNF-α -308 A was influenced by sex. We separated males and females, and examined the frequency of the OPN rs28357094 G allele in the presence and absence of TNF-α -308 A allele (Table II). There was a marked increase in rs28357094 G allele frequency in female carriers of TNF-α -308 A compared with male carriers of TNF-α -308 A (Table II, 34.7% frequency vs. 0% frequency, p = 7.2 × 10−3). In multivariate logistic regression examining the association of each of these alleles with sex in the JDM cohort, the multiplicative gene-gene interaction term showed the strongest association with sex (Table III), providing further support for a gene-gene interaction upon sex in the JDM cohort. Quantitative assessment of the attributable proportion due to interaction is not performed due to the sample size and lack of events in one category (no OPN G alleles present in the male TNF -308 A- allele category). Although the TNF-α -308 A allele has previously been associated with JDM, no similar data exist for OPN rs28357094 G. We genotyped 112 sex-matched healthy controls of European ancestry to compare allele frequencies with our JDM cohort (Table IV). There was no significant association between the OPN allele in all cases vs. all controls, and the large sex-differential in allele frequency observed in the patients with JDM at this locus was not observed in the healthy controls. Thus, the rs28357094 G allele was more frequent in female patients with JDM than in female controls, and less frequent in male patients with JDM than in male controls. A significant difference in allele frequency was observed between female patients with JDM carrying the TNF-α -308 A allele and female controls, supporting the importance of the gene-gene-sex model in the association of the OPN rs28357094 G allele with JDM.
Table 2.
Genotype and allele counts, and allele frequencies for the OPN rs28357094 SNP in male and female patients stratified by TNF-α -308 genotype
| Female TNF-α GG |
Male TNF-α GG |
Female TNF-α A- |
Male TNF-α A- |
|
|---|---|---|---|---|
|
OPN rs28357094 Genotype |
21 TT | 11 TT | 12 TT | 8 TT |
| 14 GT | 4 GT | 10 GT | 0 GT | |
| 1 GG | 0 GG | 4 GG | 0 GG | |
| Allele counts | 56 T | 26 T | 34 T | 16 T |
| 16 G | 4 G | 18 G | 0 G | |
| Allele Freq. | 0.222 G | 0.133 G | 0.346 G | 0 G |
| Odds Ratio | 1.86 (0.5–6.1) |
9.22* (1.14–74.8)* |
||
| p value | 0.41 | 7.1×10−3 | ||
Allele Freq. = frequency of the OPN rs28357094 G allele in different patient strata. TNF-α A- = TNF-α -308 promoter genotypes AA and AG, TNF-α GG = TNF-α -308 promoter genotype GG, odds ratio calculated as the odds for the presence of the OPN rs28357094 G allele in female vs. male JDM subjects within a given TNF-α -308 genotype category.
Odds ratio estimate, as standard odds ratio cannot be calculated due to zero value in male G allele category. Estimation of the odds ratio is performed by addition of 1 to each cell in the table. P values calculated using two-sided Fisher’s exact test.
Table 3.
Output from multivariate logistic regressions detecting association between OPN rs28357094 G and TNF-α -308 A alleles and sex in the JDM cohort
| Predictor Variables |
Model including each SNP as an independent predictor |
Model including each SNP and a multiplicative interaction term |
||
|---|---|---|---|---|
| OR (95% CI) |
p value | OR (95% CI) |
p value | |
| OPN rs28357094 G |
3.90 (1.26–11.99) |
0.018 | 2.07 (0.59–7.25) |
0.25 |
| TNF-α -308 A | 1.26 (0.55–2.91) |
0.59 | 0.88 (0.35–2.23) |
0.79 |
| Interaction term (OPN*TNF-α) |
- | - | 9.22* (1.14–74.8)* |
7.1×10−3* |
Results from individual predictor variables are shown, using gender as the outcome variable. Interaction term is generated by multiplying the OPN allele term (0, 1, or 2) by the TNF allele term (0, 1, or 2).
OR = odds ratio, 95% CI = 95% confidence interval
Odds ratio is an estimate, as standard odds ratio cannot be calculated due to zero value in male G allele category. Estimation of the odds ratio is performed by addition of 1 to each cell in the table. TNF-α A- = TNF-α -308 promoter genotypes AA and AG, TNF-α GG = TNF-α -308 promoter genotype GG. P value for this comparison is calculated using two-sided Fisher exact test, as the interaction term is a perfect predictor of sex and does not fit a logistic curve to allow for calculation of the p-value in the logistic regression.
Table 4.
Case-control analysis stratified by sex for the OPN rs28357094 SNP
| Subject category | rs28357094 G frequency |
OR (95% CI) |
P value |
|---|---|---|---|
| All cases | 0.224 | 1.21 (0.74–1.98) |
0.44 |
| All controls | 0.192 | ||
| Female controls | 0.185 | 0.83 (0.39–1.75) |
0.70 |
| Male controls | 0.214 | ||
| Female cases | 0.274 | 1.67 (0.96–2.91) |
0.069 |
| Female controls | 0.185 | ||
| Male cases | 0.087 | 0.35 (0.10–1.17) |
0.10 |
| Male controls | 0.214 | ||
| Female TNF-α -308 A allele carriers |
0.346 | 2.34 (1.17–4.67) |
0.021 |
| Female controls | 0.185 |
OR = Odds Ratio, 95% CI = 95% confidence interval, p values by Fisher Exact test
Discussion
In this study we demonstrate that simultaneous presence of the TNF-α -308 A allele and the OPN rs28357094 G allele was associated with increased serum IFN-α activity in untreated patients with JDM of European ancestry. Similar to OPN genetic studies in SLE (20, 25), the OPN rs28357094 SNP demonstrated significant skewing of association by sex in our study. It is interesting that the particular allele of OPN we find to be associated with sex in children with JDM is different from the one in which a sex effect is observed in SLE (rs9138), although the rs28354097 G allele has been associated with both SLE susceptibility and serologic profile within SLE (20, 30).
This sex difference in the OPN rs28357094 G allele in children with JDM was greatly accentuated in carriers of the TNF-α -308 A allele, thus illustrating a gene-gene-sex relationship between these two loci on different somatic chromosomes. The fact that these two alleles define a high serum IFN-α group of patients is of note, and further supports the hypothesis that these alleles cooperate in JDM immunopathogenesis in females. In previous work we have demonstrated that serum IFN-α activity is highest in patients with less than 1 year of symptoms (10), suggesting that IFN-α may be important in the disease initiation phase. These data suggest a pathologic synergy between these two cytokine gene polymorphisms and serum IFN-α in JDM pathogenesis, and could begin to explain some of the increased incidence of JDM in females.
This synergy between the OPN and TNF-α loci in children with JDM may be associated with one of the consequences of JDM, dystrophic calcifications. Osteopontin, a master T cell regulator (31), is also a major component of dystrophic calcifications that develop in children with JDM (32, 33). We speculate that ligation of CD44 by OPN, which decreases interleukin-10 production, may also help drive production of increased serum levels of TNF-α (34). Increased serum levels of TNF-α, associated with the TNF-308A allele, was previously identified as a contributing factor to a chronic disease course and the development of dystrophic calcifications in children with JDM (4).
In summary, this study shows the simultaneous presence of two cytokine genes promoter polymorphisms on different chromosomes that defines a subgroup of children with JDM who have high serum IFN-α activity. Furthermore, these polymorphisms interact in gene-gene-sex modality, suggesting pathogenic synergy between these two loci. This finding may begin to explain some of the increased female incidence observed in JDM.
Acknowledgments
Supported by NIH/NIAID K08 Award AI083790, NIAID Clinical Research Loan Repayment AI071651, CTSA Collaborative Pilot Grant from UL1 RR024999, Arthritis National Research Foundation Eng Tan Scholar Award, Alliance for Lupus Research, and Lupus Research Institute Novel Research Grant (T.N.); . NIAMS RO-1 AR48289, The CureJM Foundation, and Macy's Miracle Foundation (L.P).
Abbreviations
- IFN-α
interferon alpha
- JDM
juvenile dermatomyositis
- OPN
osteopontin
- TNF-α
tumor necrosis factor alpha
- SLE
systemic lupus erythematosus
- PTPN22
protein tyrosine phosphatase non-receptor type 22
- IRF5
interferon regulatory factor 5
- SNPs
single nucleotide polymorphisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Feldman BM, Rider LG, Reed AM, Pachman LM. Juvenile dermatomyositis and other idiopathic inflammatory myopathies of childhood. Lancet. 2008;371(9631):2201–2212. doi: 10.1016/S0140-6736(08)60955-1. [DOI] [PubMed] [Google Scholar]
- 2.Reed AM, Pachman L, Ober C. Molecular genetic studies of major histocompatibility complex genes in children with juvenile dermatomyositis: increased risk associated with HLA-DQA1 *0501. Hum Immunol. 1991;32(4):235–240. doi: 10.1016/0198-8859(91)90085-n. [DOI] [PubMed] [Google Scholar]
- 3.Mamyrova G, O'Hanlon TP, Monroe JB, Carrick DM, Malley JD, Adams S, et al. Immunogenetic risk and protective factors for juvenile dermatomyositis in Caucasians. Arthritis Rheum. 2006;54(12):3979–3987. doi: 10.1002/art.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pachman LM, Liotta-Davis MR, Hong DK, Kinsella TR, Mendez EP, Kinder JM, et al. TNFalpha-308A allele in juvenile dermatomyositis: association with increased production of tumor necrosis factor alpha, disease duration, and pathologic calcifications. Arthritis Rheum. 2000;43(10):2368–2377. doi: 10.1002/1529-0131(200010)43:10<2368::AID-ANR26>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301(1):5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 6.Crow MK. Type I interferon in systemic lupus erythematosus. Curr Top Microbiol Immunol. 2007;316:359–386. doi: 10.1007/978-3-540-71329-6_17. [DOI] [PubMed] [Google Scholar]
- 7.Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML, et al. Activation of the type I interferon system in primary Sjogren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52(4):1185–1195. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 8.Niewold TB, Rivera TL, Buyon JP, Crow MK. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 2008;58(2):541–546. doi: 10.1002/art.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007;13(1–2):59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niewold TB, Kariuki SN, Morgan GA, Shrestha S, Pachman LM. Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum. 2009;60(6):1815–1824. doi: 10.1002/art.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener PA, Pinkus JL, et al. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum. 2007;56(11):3784–3792. doi: 10.1002/art.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52(5):1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 13.Tezak Z, Hoffman EP, Lutz JL, Fedczyna TO, Stephan D, Bremer EG, et al. Gene expression profiling in DQA1*0501+ children with untreated dermatomyositis: a novel model of pathogenesis. J Immunol. 2002;168(8):4154–4163. doi: 10.4049/jimmunol.168.8.4154. [DOI] [PubMed] [Google Scholar]
- 14.Chen YW, Shi R, Geraci N, Shrestha S, Gordish-Dressman H, Pachman LM. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008;9:43. doi: 10.1186/1471-2172-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor KA, Abbott KA, Sabin B, Kuroda M, Pachman LM. MxA gene expression in juvenile dermatomyositis peripheral blood mononuclear cells: association with muscle involvement. Clin Immunol. 2006;120(3):319–325. doi: 10.1016/j.clim.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58(7):2113–2119. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum. 2008;58(9):2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009;10(5):487–494. doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62(2):553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: Autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182(1):34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chinoy H, Platt H, Lamb JA, Betteridge Z, Gunawardena H, Fertig N, et al. The protein tyrosine phosphatase N22 gene is associated with juvenile and adult idiopathic inflammatory myopathy independent of the HLA 8.1 haplotype in British Caucasian patients. Arthritis Rheum. 2008;58(10):3247–3254. doi: 10.1002/art.23900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, et al. Osteopontin and systemic lupus erythematosus association: a probable gene-gender interaction. PLoS ONE. 2008;3(3):e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohan A, Peter JB. Polymyositis and dermatomyositis (first of two parts) N Engl J Med. 1975;292(7):344–347. doi: 10.1056/NEJM197502132920706. [DOI] [PubMed] [Google Scholar]
- 27.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54(6):1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 28.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104(16):6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 30.D'Alfonso S, Barizzone N, Giordano M, Chiocchetti A, Magnani C, Castelli L, et al. Two single-nucleotide polymorphisms in the 5' and 3' ends of the osteopontin gene contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2005;52(2):539–547. doi: 10.1002/art.20808. [DOI] [PubMed] [Google Scholar]
- 31.Cantor H, Shinohara ML. Regulation of T-helper-cell lineage development by osteopontin: the inside story. Nat Rev Immunol. 2009;9(2):137–141. doi: 10.1038/nri2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pachman LM, Veis A, Stock S, Abbott K, Vicari F, Patel P, et al. Composition of calcifications in children with juvenile dermatomyositis: association with chronic cutaneous inflammation. Arthritis Rheum. 2006;54(10):3345–3350. doi: 10.1002/art.22158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urganus AL, Zhao YD, Pachman LM. Juvenile dermatomyositis calcifications selectively displayed markers of bone formation. Arthritis Rheum. 2009;61(4):501–508. doi: 10.1002/art.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinohara ML, Jansson M, Hwang ES, Werneck MB, Glimcher LH, Cantor H. T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc Natl Acad Sci U S A. 2005;102(47):17101–17106. doi: 10.1073/pnas.0508666102. [DOI] [PMC free article] [PubMed] [Google Scholar]