Abstract
Background
Recent genome-wide association studies (GWAS), mostly conducted among women of European ancestry, have identified 16 single nucleotide polymorphisms (SNPs) associated with breast cancer.
Methods
We evaluated these SNPs with the risk of breast cancer and further by estrogen receptor (ER) status in a population-based study of 6,498 cases and 3,999 controls in Chinese women. We also searched for novel genetic risk variants in four loci, 2q35, 5p12/MRPS30, 8q24.21, and 17q23.2/COX11, in a two-stage study. In stage I, 868 SNPs were analyzed in 2,073 cases and 2,084 controls. In stage II, 58 SNPs selected from stage I were evaluated, including 4,425 cases and 1,915 controls.
Results
Statistically significant associations (P<0.05) were observed for 8 GWAS-identified SNPs, including rs4973768 (3p24/SLC4A7), rs889312 (5q11.2MAP3K1), rs2046210 (6q25.1), rs1219648 (10q26.13FGFR2), rs2981582 (10q26.13/FGFR2), rs3817198 (11p15.5/LSP1), rs8051542 (16q12.1TOX3), and rs3803662 (16q12.1/TOX3). Two additional SNPs, rs10941679 (5p12/MRPS30) and rs13281615 (8q24.21) showed a marginally significant association. Some of these associations varied by ER status. In the fine-mapping analysis, 5 SNPs showed a consistent association with breast cancer risk in both stages: rs10169372 (2q35), rs283720 (8q24.21), rs10515083 (17q23.2/COX11), rs16955329 (17q23.2/COX11), and rs2787487 (17q23.2/COX11).
Conclusions
This study shows that approximately half of the SNPs initially reported from GWAS of breast cancer in European descendants can be directly replicated in Chinese. Our fine-mapping analyses revealed several candidates of risk variants that can be further evaluated in studies with a larger sample size.
Impact
Findings from this study may help guide future fine-mapping studies to identify causal variants for breast cancer.
Keywords: GWAS, SNP, breast cancer, association
Introduction
Breast cancer is the most common malignancy among women in the United States and many other parts of the world. Genetic factors play an important role in the etiology of breast cancer. Recently, several genome-wide association studies (GWAS) (1-8), including our own study among Chinese women in Shanghai (5), have identified multiple genetic susceptibility loci for breast cancer. With the exception of our study, all other reported GWAS have been conducted among women of European ancestry. The vast majority of the risk variants identified thus far, however, are single nucleotide polymorphisms (SNPs) that are associated with disease risk through linkage disequilibrium (LD) with the causal variants. Therefore, some risk alleles identified in Europeans may not be extrapolated to Asians given the difference in (LD patterns between these two populations. Investigation of previously reported loci in non-European populations may help to evaluate the generalizability of these initial findings and to identify causal variants. Further evaluation of previously reported loci could also help to identify additional risk variants in some of the loci, as in the case of 8q24.21 for prostate cancer risk (9-11) and 16q12 for breast cancer risk (1).
Using data from the Shanghai Breast Cancer Study, a population-based case-control study, we previously evaluated 11 SNPs identified initially in GWAS conducted in women of European ancestry (12). In this study, we evaluated four newly identified loci for breast cancer risk from recent GWAS conducted among Europeans or European Americans. The associations of all GWAS-identified SNPs were further evaluated by estrogen receptor status. Finally, we conducted analyses to explore additional independent genetic risk variants in four loci.
Materials and Methods
Study Participants
Included in the study were 6,498 cases from the Shanghai Breast Cancer Study (SBCS) and Shanghai Breast Cancer Survival Study (SBCSS), as well as 3,999 controls from the SBCS and the Shanghai Endometrial Cancer Study (SECS). The SBCS is a large, population-based, case-control study of women in urban Shanghai that has been previously described in detail (5, 13). Subject recruitment in the initial phase of the SBCS (SBCS-I) was conducted between August 1996 and March 1998. The second phase (SBCS-II) of recruitment occurred between April 2002 and February 2005. Breast cancer cases were identified through the population-based Shanghai Cancer Registry, which for the SBCS-I was supplemented by a rapid case-ascertainment system. Controls were randomly selected using the Shanghai Resident Registry. Also included in the present study were cases recruited between April 2002 and December 2006 as part of SBCSS. The controls for the SBCSS cases came from the SECS, which recruited healthy women between January 1997 and December 2003. Of the eligible participants, 1,459 cases (91.1%) and 1,556 controls (90.3%) in the SBCS-I, 1,989 cases (83.7%) and 1,918 (70.4%) controls in the SBCS-II, and 5,046 cases (80.1%) in the SBCSS and 1,212 controls (74.4%) in the SECS completed in-person interviews with structured questionnaires. Blood or buccal cell samples were collected and made available for 1,193 cases (81.8%) and 1,310 controls (84.2%) from the SBCS-I, 1,932 cases (97.1%) and 1,857 controls (96.8%) from the SBCS-II, 4,845 (96.0%) cases from the SBCSS, and 1,039 (85.7%) controls from the SECS. Because of a time overlap in subject recruitment, 1,469 breast cancer patients participated in both the SBCS-II and SBCSS, and 109 controls participated in both the SBCS-I and SECS, so that the actual total number of participants came to 3,466 cases from the SBCSS and 930 controls from the SECS. Genomic DNA was extracted using commercial DNA purification kits. Approval of the study was granted by the relevant institutional review boards in both China and the United States.
SNP selection and statistical analysis
Four loci reported from studies conducted among Europeans or European Americans, including 2q35, 5p12/MRPS30, 8q24.21, and 17q23.2/COX11, were selected to identify additional SNPs that may be associated with breast cancer in our Chinese population. These four loci were selected because the initially-reported SNPs in each of these loci did not show an apparent association with the overall risk of breast cancer in the Chinese population (Table 2).
Table 2.
Association of breast cancer risk with 16 SNPs identified from previous GWA studies in all SBCS samples
| SNP | Chr/genea | No of cases |
No of controls |
Risk allele freq | All women | ER positive | ER negative | Pe | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | |||||
| rs11249433 | 1p11.2/NOTCH2 | 2044 | 2054 | 0.03 | 0.03 | 1.19(0.92-1.54) | 0.18 | 1.22(0.90-1.66) | 0.19 | 1.10(0.75-1.61) | 0.63 | 0.61 |
| rs13387042 | 2q35/unknown | 2951 | 3006 | 0.12 | 0.11 | 1.03(0.92-1.16) | 0.56 | 1.06(0.93-1.22) | 0.38 | 0.97(0.82-1.16) | 0.77 | 0.32 |
| rs4973768 | 3p24/SLC4A7 | 6163 | 3904 | 0.20 | 0.18 | 1.12(1.04-1.21) | 0.003 | 1.14(1.05-1.25) | 0.002 | 1.05(0.94-1.16) | 0.38 | 0.28 |
| rs10941679 | 5p12/ MRPS30 | 2950 | 2986 | 0.52 | 0.50 | 1.07(0.99-1.15) | 0.07 | 1.08(0.99-1.18) | 0.10 | 0.99(0.89-1.11) | 0.88 | 0.14 |
| rs889312 | 5q11.2/ MAP3K1 | 2924 | 2997 | 0.53 | 0.52 | 1.07(0.99-1.15) | 0.08 | 1.10(1.01-1.20) | 0.03 | 1.08(0.97-1.20) | 0.17 | 0.83 |
| rs2180341 | 6q22.33/ECHDC1 | 3018 | 2927 | 0.25 | 0.26 | 0.94(0.86-1.02) | 0.13 | 0.91(0.83-1.01) | 0.08 | 0.94(0.83-1.07) | 0.34 | 0.70 |
| rs2046210 | 6q25.1/unknown | 6425 | 3948 | 0.42 | 0.36 | 1.28(1.21-1.36) | 1.8×10−15 | 1.25(1.16-1.34) | 6.6×10−10 | 1.35(1.25-1.47) | 6.9×10−13 | 0.01 |
| rs13281615 | 8q24.21/unknown | 2945 | 2981 | 0.52 | 0.50 | 1.06(0.98-1.14) | 0.13 | 1.03(0.95-1.13) | 0.46 | 1.14(1.02-1.27) | 0.02 | 0.10 |
| rs1219648 | 10q26.13/FGFR2 | 6263 | 3693 | 0.42 | 0.39 | 1.15(1.08-1.22) | 9.2×10−6 | 1.20(1.12-1.29) | 3.2×10−7 | 1.07(0.98-1.16) | 0.11 | 0.001 |
| rs2981582 | 10q26.13/FGFR2 | 6279 | 3688 | 0.35 | 0.32 | 1.14(1.07-1.22) | 5.4×10−5 | 1.17(1.09-1.26) | 2.1×10−5 | 1.09(1.00-1.19) | 0.06 | 0.07 |
| rs3817198 | 11p15.5/LSP1 | 6435 | 3839 | 0.13 | 0.12 | 1.11(1.01-1.21) | 0.03 | 1.12(1.01-1.24) | 0.03 | 1.11(0.98-1.25) | 0.10 | 0.86 |
| rs999737 | 14q24.1/RAD51L1 | 2041 | 2054 | 0.003 | 0.001 | 1.78(0.71-4.45) | 0.22 | 1.33(0.41-4.30) | 0.63 | 1.81(0.45-7.32) | 0.40 | 0.78 |
| rs8051542 | 16q12.1/ TOX3 | 6158 | 3658 | 0.20 | 0.18 | 1.11(1.03-1.20) | 0.006 | 1.13(1.04-1.24) | 0.006 | 1.06(0.95-1.17) | 0.31 | 0.34 |
| rs12443621 | 16q12.1/ TOX3 | 2954 | 2997 | 0.43 | 0.43 | 0.99(0.92-1.06) | 0.75 | 1.00(0.92-1.10) | 0.92 | 0.95(0.85-1.06) | 0.35 | 0.32 |
| rs3803662 | 16q12.1/ TOX3 | 6345 | 3795 | 0.68 | 0.65 | 1.12(1.05-1.19) | 4.5×10−4 | 1.13(1.05-1.21) | 0.001 | 1.08(0.99-1.18) | 0.09 | 0.29 |
| rs6504950 | 17q23.2/COX11 | 6387 | 3909 | 0.92 | 0.92 | 1.00(0.89-1.11) | 0.97 | 1.00(0.88-1.14) | 0.97 | 0.97(0.83-1.12) | 0.64 | 0.83 |
Bolded regions were selected for finemapping
Test allele/reference allele.
frequency in cases/controls.
Adjusted for age and education and study stage.
P for heterogeneity calculated from case-only analyses
In each of these four loci, a region (± 100 kb) flanking the initially-reported SNP was selected. The initially selected region was extended according to the following two scenarios: a) if the LD block, including the initially-reported SNP, extended outside the 200 kb region, then the whole LD block was included, or b) if the 100 kb flanking region contained part of a known gene, the whole gene was included. Using these criteria, the following four regions were investigated: 44642255-44996680 (354 kb) for 5p12 (rs10941679), 50311470-50628909 (317 kb) for 17q23.2 (rs6504950), and a 200 kb region for 2q23 (rs13387042) and 8q24.21 (rs13281615), based on NCBI Build 36.
Stage I analyses were conducted primarily based on the GWAS data obtained using Affymetrix SNP 6.0 arrays. SNPs not found on the array were imputed using the program MACH with the HapMap II Asian data (release 22) as a reference. Association analysis for each SNP was performed by logistic regression, and imputation uncertainty was taken into account by using the program MACH2DAT. Within each region, the SNP identified in previous GWAS was adjusted in the logistic regression model. A total of 868 SNPs with a MAF ≥0.05 were analyzed, including 241 directly genotyped and 627 imputed SNPs. Of these, 32 SNPs had low imputation quality (quality score <0.9), and 26 SNPs showed significant association with breast cancer at P ≤0.05 after adjusting for the initially-reported SNP. A total of 35 tagging SNPs were selected to cover these 58 SNPs, with pairwise r2≥0.8 using the HapMap Asian data as reference. Of these 35 tagging SNPs, 32 were successfully genotyped in Stage II samples, including 4,425 cases and 1,915 controls. Of the 32 successfully typed SNPs, five were significantly associated with breast cancer in Stage II samples and showed low imputation quality in Stage I. They were directly genotyped in Stage I samples, which we referred to as Stage III in this study. Logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for each SNP in association with breast cancer risk after adjusting for age, education and BMI. The results did not change appreciably with or without these potential confounding factors. Heterogeneity between the associations of SNPs with ER positive and ER negative diseases was assessed using logistic regression analyses restricted to cases (case only analyses) with the ER status as the outcome variable. P-values based on 2-tailed tests are presented. All analyses were performed using SAS version 9.1 (SAS Institute).
Genotyping methods
Genotyping using the Affymetrix GeneChip Mapping 500K Array Set and the Affymetrix Genome-Wide Human SNP Array 6.0 has been described previously (5). Among the 16 SNPs reported in previous GWAS, 4 SNPs, rs2180341 (6q22.33/ECHDC1), rs3817198 (11p15.5/LSP1), rs3803662 (16q12.1/ TOX3), and rs2046210 (6q25.1/unknown), were included on both the Affymetrix SNP Array 6.0 and the GeneChip Mapping 500K Array Set. Therefore, genotyping data for these 4 SNPs were available for 4,157 participants. Three SNPs, rs1219648 (10q26.13/FGFR2), rs2981582 (10q26.13/FGFR2), and rs8051542 (16q12.1/TOX3), were included only on Affymetrix 6.0 and not on Affymetrix 500K, thus, genotyping data were available for only 3,866 GWAS participants who were genotyped by Affymetrix 6.0. Of the remaining participants not included in the genotyping using the Affymetrix SNP arrays, these 7 SNPs were genotyped using iPLEX™ Sequenom MassARRAY® platform. The 4 recently reported SNPs, rs11249433 (1p11.2/NOTCH2), rs4973768 (3p24/SLC4A7), rs999737 (14q24.1/RAD51L1), and rs6504950 (17q23.2/COX11), were not included on the Affymetrix 6.0 array and were also genotyped using Sequenom. The remaining 5 SNPs, rs13387042 (2q35/unknown), rs10941679 (5p12/MRPS30), rs889312 (5q11.2/MAP3K1), rs13281615 (8q24.21/unknown), and rs12443621 (16q12.1/TOX3) were genotyped using the TaqMan allelic discrimination assay (Applied Biosystems).
In Stages II and III, the iPLEX™ Sequenom MassARRAY® platform was used for genotyping. On each 96-well plate, two negative controls, two blinded duplicates, and two samples from the HapMap project were included. The mean consistency rates were 98.2% for the blinded duplicates and 99.2% compared with data from HapMap.
Results
The distribution of demographic characteristics and known breast cancer risk factors for cases and controls are shown in Table 1. An elevated risk of breast cancer was consistently observed for all known major breast cancer risk factors, including family history of breast cancer, prior history of benign breast disease, physical inactivity, early onset of menarche, late onset of menopause, late age at first live birth, high body mass index (BMI), and high waist-to-hip ratio (WHR).
Table 1.
Distribution of demographic characteristics and known breast cancer risk factors for cases and controls included in the study
| Demographic Factorsa | Stage I |
Stage II |
||
|---|---|---|---|---|
| Cases (n=2,073) |
Controls (n=2,084) |
Cases (n=4,425) |
Controls (n=1,915) |
|
| Age (yr) | 49.3 ± 8.3 | 49.4 ± 8.5 | 53.9 ± 10.2 | 52.8 ± 9.2b |
| Education ≥ high school (%) | 46.4 | 44.4 | 55.0 | 44.9b |
| Reproductive Risk Factorsa | ||||
| Age at menarche (yr) | 14.5 ± 1.7 | 14.7 ± 1.8b | 14.4 ± 1.7 | 14.7 ± 1.8b |
| Postmenopausal (%) | 38.7 | 41.5 | 51.1 | 55.2b |
| Age at menopausec | 48.4 ± 4.5 | 47.7 ± 4.8b | 49.0 ± 4.3 | 48.8 ± 4.1 |
| Number of live births | 1.4 ± 0.8 | 1.5 ± 0.8b | 1.5 ± 0.9 | 1.6 ± 1.0b |
| Age at first live birth (yr)d | 26.5 ± 3.9 | 26.0 ± 3.8b | 26.8 ± 3.9 | 25.5 ± 3.9b |
| Used estrogen replacement therapy (%) | 3.1 | 2.4 | 5.7 | 3.7b |
| Other Risk Factorsa | ||||
| First-degree relative with breast cancer (%) | 4.4 | 3.0b | 5.6 | 2.4b |
| Ever diagnosed with breast fibroadenoma (%) | 9.7 | 5.5b | 10.0 | 5.5b |
| Body mass index | 23.8 ± 3.3 | 23.3 ± 3.4b | 24.0 ± 3.4 | 23.6 ± 3.4b |
| Waist-to-hip ratio | 0.82 ± 0.06 | 0.81 ±0.06b | 0.83 ± 0.05 | 0.82 ± 0.06b |
Unless otherwise specified, mean ± sd are presented.
P ≤ 0.05 for case-control comparisons.
Among postmenopausal women.
Among parous women.
Among the 16 SNPs identified in previous GWAS, significant associations (P<0.05) were observed at 8 SNPs: rs4973768 (3p24/SLC4A7), rs889312 (5q11.2/MAP3K1), rs2046210 (6q25.1/unknown), rs1219648 (10q26.13/FGFR2), rs2981582 (10q26.13/FGFR2), rs3817198 (11p15.5/LSP1), rs8051542 (16q12.1/TOX3), and rs3803662 (16q12.1/TOX3). Two additional SNPs, rs10941679 (5p12/MRPS30), and rs13281615 (8q24.21/unknown), showed an association of borderline significance (P≤0.15) (Table 2). Interestingly, the association with rs13281615 was statistically significant for ER negative breast cancer. Two other SNPs have a very low MAF in Chinese: 3% for rs11249433 (1p11.2/NOTCH2) and 0.2% for rs999737 (14q24.1/RAD51L1). Therefore, the statistical power to detect a significant association in this study is low.
Although no overall association of breast cancer was found for rs13281615 (8q24.21/unknown), analyses by ER status revealed a statistically significant association with ER negative tumors (P=0.02). With the exception of rs13281615 and rs2046210 (6q25.1/unknown), breast cancer associated SNPs, in general, showed a stronger association with ER positive tumor than ER negative tumor and the difference was statistically significant for rs1219648 (10q26.13/FGFR2).
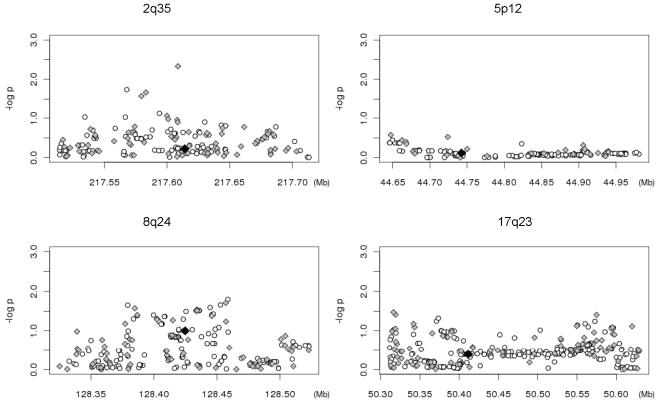
Four loci, including rs13387042 (2q35/unknown), rs10941679 (5p12/MRPS30), rs13281615 (8q24.21/unknown), and rs6504950 (17q23.2/COX11), were further investigated to identify potential novel breast cancer risk variants in Chinese women. Stage I data for these four loci were extracted from the GWAS data of 2,073 cases and 2,084 controls. In these four regions, a total of 241 SNPs passed our quality control (QC) protocol (5), with a call rate ≥95%, a concordance rate ≥95% among duplicated samples, and a MAF ≥0.05. Another 627 SNPs were successfully imputed (with a quality score ≥0.9) by using the program MACH with the HapMap Asian data as the reference. Among these 868 SNPs, 30 SNPs showed an association at P ≤0.05, including 3 SNPs in the region of 2q35, 23 in 8q24.21, and 4 in 17q23.2. After adjusting for the reported SNPs in each locus, 26 of these 30 SNPs still showed an association with breast cancer at P ≤0.05 (Figure 1). In these 4 loci, 32 SNPs on HapMap were imputed with low quality (quality score <0.9) and these SNPs along with SNPs showing an association with a P ≤0.05 26were selected for further evaluation. A total of 35 SNPs were selected to tag these 58 SNPs for Stage II validation.
Figure 1.
Schematic view of genetic association between SNPs in the four loci regions in GWAS and breast cancer risk. Results (-log10P) are shown for directly genotyped (diamonds) and imputed (circles) SNPs. SNPs reported in previous GWAS are highlighted in black.
In Stage II samples, among the 32 successfully genotyped SNPs, SNP rs12949538, located in 17q23.2/COX11, was significantly associated with breast cancer risk with an OR (95% CI) 0.84 (0.75- 0.94) at P =0.002. The association direction, however, was contrary to results from the GWAS data in Stage I. In Stage II, another 5 SNPs, including rs7703618 (5p12/MRPS30), rs7003345 (8q24.21/unknown), rs11986916 (8q24.21/unknown), rs16955329 (17q23.2/COX11), and rs2958919 (17q23.2/COX11), were significantly associated with breast cancer risk at P ≤0.05 (Table 3). All 5 SNPs showed an imputation quality score <0.9 in Stage I. In order to validate the results observed in Stage II, these SNPs were directly genotyped in Stage III samples. None of these five SNPs, however, showed significant associations in Stage III (Table 3).
Table 3.
Association of breast cancer risk with all SNPs genotyped for finemapping in Stage II samples
| Region/Ge ne |
SNP | chr | position | Stage I (2,073 cases/ 2,084 controls) |
Stage II (4,425 cases/ 1,915 controls) |
Stage III (2,073 cases/ 2,084 controls) |
Combined (6,498 cases/ 3,999 controls) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scorea | OR (95% CI)b | Pb | OR (95% CI)b | Pb | OR (95% CI)b | Pb | OR (95% CI)b | Pb | ||||
| 2q35/ unknown |
rs1882420 | 2 | 217559754 | 0.88 | 1.02(0.87-1.20) | 0.77 | 1.06(0.94-1.18) | 0.34 | 1.04(0.95-1.14) | 0.36 | ||
| rs16856795 | 2 | 217561403 | 0.89 | 1.32(1.04-1.67) | 0.02 | 1.02(0.89-1.17) | 0.80 | 1.09(0.96-1.22) | 0.18 | |||
| rs2542197 | 2 | 217564437 | 0.68 | 0.97(0.80-1.18) | 0.78 | 1.01(0.93-1.11) | 0.76 | 1.01(0.93-1.09) | 0.87 | |||
| rs10169372 | 2 | 217579594 | 1.00 | 1.26(1.04-1.54) | 0.02 | 1.06(0.88-1.27) | 0.55 | 1.14(1.00-1.31) | 0.05 | |||
| rs10177578 | 2 | 217593904 | 0.99 | 0.85(0.71-1.02) | 0.07 | 0.97(0.84-1.13) | 0.73 | 0.92(0.82-1.03) | 0.16 | |||
| rs7579306 | 2 | 217652872 | 0.79 | 1.14(0.88-1.48) | 0.30 | 0.95(0.85-1.06) | 0.37 | 0.98(0.89-1.09) | 0.70 | |||
| rs13013872 | 2 | 217664780 | 0.88 | 0.97(0.88-1.08) | 0.60 | 1.03(0.96-1.12) | 0.40 | 1.01(0.95-1.08) | 0.72 | |||
| rs12466929 | 2 | 217688678 | 0.60 | 0.95(0.80-1.12) | 0.52 | 0.96(0.89-1.04) | 0.29 | 0.96(0.89-1.03) | 0.21 | |||
| rs10173363 | 2 | 217692905 | 0.86 | 0.98(0.87-1.10) | 0.71 | 0.97(0.89-1.06) | 0.51 | 0.97(0.91-1.04) | 0.45 | |||
|
| ||||||||||||
| 5p12/ MRPS30 |
rs12652273 | 5 | 44663762 | 0.81 | 0.99(0.84-1.17) | 0.89 | 0.97(0.87-1.09) | 0.64 | 0.98(0.89-1.08) | 0.65 | ||
| rs7703618 | 5 | 44950336 | 0.85 | 1.00(0.89-1.12) | 1.00 | 0.87(0.79-0.94) | 9.8×10−4 | 1.02( 0.93- 1.13) | 0.69 | 0.94(0.88-1.00) | 0.05 | |
|
| ||||||||||||
| 8q24.21/ unknown |
rs283738 | 8 | 128328585 | 0.85 | 0.92(0.75-1.13) | 0.44 | 1.02(0.89-1.17) | 0.75 | 0.99(0.88-1.11) | 0.85 | ||
| rs7003345 | 8 | 128336959 | 0.89 | 1.00(0.90-1.11) | 1.00 | 0.91(0.84-0.99) | 0.03 | 1.03( 0.93- 1.13) | 0.57 | 0.96(0.90-1.03) | 0.25 | |
| rs10956354 | 8 | 128348456 | 0.83 | 0.94(0.84-1.06) | 0.34 | 0.95(0.87-1.04) | 0.29 | 0.95(0.89-1.02) | 0.17 | |||
| rs11994592 | 8 | 128376246 | 0.81 | 0.98(0.87-1.10) | 0.69 | 1.03(0.95-1.12) | 0.46 | 1.01(0.95-1.09) | 0.68 | |||
| rs283718 | 8 | 128376264 | 0.78 | 0.95(0.84-1.08) | 0.43 | 0.99(0.91-1.08) | 0.85 | 0.98(0.91-1.05) | 0.53 | |||
| rs283720 | 8 | 128379147 | 0.95 | 0.89(0.81-0.98) | 0.02 | 0.95(0.88-1.03) | 0.26 | 0.93(0.87-0.99) | 0.02 | |||
| rs283704 | 8 | 128384764 | 0.99 | 0.88(0.79-0.98) | 0.02 | 0.99(0.90-1.09) | 0.87 | 0.94(0.87-1.01) | 0.10 | |||
| rs1949808 | 8 | 128463720 | 0.68 | 0.97(0.86-1.10) | 0.64 | 1.05(0.97-1.14) | 0.19 | 1.03(0.97-1.10) | 0.36 | |||
| rs896324 | 8 | 128465694 | 0.74 | 1.01(0.82-1.25) | 0.92 | 1.04(0.93-1.17) | 0.45 | 1.04(0.94-1.14) | 0.48 | |||
| rs10441525 | 8 | 128472135 | 0.79 | 1.04(0.82-1.32) | 0.74 | 0.96(0.86-1.08) | 0.52 | 0.98(0.88-1.08) | 0.65 | |||
| rs7844673 | 8 | 128472696 | 0.85 | 0.97(0.71-1.31) | 0.82 | 1.05(0.86-1.28) | 0.61 | 1.02(0.87-1.20) | 0.79 | |||
| rs10956365 | 8 | 128473069 | 0.76 | 0.99(0.77-1.27) | 0.91 | 0.96(0.86-1.07) | 0.46 | 0.96(0.87-1.07) | 0.49 | |||
| rs11986916 | 8 | 128488689 | 0.74 | 0.99(0.79-1.24) | 0.94 | 1.14(1.02-1.28) | 0.02 | 1.00( 0.88- 1.14) | 0.97 | 1.08(0.99-1.17) | 0.09 | |
|
| ||||||||||||
| 17q23.2/ COX11 |
rs10515083 | 17 | 50319056 | 0.99 | 0.89(0.80-0.99) | 0.03 | 0.94(0.85-1.03) | 0.18 | 0.92(0.86-0.98) | 0.02 | ||
| rs16955329 | 17 | 50324127 | 0.88 | 1.06(0.96-1.17) | 0.24 | 1.10(1.02-1.19) | 0.02 | 1.06(1.00-1.12) | 0.05 | |||
| rs2958919 | 17 | 50324600 | 0.83 | 1.00(0.89-1.14) | 0.95 | 0.88(0.81-0.96) | 0.005 | 0.94(0.88-1.00) | 0.06 | |||
| rs16955339 | 17 | 50326059 | 0.80 | 0.93(0.79-1.09) | 0.36 | 0.94(0.85-1.04) | 0.22 | 1.01( 0.93- 1.10) | 0.80 | 0.93(0.86-1.02) | 0.12 | |
| rs8067139 | 17 | 50331318 | 0.88 | 1.00(0.90-1.10) | 0.93 | 1.04(0.96-1.13) | 0.34 | 1.02(0.96-1.09) | 0.50 | |||
| rs12949538 | 17 | 50365669 | 0.99 | 1.14(1.00-1.31) | 0.04 | 0.84(0.75-0.94) | 0.002 | 0.96(0.88-1.05) | 0.37 | |||
| rs2787487 | 17 | 50564381 | 1.00 | 1.08(0.99-1.18) | 0.08 | 1.04(0.96-1.13) | 0.32 | 1.06(1.00-1.12) | 0.06 | |||
| rs2529506 | 17 | 50566176 | 0.86 | 0.95(0.87-1.05) | 0.33 | 0.94(0.87-1.01) | 0.10 | 1.02( 0.93- 1.13) | 0.64 | 0.94(0.89-1.00) | 0.06 | |
imputation quality score from MACH
Adjusted for age and education.
In the analysis of combined data from Stage II and Stage I/III, 6 SNPs, including rs10169372 (2q35/unknown), rs7703618 (5p12/MRPS30), rs283720 (8q24.21/unknown), and 3 SNPs located in 17q23.2/COX11 (rs10515083, rs2787487, and rs16955329), showed an association with breast cancer risk, including 5 SNPs that showed a consistent association in both study stages (Table 3). Analyses stratified by ER status showed that all of these 5 SNPs showed stronger associations with ER positive tumors than ER negative tumors, although the heterogeneity test was statistically significant only for SNP rs16955329 (Table 4).
Table 4.
Associations of SNP rs10515083 with breast cancer risk, stratified by menopausal, ER and PR status
| SNP | population | Stage I | Stage II | Stage I and II combined | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI)a | Pa | OR (95% CI)a | Pa | OR (95% CI)a | Pa | Pb | ||
| rs10169372 | ||||||||
| ER positive vs controls | 1.33(1.05- 1.68) | 0.02 | 1.14(0.94- 1.38) | 0.19 | 1.21(1.04- 1.40) | 0.02 | 0.08 | |
| ER negative vs controls | 1.16(0.86- 1.57) | 0.33 | 0.95(0.76- 1.20) | 0.67 | 1.02(0.85- 1.23) | 0.80 | ||
| rs283720 | ||||||||
| ER positive vs controls | 0.85(0.76- 0.96) | 0.01 | 0.95(0.87- 1.04) | 0.30 | 0.92(0.85- 0.98) | 0.01 | 0.54 | |
| ER negative vs controls | 0.92(0.80- 1.06) | 0.27 | 0.96(0.87- 1.06) | 0.43 | 0.95(0.87- 1.03) | 0.19 | ||
| rs10515083 | ||||||||
| ER positive vs controls | 0.89(0.78- 1.01) | 0.07 | 0.93(0.84- 1.03) | 0.14 | 0.91(0.84- 0.99) | 0.02 | 0.38 | |
| ER negative vs controls | 0.95(0.81- 1.11) | 0.54 | 0.95(0.85- 1.07) | 0.39 | 0.95(0.87- 1.04) | 0.30 | ||
| rs2787487 | ||||||||
| ER positive vs controls | 1.11(1.00- 1.24) | 0.05 | 1.03(0.95- 1.13) | 0.46 | 1.06(1.00- 1.14) | 0.07 | 0.58 | |
| ER negative vs controls | 1.04(0.91- 1.19) | 0.57 | 1.04(0.94- 1.15) | 0.47 | 1.04(0.96- 1.12) | 0.35 | ||
| rs16955329 | ||||||||
| ER positive vs controls | 1.15(1.03- 1.30) | 0.02 | 1.10(1.01- 1.20) | 0.03 | 1.10(1.02- 1.17) | 0.01 | 0.04 | |
| ER negative vs controls | 0.83(0.72- 0.96) | 0.01 | 1.09(0.99- 1.20) | 0.09 | 0.99(0.91- 1.07) | 0.79 | ||
P for association with breast cancer risk, adjusted for age and education.
P for heterogeneity calculated from case-only analyses
Discussion
In the present study, of the 14 independent variants identified in GWAS conducted among women of European ancestry (excluding rs2981582 in 10q26.13/FGFR2 and rs2046210 (6q25.1/unknown), which were initially identified in a Chinese population), 8 SNPs showed an association consistent with that observed in women of European ancestry, and the per allele ORs were either statistically significant [rs4973768 (3p24/SLC4A7), rs889312 (5q11.2/MAP3K1), rs1219648 (10q26.13/FGFR2), rs3817198 (11p15.5/LSP1), rs8051542 (16q12.1/TOX3), rs3803662 (16q12.1/ TOX3)], or marginally significant [rs10941679 (5p12/MRPS30), and rs13281615(8q24.21)]. Analyses by ER status showed that the association of breast cancer for some SNPs may differ by ER status. Our fine-mapping analyses revealed several promising candidates that could be further evaluated. Overall, the results from this study provide further evidence for the association of GWAS identified SNPs in relation to breast cancer risk in non-European populations.
SNPs rs11249433 (1p11.2/NOTCH2) and rs999737 (14q24.1/RAD51L1) have a very low MAF in Chinese (3.0% and 0.2%, respectively). Intriguingly, the MAFs for these SNPs are quite high in European populations, 42.5% for rs11249433 (1p11.2/NOTCH2) and 26.1% for rs999737 (14q24.1/RAD51L1). Therefore, the genetic architectures in these two loci between Chinese and Europeans are quite different. For the other 4 SNPs, we found either a null or very weak association, rs13387042 (2q35/unknown), rs12443621 (16q12.1/TOX3), rs6504950 (17q23.2/COX11) or an association that was the opposite of that observed previously [rs2180341 (6q22.33/ECHDC1)]. With the sample size of the current study we have 80% of statistical power to detect an OR as small as 1.13, 1.08, 1.14, and 1.09 for SNPs rs13387042 (2q35/unknown), rs12443621 (16q12.1/TOX9), rs6504950 (17q23.2/COX11), and rs2180341 (6q22.33/ECHDC1). Therefore, we could reasonably conclude that these 4 SNPs are not strongly associated with breast cancer risk in Chinese. Stratification analyses by ER status for these 4 SNPs did not show any association consistent with that observed in women of European ancestry.
Previous studies among women of European origins showed that the association of breast cancer with rs1219648 (10q26.13/FGFR2), rs10941679 (5p12/MRPS30), and rs889312 (5q11.2/MAP3K1) was stronger in ER-positive than ER-negative tumor (8, 14, 15). Results from this study were in general consistent with previous findings for these SNPs, although the test for heterogeneity was statistically significant for rs1219648 (10q26.13/FGFR2) with P=0.001. We found that rs13281615 (8q24.21/unknown) was more related to ER-negative than ER positive cancer, a finding that was inconsistent with that from a previous study among women of European ancestry (14). The reason for this inconsistency is unknown. As reported previously (5), rs2046210 (6q25.1/unknown) was found to be more closely related to ER-negative than ER-positive breast cancer. This association in non-Chinese women remains to be evaluated.
SNP rs13387042 at 2q35 was originally associated with breast cancer, especially estrogen-receptor-positive cancer, in a study conducted among Europeans (3). This SNP lies in a 90-kb high LD region that contains neither known genes nor non-coding RNAs (3). Recently, this SNP was investigated in approximately 30,000 cases and 30,000 controls from 25 studies in the Breast Cancer Association Consortium (BCAC) (16). A significant association was observed in Europeans with an OR (95% CI) of 1.12 (1.09 - 1.15), which is much smaller than that originally observed of 1.20 (1.14 - 1.26). A significant association with this SNP was also observed in our previous study of African-American women, which included 810 cases and 1,784 controls (17). However, no significant association has been observed in Asian populations (3, 12, 16).
SNP rs12443621 is located in 16q12.1, a region where two additional genetic risk variants for breast cancer (rs8051542 and rs3803662) were reported previously in a study conducted among women of European ancestry (1). Recently, we identified a functional genetic variant (rs4784227) at this chromosome region for breast cancer risk (18). In the present study, the other two reported SNPs, rs3803662 and rs8051542, showed significant associations consistent with that observed in women of European ancestry. The LD pattern of this region in Asians is very different from the pattern found in European descendents. For example, there is no LD between rs12443621 and rs3803662 (r2=0.04) in Chinese, but there is moderate LD (r2=0.3) in Europeans.
SNP rs6504950 at 17q23.2 did not show a significant association in the present study; this finding was consistent with the results in Asians in the original GWAS (7) that discovered this SNP. No statistically significant association was observed in Asian women, although the per-allele OR was very similar: 0.96 (0.82–1.12) for Asians and 0.95 (0.93–0.98) for Europeans(7). The genetic architecture in this locus differs considerably across populations, for example, the MAF is 8% in Chinese and 30% in Europeans.
SNP rs2180341 was originally discovered in the Ashkenazi Jewish population (19). Later, it was replicated in an additional 487 Ashkenazi Jewish breast cancer cases and in a European American population of 1,466 breast cancer cases and 1,467 controls (20). There was no data available for Asians. In the present study, we observed a borderline significant association with ER positive tumors; however, the association was opposite to the original finding in the Ashkenazi Jewish population.
There are some potential explanations for the failure of direct replication of the loci identified in Europeans or European Americans. One possibility is that, in the Chinese populations, no common SNPs exist in the regions that are associated with breast cancer. It is possible that other common SNPs in these regions have not been reported and thus were not included in the current study. It is also possible that some other types of variants located in these regions, such as copy number variation (CNV), small insertion-deletion (indel) polymorphisms, or rare variants, are associated with breast cancer. Additionally, Asian women might have different lifestyles or environmental exposures that may mask the effect of these SNPs in breast cancer risk. Genetic interactions with other SNPs that differ in frequency between populations could also manifest as effect heterogeneity.
In an attempt to identify risk variants for breast cancer in regions where the original GWAS-identified SNP showed no apparent association with breast cancer risk, we performed fine-mapping for 4 breast cancer susceptibility loci, 2q35, 5p12, 8q24.21, and 17q23.2. We investigated the associations for all 868 SNPs on HapMap, covering at least a 200 kb region for each locus in a total sample size of over 10,000 subjects. All SNPs were either imputed with high quality or directly genotyped. A total of 5 SNPs, including rs10169372 (2q35/unknown), rs283720 (8q24.21/unknown), rs10515083 (17q23.2/COX11), rs16955329 (17q23.2/COX11), and rs2787487 (17q23.2/COX11), showed a consistent association with breast cancer risk in both stages. Although the associations with these SNPs in the combined analyses all reach a nominal significance level, they were not significant after adjusting for multiple comparisons. Nevertheless, these SNPs are good candidates for future studies. One limitation for this finemapping work is that SNPs not included in HapMap were not investigated. It would be helpful to sequence the targeted region for future studies to discover variants not included in the HapMap database.
In summary, we have now evaluated 14 independent SNPs that were initially reported in Europeans or European Americans. Eight of these SNPs showed strong evidence of association with breast cancer risk (statistically significant or marginally significant with an association consistent with those seen in previous GWAS), which brings the total number of GWAS-identified SNPs in Chinese populations to nine. We searched for additional independent genetic risk variants in four GWAS-mapped loci, in which the reported SNPs showed no apparent associations in Chinese. Several SNPs in these regions showed a statistically significant association with breast cancer risk. Although these associations were not statistically significant after adjusting for multiple comparisons, they may be good candidates for future studies. Additional in-depth, fine-mapping studies with large sample sizes may be needed to fully evaluate these regions and to identify potential risk variants for breast cancer in Asian women.
Acknowledgement
This research was supported in part by National Institutes of Health grants (R01CA124558, R01CA64277, R01CA90899, R01CA92585, R01CA122756, and R01CA137013). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors wish to thank the study participants and research staff for their contributions and commitment to this project, Regina Courtney and the late Qing Wang for DNA preparation, and Brandy Venuti for clerical support in the preparation of this manuscript. Sequenom genotyping was carried out at Proactive Genomics, Winston-Salem, NC. Sample preparation and genotyping using Affymetrix chips and TaqMan platform were conducted at the Survey and Biospecimen and Functional Genomic that are supported in part by the Vanderbilt-Ingram Cancer Center (P30 CA68485).
Reference List
- (1).Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- (4).Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- (9).Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- (11).Ghoussaini M, Song H, Koessler T, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zheng W, Wen W, Gao YT, et al. Genetic and Clinical Predictors, Risk Stratification, and Targeted Prevention of Breast Cancer. J Natl Cancer Inst. 2010 doi: 10.1093/jnci/djq170. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gao YT, Shu XO, Dai Q, et al. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- (14).Garcia-Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007 doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- (16).Milne RL, Benitez J, Nevanlinna H, et al. Risk of estrogen receptor-positive and - negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009;101:1012–8. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zheng W, Cai Q, Signorello LB, et al. Evaluation of 11 Breast Cancer Susceptibility Loci in African-American Women. Cancer Epidemiol Biomarkers Prev. 2009;18:2761–4. doi: 10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Long JR, Cai Q, Shu XO, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: Results from the Asia Breast Cancer Consortium. PLoS.Genet. 2010 doi: 10.1371/journal.pgen.1001002. in press. Ref Type: Journal (Full) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kirchhoff T, Chen ZQ, Gold B, et al. The 6q22.33 locus and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2009;18:2468–75. doi: 10.1158/1055-9965.EPI-09-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]