Abstract
The coding sequence of a protein must contain the information required for the canonical amino acid sequence. However, the redundancy of the genetic code creates potential for embedding other types of information within coding regions as well. In a genome-wide computational screen for functional motifs within coding regions based on evolutionary conservation, highly conserved motifs included some expected motifs, some novel motifs and coding region target sites for known microRNAs, which are generally presumed to target 3’ untranslated regions (UTRs) (www.SiteSifter.org). We report here an analysis of published proteomics experiments that further support a functional role for coding region microRNA binding sites, though the effects are weaker than for sites in the 3’ UTR. We also demonstrate a positional bias with greater conservation for sites at the end of the coding region, and the beginning and end of the 3’ UTR. An increased effectiveness of microRNA binding sites at the 3’ end of transcripts could reflect proximity to the poly(A) tail or interactions with the 5’ terminal 7mGpppN “cap”, which is physically adjacent to this region once the message is circularized. The effectiveness of 3’ UTR sites could reflect a cooperative role for RNA binding proteins. Finally, increased microRNA conservation near the stop codon suggests to us the possible involvement of proteins that execute nonsense-mediated decay, since this process is activated by tagging of termination codons with factors that induce transcript degradation.
Keywords: microRNA, coding region, evolutionary conservation, RNA binding protein, nonsense mediated decay
The redundancy of the genetic code implies that the selection of a specific triplet codon from several options can be optimized to embed additional information. For instance, splice start and end positions, binding sites for structural proteins like histones, and functional motifs that regulate the amount of RNA transcribed from specific genes could all be encoded within the coding region. Indeed, among possible genetic codes, the universal code has been shown to be nearly optimal for incorporating embedded information.1
We performed a computational analysis to identify potential functional motifs within coding regions based on extreme evolutionary conservation in a multiple species alignment of 17 genomes.2 By quantifying conservation at the DNA sequence level over and above that required for amino acid level conservation, we identified motifs that were selected for during evolution (www.SiteSifter.org). Among the highly conserved motifs was the recognition site for Tra2β, an exonic splicing factor also identified in previous reports of conserved coding region motifs.3 Several novel motifs were also identified that may play an as yet unknown functional role.
Other highly conserved motifs represent binding sites for microRNAs. We describe below the evidence that microRNAs target coding regions as well as 3’ UTRs. We discuss more recent proteomics experiments that allow an analysis of the efficacy of microRNA binding sites in coding regions and 3’ UTRs and consider reasons for the higher efficacy of sites within 3’ UTRs compared with sites in coding regions, including a potential role for RNA binding proteins as facilitators of microRNA function. We also present an analysis of the efficacy of sites based on their position within the transcript and suggest that the microRNA and nonsense-mediated decay pathways may converge.
microRNAs target coding regions
Highly conserved functional motifs in coding regions include multiple target sites for microRNAs.2 microRNAs are short (21–23 nt) RNAs that target specific mRNAs by binding to them through a complementary 5’ “seed” sequence (at bp 2–8 of the microRNA) and decreasing protein levels of the target sequence as part of the RNA-induced silencing complex (RISC).4, 5 The base pairing complementarity of the microRNA can guide the endoribonuclease Argonaute (Ago) to direct target degradation, or alternatively, to mediate translational repression.6–8 microRNAs can also result in translocation of target mRNAs to P bodies, sites of degradation for cellular mRNAs.9–11
In plants, long stretches of complementarity between the microRNA and its target result in endonucleolytic cleavage of the double stranded RNA (dsRNA) in a manner similar to siRNAs.12 Targeting of coding regions is common in plants.13 In animals, most targeting by microRNAs results in imperfect duplexes and occurs in the 3’ UTRs of targeted transcripts.14–16 Our analysis suggested that evolutionarily conserved targeting occurs in coding regions as well. Some other bioinformatic analyses of microRNA targeting have also concluded that there is likely targeting of microRNAs in coding regions either based on evolutionary conservation17–19 or on RNA folding patterns.20, 21
Several instances of microRNAs targeting coding regions have been demonstrated experimentally. The let-7 microRNA downregulates the microRNA processing enzyme, Dicer,2 leading to a negative feedback loop similar to that previously reported in plants.22–24 miR-148 was shown to repress DNA methyltransferase 3b expression through a coding region site.25 An evolutionarily conserved miR-126 recognition site was identified within the homeobox domain of the HoxA9 gene, and modulation of miR-126 in immortalized bone marrow cells led to regulation of HoxA9 levels.26 Increased p16 expression with replicative senescence was associated with decreased miR-24 expression, and miR-24 was found to repress p16 expression primarily through a coding region recognition site.27 Finally, five functional coding region target sites for the miR-296, miR-470 and miR-134 microRNAs were discovered in the nanog, oct4 and sox2 transcription factors, with functional consequences for differentiation of mouse embryonic stem cells.21 Thus, there are now multiple independent studies with a number of microRNAs demonstrating a functional role for target sites in coding regions.
From an evolutionary perspective, 3’ UTRs and coding regions experience very different types of selective pressure. In 3’ UTRs, microRNA binding sites can be created and then mutated away if the microRNA-target interaction that results does not confer a selective advantage. For sites within 3’ UTRs the creation and destruction of the site can occur without constraints related to the function of an encoded protein. In fact, the vast majority of target recognition sites in 3’ UTRs may have little functional effect and may be rapidly eliminated by evolution.28 For microRNA recognition sites in coding regions, a point mutation that creates a new site could be selected against if the encoded protein functions less efficiently than without the mutation. For this reason, new sites might not become as prevalent within a population. In addition, microRNA binding sites that are present within coding regions but for which the microRNA-mRNA interaction is neutral, might be less likely to be destroyed by evolution as sites within 3’ UTRs, since some of the possible mutations that would destroy the site might affect the functionality of the encoded protein. Yet, there is evidence that sites might be created and eliminated over relatively short evolutionary time frames in coding regions as well. In the study by Tay and colleagues, which did not rely on evolutionary conservation for the identification of potential binding sites, four of five validated coding region microRNA recognition sites identified in mouse were not present in humans or rhesus.21
Localizing microRNA binding sites within the coding region may even have specific advantages for the cell compared with localizing them within the 3’ UTR. In some instances, a microRNA binding site in the coding region may ensure that a specific microRNA biding site is always present. The existence of alternative polyadenylation sites suggests that 3’ UTRs may differ under different conditions and in some cases, microRNA binding sites would be eliminated. For instance, a large number of transcripts utilize different polyadenylation sites in proliferating versus quiescent lymphocytes.29 For sites in 3’ UTRs, there is thus another opportunity for regulation of microRNA function. In contrast, microRNA binding sites in the coding region would be present regardless of the polyadenylation signal utilized for a given transcript, making it less susceptible to changes with cellular state.30 In other instances, coding region sites may provide an opportunity for another level of regulation based on whether they are included or excluded from the final transcript during splicing. In this way, binding sites in coding regions could allow the targeting of specific splice variants since the binding site could be present in an exon that is present in some but not all isoforms.21,31
Coding region microRNA target sites are functional, but less effective than 3’ UTR sites
Recent experiments have allowed further analysis of the functional role of microRNA binding sites in coding regions. Rajewsky and colleagues analyzed the effects of microRNAs on global protein output by treating HeLa cells with a locked nucleic acid (LNA) directed against let-7 followed by stable isotope labeling and mass spectrometry based-proteomics.29 We analyzed their data to determine the cumulative fold change induced by inhibition of endogenous let-7 with LNAs. As shown in Figure 1, let-7 seed matches present in 3’ UTRs have a relatively modest effect on the protein levels of target genes. The subset of seed matches in the 3’ UTR that are evolutionarily conserved have stronger effects. Coding region seed matches are clearly functional, as the cumulative distribution of their effect is statistically distinguishable from the distribution of effects for no site, but they are not as effective as sites in 3’ UTRs. Among coding region seed matches, those that are evolutionarily conserved have a stronger effect. A similar proteomics approach was applied to samples in which microRNAs were overexpressed,29 and our analysis reveals the same trends (data not shown).
Figure 1. Coding region microRNA target sites are functional, but less effective than 3’ UTR sites.
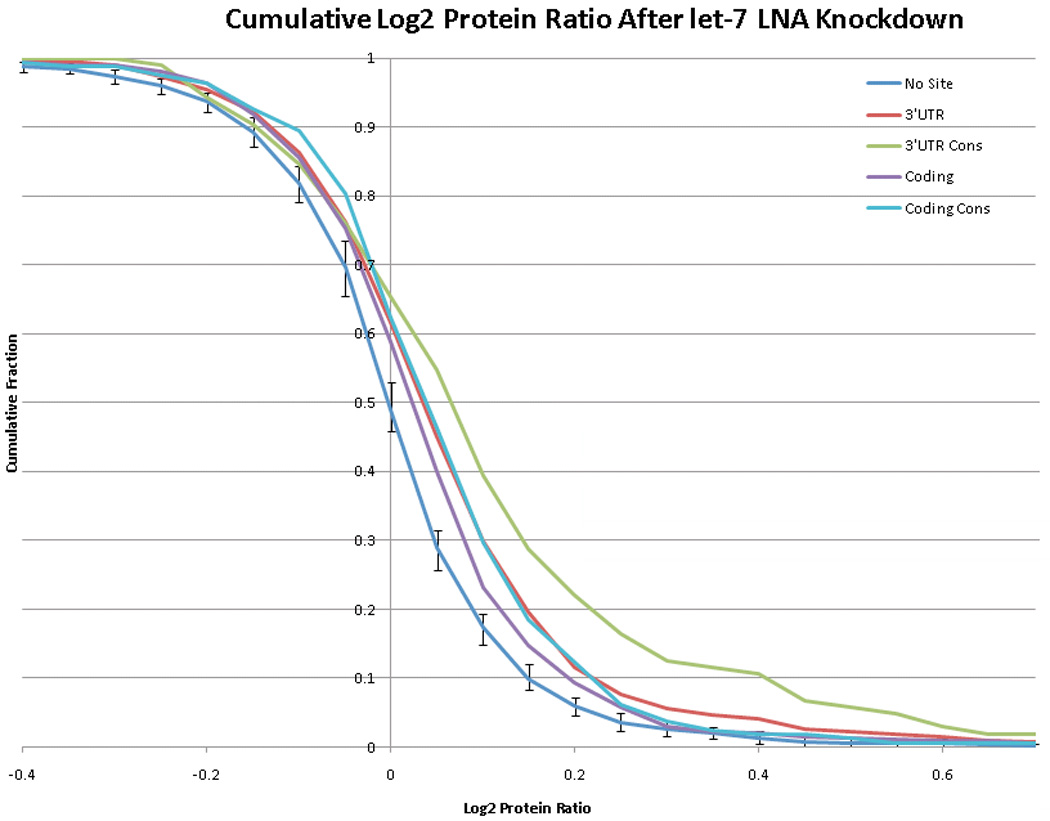
Protein levels were determined for 2634 proteins in HeLa cells that were either mock-treated or treated with a LNA that targets the let-7 microRNA using pulse SILAC labeling followed by mass spectrometry.29 The cumulative fraction of proteins with a given ratio of protein level in mock-treated versus let-7 LNA-treated cells is plotted for proteins with no target site, a 3’ UTR target site, an evolutionarily conserved 3’ UTR target site, a coding region site or an evolutionarily conserved coding region site. The number of genomes in which a site was conserved was summed across all occurrences of the let-7 target site in each region, and the highest-scoring 25% were designated as "conserved." In order to define error bars for the no site distribution, a subset of 649 proteins without a let-7 binding site was randomly selected 100 times and the 5th percentile highest and lowest values were plotted as the error. Coding region target sites were clearly functional, as the distribution of effect sizes is statistically significantly different from the distribution for proteins with no recognition site (Wilcoxon p-value < 10−5). However, the presence of a recognition site within the coding region confers less repression than the presence of a 3’ UTR site on average. For both the coding region and the 3’ UTR, evolutionarily conserved sites tend to confer a stronger repressive effect than all sites.
While coding region microRNA binding sites can be effective, sites within 3’ UTRs are clearly preferred. The reasons for this are unclear and the data are contradictory. In some studies, microRNA binding sites artificially introduced into the coding region or 5’ UTR have been shown to function efficiently,21, 30 although other studies have reached the opposite conclusion.31 One proposed model is that coding regions are effectively targeted when there is a large degree of complementarity resulting in exonucleolytic cleavage, but that inhibition of translation is less effective in coding regions than 3’ UTRs. An explanation that has been put forth is that coding regions are constantly being traversed by ribosomes, which could destabilize interaction with the RISC complex.31 Clearly, microRNAs in plants target coding regions effectively.13, 32 In this case, long stretches of perfect base pairing direct RISC-mediated dsRNA cleavage. Short interfering RNAs (siRNAs) in animals can also target coding regions for RISC-mediated endonucleolytic degradation through complementary interactions. The efficacy of such interactions could reflect the more extensive Watson-Crick base-pairing of siRNAs than microRNAs, or that siRNAs require the functional interaction with the target to persist for a shorter time in order to mediate an effect.33 In either case, siRNA-type interactions might be less likely to be disturbed by traversing ribosomes.
The patterns of base pairing between microRNAs and target mRNAs could support this hypothesis.2 For target sites in 3’UTRs, evolutionarily conserved binding sites are more likely to have a “bulge”—nucleotides immediately after the seed sequence that do not base pair with the microRNA—as previously reported.34 Conserved microRNA target sites within coding regions, on the other hand, are not more likely to contain a bulge at this position. For coding region targets, base pairing across the entire length of the microRNA-target interaction was associated with evolutionary conservation. The results suggest that coding region targets might rely more heavily on mechanisms in which perfect base pairing results in degradation of double stranded RNA.
This conclusion is further supported by an experiment in which a microRNA target site was introduced into the 3’ UTR of a reporter designed so that a single upstream nonsense to sense substitution allowed the same microRNA target site to be either in the coding region or the 3’ UTR.31 A perfectly matching microRNA or a microRNA containing a bulge after the seed was introduced. When the target sites were in the coding region, the perfectly matched microRNA but not the bulging microRNA exhibited repressive activity, whereas both microRNAs effectively targeted sites localized in the 3’ UTR. These findings support the hypothesis that siRNA-mediated cleavage mechanisms may be intact in coding regions, while microRNA-mediated translational repression is less effective. The authors also provide some evidence that the distinction reflects interference from passing ribosomes.31 They introduced a nine amino acid cluster of rare codons upstream of the coding region microRNA target. The rare codons were expected to cause ribosomal pausing, and under these conditions, the microRNA regained its effectiveness at inhibiting translation. On the other hand, in a study of developing zebrafish, a construct containing two let-7 coding region sites with bulges was functionally effective in producing a severe slowing of development.35 Further, in the analysis of microRNA targets in coding regions by Tay and colleagues, the effects on protein level were consistently stronger than the effects on transcript abundance, which were often modest.21 Finally, Dicer, which contains coding region let-7 binding sites, is particularly strongly inhibited at the translational level by let-7.29 Thus, there is at this point some evidence that coding region microRNA target sites are effective for nuclease activity and less effective for translational inhibition, but there are data to the contrary as well.
Sites at the 3’ end of transcripts are particularly effective
To explore possible mechanisms of microRNA function in coding regions and 3’ UTRs, we analyzed the pattern of evolutionary conservation of microRNA target sites based on their position within the sequence (Figure 2). We discovered that microRNA target sites at the very 3’ end of transcripts are more likely to be highly evolutionarily conserved, consistent with previous studies reporting that sites in this location are more likely to be functional.34, 36 The increased effectiveness of these sites may reflect their proximity to portions of the transcript that are important for microRNA function. As one example, target sites at the very 3’ end of transcripts are localized close to the poly(A) tail. microRNAs can induce deadenylation and thereby accelerate turnover of targeted transcripts.17, 37–42 Indeed, some previous studies have indicated that the poly(A) tail is essential for microRNA-mediated repression, and target sites near the end of the poly(A) tail might execute this function more effectively.43, 44 However, deadenylation alone is clearly not the only mechanism of microRNA-mediated repression of translation since mRNAs without poly(A) tails can still be repressed by microRNAs,40, 41, 45, 46 and some targets are repressed without changes in their degradation rates.29, 47 Nevertheless, if proximity to the poly(A) tail facilitates the ability of the RISC complex to induce deadenylation, a bias toward highly conserved and functional sites at the end of the 3’ UTR would be expected.
Figure 2. Target site conservation by location within the coding region or 3’ UTR.
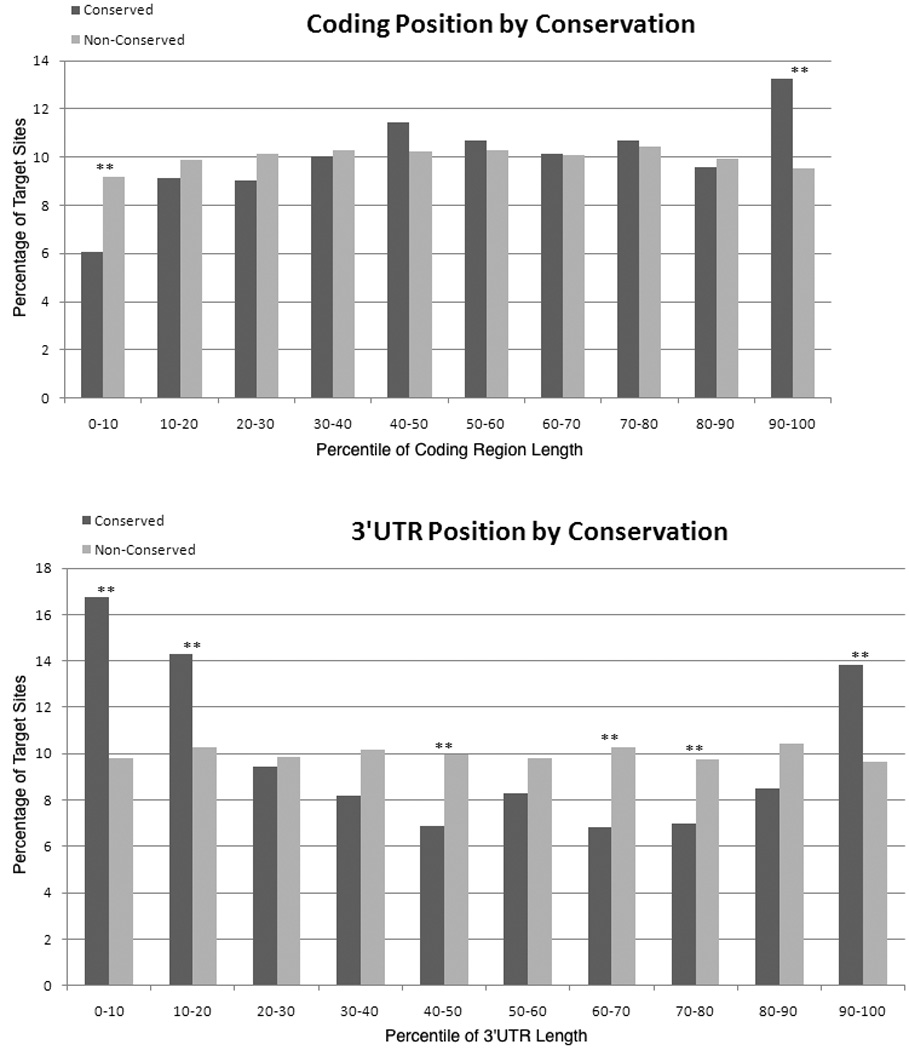
The location of microRNA binding sites was determined for all highly conserved microRNA families (as listed in TargetScan version 5.0) using a modified version of SiteSifter (www.sitesifter.org). Target sites were grouped based on their location within the coding region or 3’ UTR such that target sites within the first 10% of the coding region or 3’ UTR were considered together, target sites within the second 10% of the coding region or 3’ UTR were considered together, etc. For target sites within each group, the fraction that are well-conserved (black bars), defined as 14+ genomes for coding region target sites and 13+ for 3'UTR target sites, and the fraction that are poorly-conserved (empty bars), defined as the remainder of sites, are plotted. Conservation was determined based on a 17-genome alignment available from the University of California at Santa Cruz. A statistically significant enrichment for evolutionarily conserved sites is observed at the end of the coding region and at the beginning and end of the 3’ UTR. In the middle of the 3’ UTR, sites are more likely not to be evolutionarily conserved. Statistically significant differences are indicated with asterisks and reflect a p value less than 0.01 using a chi-squared test after correcting for multiple hypothesis testing.
A number of studies also support a model in which microRNAs affect translation initiation. Some of these studies suggest that microRNAs inhibit translation of targeted transcripts by interacting with the 5’ terminal modified guanine cap that binds to the poly(A) tail after circularization to achieve efficient translation of transcripts through multiple rounds of ribosome loading.6, 48, 49 Several reports suggest that the cap is necessary for microRNA function.6, 43–45, 49–51 Similarly, reports of fewer loaded ribosomes on microRNA-targeted transcripts also suggest an important role for the cap.6, 45, 52–54 Other studies, however, have concluded the opposite,30, 46, 55–57 and thus the importance of interactions between microRNAs and the cap remains controversial.58 Localization of microRNA binding sites close to the 3’ end of the transcript would place the RISC complex physically close to the cap after circularization.
microRNA-mediated effects on the poly(A) tail and the cap are not mutually exclusive. In fact, microRNAs likely mediate their effects through multiple mechanisms. For instance, when translation is inhibited with cycloheximide, some microRNA targets are stabilized while others continue to be degraded in a microRNA-dependent manner.59 The results suggest that some mechanisms of microRNA targeting directly affect transcript abundance without reliance on translational inhibition, possibly through deadenylation or endonucleolytic cleavage, while other mechanisms are more effective at inhibiting translation through interference with cap-mediated ribosomal initiation.60 In Drosophila, these two functions seem to be separated into two different Ago proteins.60 Drosophila Ago1 functions via target deadenylation (and at a distinct step after translation initiation) while Drosophila Ago2 is an endonuclease and can repress translation initiation by interfering with interactions at the message cap. However, in global analyses of the roles of microRNA function in mammalian cells, a close correlation between changes in mRNA levels and protein level has been observed, and the authors have conjectured that the mechanisms for transcript degradation and translational inhibition are linked.29, 47, 61 A microRNA binding site at the very end of the 3’ UTR might allow RISC to affect both the poly(A)tail and the cap, and could help to explain why these sites are so effective.
RNA binding proteins and microRNA function
The increased effectiveness of microRNA binding sites in 3’ UTRs compared with coding regions could reflect mechanisms that specifically target the RISC complex to the 3’ UTR. For instance, there may be proteins that either target microRNAs to 3’ UTRs or that are themselves localized to 3’ UTRs and that interact with and enhance the effectiveness of the RISC complex. RNA binding proteins may play this role. Some studies have reported a cooperative role for RNA binding proteins and RISC in microRNA function.62–66 Interestingly, other studies have indicated that RNA binding proteins and microRNAs may compete for binding sites,53, 67 even within coding regions.68 One series of experiments demonstrating cooperation between RNA binding proteins and microRNAs focused on miR-16, which contains an 8 base pair region that resembles the AU-rich elements (AREs) often found in 3’ UTRs.65 Both miR-16 and the ARE binding protein tristetrapolin were required for the instability of ARE-containing transcripts. In this example, the microRNA interacted directly with an ARE in the 3’ UTR and both RNA binding protein and microRNAs promoted RISC-mediated transcript degradation.
Similarly, the RNA binding protein HuR, which also binds AREs in 3’ UTRs, was reported to work cooperatively with the let-7 microRNA to reduce expression of the c-Myc transcript.63 In this case, HuR and let-7 bound to close but distinct sites in the 3’ UTR. HuR was required for let-7 binding to the c-Myc 3’ UTR and for let-7-mediated repression of c-Myc expression. Conversely, let-7 binding was required for HuR to repress c-Myc expression. The authors proposed that HuR binding results in a change in the local secondary structure of the RNA that unmasks the let-7 binding site. Such a mechanism would require close proximity of RNA binding proteins and microRNA target sites, and might also help to explain the relative importance of 3’ UTR binding sites. Target sites within coding regions would be less likely to take advantage of such mechanisms as they are less likely to contain AU-rich regions targeted by RNA binding proteins. If the location of ARE's adjacent to microRNAs does facilitate microRNA function, this might help to explain the observation that microRNA binding sites located within AU-rich regions are particularly effective.34 Of course, the location of microRNAs within AU-rich stretches could reflect other considerations instead of or in addition to RNA binding. For instance, AU-rich sequences might intrinsically have less secondary structure, thereby facilitating interaction with the RISC complex. However an important role for RNA binding proteins in the increased efficacy of 3’ UTR microRNA binding sites might represent more than simply making microRNA binding sites accessible, since ribosome traversal would be expected to disrupt secondary structure and make microRNA binding sites accessible in coding regions, at least temporarily.
Is there a role for nonsense-mediated decay (NMD) in microRNA function?
In addition to enrichment at the very 3’ end of the transcript, we discovered that there is another position within the transcript where there is enrichment for evolutionarily conserved target sites —the position in the transcript at the junction between the end of the coding region and the beginning of the 3’ UTR (Figure 2). This finding is consistent with bioinformatic analyses of functional microRNA binding sites.34 It is also consistent with high throughput sequencing results for argonaute-bound ribonucleoprotein complexes.36 In these experiments, there was a clear enrichment in argonaute binding at the 3’ end of transcripts and near the stop codon. While many of the expected mechanisms of mRNA degradation might be consistent with enrichment at the 3' end of the 3' UTR, enrichment near the stop codon is not easily explained by the mechanisms generally considered for microRNA function.69
We wonder whether the localization of conserved microRNA binding sites near the stop codon indicates that the microRNA pathway cooperates with NMD, a quality control mechanism found in all eukaryotic species studied to date.70 NMD ensures that transcripts with a premature termination codon, which would result in a truncated and potentially dominant negative protein product, are identified and degraded.71 It has become increasingly clear that NMD not only identifies rare transcripts with mutations or splicing errors, but can also regulate the steady state level of mRNAs that do not contain premature temination codons.72–75 Genome-wide screens in budding yeast, Drosophila and human cells reveal that NMD may regulate as much as ~3–10% of the transcriptome.73–75
NMD in mammals is induced when a termination codon is positioned more than 50–55 nucleotides upstream of an exon-exon boundary.76 During the first round of translation, the ribosome stalled at a premature termination codon recruits Upf1 to eukaryotic release factor 3.77 Subsequent interaction with proteins that mark the exon-exon boundary results in the recruitment of Upf2 and Upf3,78, 79 phosphorylation of Upf1 and activation of its ATPase and helicase activities.80, 81 The transcripts thus targeted are then degraded in the 5’ to 3’ direction by decapping and exonucleases, as well as in the 3’ to 5’ direction.82–84 Some of the same decapping and degradative enzymes have been implicated in both microRNA-mediated degradation and NMD.62, 72, 85–88 Further, transcript degradation in the case of both microRNAs and NMD occurs in P bodies.11, 83, 89
A link between microRNAs and the NMD pathway90–92 could help to explain the distribution of evolutionarily conserved microRNA binding sites. If the RISC complex recruits Upf1 to the termination codon of targeted transcripts, it could initiate an NMD-type pathway that would lead to reduced levels of targets. Support for this model comes from links between NMD and microRNAs in C. elegans91 and Arabidopsis92, as well as a recent paper in which the NMD protein Upf1 was discovered to contribute to microRNA silencing.90 Upf1 knockdown resulted in de-repression of microRNA targets in cases in which there were mismatches between the microRNA and the target, but not in cases of perfect base pairing. Overexpression of Upf1, but not a helicase domain mutant, resulted in down-regulation of microRNA targets. Further, co-immunoprecipitation with tagged Upf1 and Argonaute proteins revealed an interaction between Upf1 and Ago1 and between Upf1 and Ago2 even in the presence of RNase.90 A previous analysis of Ago-associated proteins using antibodies to endogenous Ago identified Upf1 among the proteins associated with Ago1 (but not Ago2).93 Further, when expressed in the presence of Ago proteins, Upf1 co-localized with Ago1 and Ago2 in P bodies.90 While Jin and colleagues discovered that there was a functional effect of Upf1 knockdown, they did not observe an effect on microRNA function when Upf2 was knocked down, suggesting that the way in which Upf1 affects target degradation in the context of microRNAs is not though the canonical NMD pathway. However, recent studies have provided some evidence that NMD may occur independent of Upf2 and Upf3,80, 94–97 suggesting that an alternative Upf1-dependent, Upf2-independent pathway could be employed. Thus, there is evidence that some of the same molecules might be involved in NMD and in miRNA-mediated gene regulation. On the other hand, this conclusion is not supported by a study describing a small molecule inhibitor of nonsense-mediated decay.98 This molecule, which stabilizes the phosphorylated form of Upf1, did not affect microRNA function. Thus, the potential role of the NMD pathway in microRNA function is not yet clear. If NMD molecules are involved, and if microRNAs are consequently more effective when localized near the stop codon, this might explain the increased evolutionary conservation observed for microRNA binding sites at the end of the coding region and at the beginning of the 3’ UTR.
Conclusion
Evidence thus far supports the conclusion that the coding regions of genes can contain additional information besides the amino acid sequence of the encoded protein, including functional microRNA binding sites. Many questions remain. Do all of the mechanistic pathways that can result from a microRNA-target interaction in the 3’ UTR also occur for coding region target sites? What are there advantages for a microRNA coding sites being present in the 3’ UTR? What are the advantages for a microRNA binding site being present in the coding region? How does the cell exploit these advantages? What is the role of RNA binding proteins in microRNA function and do RNA binding proteins contribute to the effectiveness of microRNAs? At what rate do microRNA-target interactions appear and disappear in populations and how does this differ for coding versus 3’ UTR target sites? Do microRNA-target interactions have different physiological roles within cells depending on the location of the microRNA recognition site within the target transcript?
Acknowledgments
The authors wish to thank Eric Suh (Princeton University), Aster Legesse-Miller (Princeton University), Elizabeth Johnson (Princeton University), Joshua Bloom (Princeton University), Peng Jiang (Princeton University), Daniel Wolle (Princeton University), Leonid Kruglyak (Princeton University), David Botstein (Princeton University) and the rest of the Coller Lab for sharing unpublished data and helpful discussions. This research was supported by PhRMA Foundation grant 2007RSGl9572, NIH/NIGMS 1R01 GM081686, NIH/NIGMS 1R01 GM086465, NCI training grant 5T32 CA009528, and NIGMS Center of Excellence grant P50 GM071508. HAC is the Milton E. Cassel scholar of the Rita Allen Foundation.
Abbreviations and acronyms
- UTR
untranslated region
- RISC
RNA-induced silencing complex
- Ago
Argonaute
- dsRNA
double stranded RNA
- LNA
locked nucleic acid
- siRNAs
short interfering RNAs
- ARE
AU-rich element
- NMD
nonsense-mediated decay
Contributor Information
Joshua J. Forman, Department of Molecular Biology, Princeton University, jforman@post.harvard.edu
Hilary A. Coller, Department of Molecular Biology, Princeton University, hcoller@princeton.edu.
Literature Cited
- 1.Itzkovitz S, Alon U. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res. 2007;17:405–412. doi: 10.1101/gr.5987307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H, Blanchette M. Detecting non-coding selective pressure in coding regions. BMC Evol Biol. 2007;7 Suppl 1:S9. doi: 10.1186/1471-2148-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Yang L, Li H, Li L, Chen J. Residues that affect human Argonaute2 concentration in cytoplasmic processing bodies. Biochem Biophys Res Commun. 2009;378:620–624. doi: 10.1016/j.bbrc.2008.11.098. [DOI] [PubMed] [Google Scholar]
- 11.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 12.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 16.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 24.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 25.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, et al. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 29.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 30.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 38.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 39.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 44.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 46.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 47.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, et al. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 49.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 50.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 51.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 54.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 56.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 57.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 58.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 59.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 61.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 65.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 66.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 68.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 70.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 71.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 72.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5' to 3' mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 74.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 75.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 77.Behm-Ansmant I, Izaurralde E. Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- 78.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 82.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 83.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 84.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3'-->5' degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 85.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 86.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin H, Suh MR, Han J, Yeom KH, Lee Y, Heo I, et al. Human UPF1 modulates small RNA-induced mRNA down-regulation. Mol Cell Biol. 2009 doi: 10.1128/MCB.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Domeier ME, Morse DP, Knight SW, Portereiko M, Bass BL, Mango SE. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- 92.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 93.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 95.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 96.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 98.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]


