Abstract
Complete folding is not a prerequisite for protein function, as disordered and partially folded states of proteins frequently perform essential biological functions. In order to understand their functions at the molecular level, we utilized diverse experimental measurements to calculate ensemble models of three non-homologous, intrinsically disordered proteins: I-2, spinophilin and DARPP-32, which bind to and regulate protein phosphatase 1 (PP1). The models demonstrate that these proteins have dissimilar propensities for secondary and tertiary structure in their unbound forms. Direct comparison of these ensemble models with recently determined PP1 complex structures suggests a significant role for transient, pre-formed structure in the interactions of these proteins with PP1. Finally, we generated an ensemble model of partially disordered I-2 bound to PP1 that provides insight into the relationship between flexibility and biological function in this dynamic complex.
Highlights.
First ensemble comparison of three different IDPs that bind the same target, PP1
Ensemble models of unbound PP1 regulators show diverse transient 2° & 3° structure
Free and bound state similarities suggest pre-formed structure is important
Model of partially disordered PP1:I-2 complex provides insight into function
Introduction
In recent years, many exceedingly flexible proteins with important biological functions, known as intrinsically disordered proteins (IDPs), have been described (Tompa, 2002; Wright and Dyson, 1999; Dunker et al., 2002; Dunker et al., 2001; Dunker and Obradovic, 2001; Iakoucheva et al., 2002; Uversky, 2002a, b; Uversky et al., 2000). IDPs lack the typical hydrophobic cores that generally stabilize folded proteins, and their highly dynamic nature prevents their description as single, rigid structures. However, structural studies of a number of IDPs indicate that they are not random-coil polymers; rather they often have preferences for transient secondary and tertiary structure elements. It has been proposed that pre-organization and spatial restriction in IDPs expose primary contact sites to enable faster and more effective binding to target molecules (Cheng et al., 2007; Csizmok et al., 2005; Mohan et al., 2006; Oldfield et al., 2005; Tompa, 2005; Vacic et al., 2007). Binding is often accompanied by stabilization of transient secondary structure or folding-upon-binding transitions (Dyson and Wright, 2002). However, the abundance and diversity of transient structural elements in IDPs and their importance in binding interactions are still unclear. In addition, it is uncertain whether functionally related IDPs (e.g. IDPs that bind identical targets) show conservation of functional motifs with conserved structural features, as is commonly seen in folded proteins.
Protein phosphatase 1 (PP1) is a major serine/threonine phosphatase with roles in diverse cellular processes such as muscle contraction and cell signaling (Cohen, 2002). The activity of PP1 is controlled by two types of proteins: targeting proteins, which direct PP1 to specific subcellular locations and alter its substrate specificity, and inhibitor proteins. Nearly all PP1 regulators interact with PP1 via a common “RVxF” motif (Bollen, 2001). However, this motif can be found in one third of all eukaryotic proteins, many of which do not interact with PP1, indicating that, although it is necessary for the interaction with PP1, it alone is not sufficient to provide specificity for PP1 binding (Wakula et al., 2003). Biochemical data (Bollen, 2001; Cohen, 2002) and structures of PP1 in complex with its targeting proteins MYPT1 (Terrak et al., 2004), spinophilin (Ragusa et al., 2010) and the inhibitor protein inhibitor-2 (I-2) (Hurley et al., 2007) show that PP1 regulators interact with multiple sites on PP1. Furthermore, both types of PP1 regulators are known to contain intrinsically disordered regions (Dancheck et al., 2008; Ragusa et al., 2010). The flexible nature of these IDPs allows them to wrap around PP1, forming contacts with distal sites and burying large surface areas (Ragusa et al., 2010; Terrak et al., 2004). While multiple, distant interaction sites on PP1 provide some specificity in the interaction with its intrinsically disordered regulators, it is unclear whether specificity is enhanced by pre-formed structural motifs present in the IDPs.
Here we describe the comprehensive structural analysis of three PP1 regulators. I-2, one of the most ancient PP1 regulatory proteins, is ubiquitously expressed from yeast to humans (Ceulemans et al., 2002; Li et al., 2007) and is important for the proper regulation of cell division (Wang et al., 2008). DARPP-32 (dopamine- and cyclic AMP-regulated phosphoprotein with molecular weight of 32 kDa) is a pseudo-substrate inhibitor of PP1 that requires phosphorylation on Thr34 to become a nanomolar PP1 inhibitor. It has been established as an essential link between the neuronal dopaminergic and glutaminergic signaling pathways. In this position it plays a critical regulatory role for numerous neurological diseases and a large number of drugs of abuse (Nairn et al., 2004). Finally, spinophilin is a PP1-targeting protein (Sarrouilhe et al., 2006). The PP1:spinophilin holo-enzyme dephosphorylates Ser845 on the GluR1 AMPA receptor subunit and regulates the closed/open state of the AMPA receptor. It is therefore essential for the regulation of long-term potentiation (Hsieh-Wilson et al., 1999; Morishita et al., 2001; Watanabe et al., 2001). Furthermore, spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist, playing an important role in the organization of the neuronal cytoskeleton (Bielas et al., 2007).
Previously we reported nuclear magnetic resonance (NMR) spectroscopy data on the dynamics and transient secondary and tertiary structural preferences of I-2 and DARPP-32 (Dancheck et al., 2008) and the PP1-binding domain of spinophilin (Ragusa et al., 2010). Here we have investigated the relationship between transient structure and the literature reported function in these three PP1 regulators by calculating detailed ensemble models of them using the program ENSEMBLE (Choy and Forman-Kay, 2001; Marsh and Forman-Kay, 2009; Marsh et al., 2007). These models demonstrate that functionally related IDPs can have diverse structural properties. In addition, the availability of PP1-bound crystal structures for I-2 and spinophilin (Hurley et al., 2007; Ragusa et al., 2010) allows for the direct comparison of the free and bound states of these proteins. This has enabled us to investigate the role of pre-formed transient structure in the molecular recognition of PP1. Finally, we have calculated an ensemble model of the PP1:I-2 complex, in which 75% of I-2 remains disordered, providing insight into both the function of I-2 and the structural properties of highly dynamic complexes.
Results
Calculation of unbound intrinsically disordered ensemble models
ENSEMBLE was used to calculate models of I-2 residues 9–164 (I-29–164), the spinophilin PP1-binding domain residues 417–494 (spinophilin417–494) and DARPP-32 residues 1–118 (DARPP-321–118). Three independent ensembles were calculated for each system, with the final ensembles containing 10–24 structures each according to the simplest-ensemble approach, in which the smallest number of conformers possible are fit to the experimental restraints (Marsh and Forman-Kay, 2009). The experimental restraints included NMR chemical shifts, distance restraints derived from PRE measurements, 15N R2 relaxation rates and hydrodynamic radii (Rh) from dynamic light scattering (Dancheck et al., 2008; Ragusa et al., 2010). These four different experimental restraint types report on diverse structural properties including secondary and tertiary structure and overall compaction. Table 1 shows the number of experimental restraints used for each system, the agreement between these experimental restraints and the values predicted from the final calculated ensembles. Table 2 shows the general structural properties of the ensembles compared to reference “coil” ensembles (Feldman and Hogue, 2000; Marsh and Forman-Kay, 2009). Plots comparing our experimental measurements to the ensemble-predicted values are shown in the Supporting Information (Figures S1–S7).
Table 1.
List of experimental restraints used in ENSEMBLE calculations and agreement between experimental and predicted values. See also Figures S1–S7.
| Restraint type | I-29–164 | Spinophilin417–494 | DARPP-321–118 | I-2:PP1 complex | ||||
|---|---|---|---|---|---|---|---|---|
| # | RMSD | # | RMSD | # | RMSD | # | RMSD | |
| 13Cα chemical shifts | 151 | 0.35 ppm | 77 | 0.35 ppm | 112 | 0.34 ppm | 100 | 0.33 ppm |
| 13Cβ chemical shifts | 143 | 0.38 ppm | 74 | 0.39 ppm | 107 | 0.39 ppm | 94 | 0.33 ppm |
| 13C′ chemical shifts | 151 | 0.41 ppm | 76 | 0.41 ppm | 105 | 0.42 ppm | 100 | 0.40 ppm |
| 13Hα chemical shifts | 151 | 0.08 ppm | 75 | 0.08 ppm | 112 | 0.08 ppm | 100 | 0.08 ppm |
| 13HN chemical shifts | 139 | 0.17 ppm | 67 | 0.17 ppm | 99 | 0.16 ppm | 93 | 0.17 ppm |
| PREs | 631 | 0.99 Å | 42 | 0.86 Å | 437 | 0.70 Å | 514 | 20.5 Åb |
| Rh | 1 | 1.9 Å | 1 | 1.9 Å | 1 | 1.8 Å | 0 | - |
| 15N R2a | 141 | 0.85 | 67 | 0.62 | 102 | 0.68 | 92 | 0.68 |
The 15N R2 restraints are applied by enforcing a correlation between experimental values and contacts within the ensemble and thus these values represent a Pearson correlation, not an RMSD.
The PRE restraints were utilized in a very different way in the I2:PP1 complex than for the unbound intrinsically disordered ensembles which accounts for this large RMSD value – see the Methods for more details.
Table 2.
General properties of calculated unbound intrinsically disordered ensembles and TraDES ‘Coil’ ensembles. Average values and standard deviations for three independently calculated ensembles and 100 TraDES ‘Coil’ ensembles are presented. See also Figures S1–S7.
| I-29–164 | Spinophilin417–494 | DARPP-321–118 | ||||
|---|---|---|---|---|---|---|
| Calculated | Coil | Calculated | Coil | Calculated | Coil | |
| Number of structures | 21.00 ±1.63 | 21 ± 0 | 10.33 ±0.47 | 11 ± 0 | 14.00 ±0.82 | 14 ± 0 |
| Radius of gyration (Å) | 34.59 ±0.98 | 36.34 ±1.57 | 16.39 ±0.33 | 25.71 ±1.68 | 28.28 ±0.52 | 31.94 ±2.03 |
| Hydrodynamic radius (Å) | 33.09 ±0.08 | 34.16 ±0.63 | 19.92 ±0.05 | 24.89 ±0.63 | 28.09 ±0.06 | 30.15 ±0.74 |
| Solvent accessible surface area (Å2) | 17259 ±133 | 18326 ±200 | 8267 ± 155 | 9213 ±144 | 12595 ± 57 | 13783 ±213 |
Unbound ensembles possess secondary structure of varying similarity to the PP1-bound complexes
The residue-specific secondary structure content of the ensembles, including the fraction of residues within the broad α, left-β and right-β regions of Ramachandran space (previously defined (Marsh and Forman-Kay, 2009)) and the fraction of residues identified as α-helical by STRIDE (Frishman and Argos, 1995) are presented in Figure 1. The left-β region includes φ angles <−100°, typically associated with β-strand formation, while the right-β region includes φ angles ≥ −100°, referred to as the polyproline II (PPII) region. Secondary structure elements identified in the PP1-bound complexes of I-2 and spinophilin are shown at the top of the plots.
Figure 1.
Secondary structure content of calculated ensembles for (a) I-29–164, (b) spinophilin417–494 and (c) DARPP-321–118. Lines represent the α-helix populations (green), populations in the broad α (blue), left-β (red) and right-β (purple) regions of the Ramachandran diagram. All values are plotted with a three-residue smoothing. Secondary structures observed in the bound-state crystal structures of I-2 and spinophilin are shown above the plots with α-helices in green, βstrands in red and other observed regions in blue.
The most notable secondary structure element observed in the three ensembles is the ~70% populated α-helix in I-29–164 spanning residues 130–142. This α-helix corresponds directly to an α-helix present in the PP1:I-2 complex, which folds over the PP1 active site and is important for PP1 inhibition. Interestingly, two other α-helices (residues 47–54 and 153–164) seen in the PP1:I-2 complex are not extensively populated in the unbound I-29–164 ensembles, demonstrating that transient secondary structure formation in free states is not necessary for all helices observed in bound complexes. Conversely, two transient α-helices are observed from residues 36–40 (~20% populated) and 97–105 (~35% populated) that are in regions of I-2 that are not visible in the PP1:I-2 complex structure.
Spinophilin417–494 has less secondary structure than I-29–164. However, a ~25% populated α-helix (residues 477–487) corresponds perfectly to the only α-helix present in the PP1-bound spinophilin structure. In addition to this helix, two β-strands (residues 430–434 and 456–460) are also observed in PP1-bound spinophilin. Interestingly, a peak in the left-β (i.e. β-strand region) plot of spinophilin (residues 456–461) corresponds very well with this second β-strand. However, there is no significant peak in the left-β plot corresponding to the first β-strand (residues 430–434). Thus, while the region corresponding to the second β-strand tends to sample strand-like Ramachandran angles, there is no evidence for hydrogen-bonded β-sheet formation in disordered spinophilin417–494.
Finally, for DARPP-321–118, a ~30% populated α-helix from residues 22–29 and a 10% populated α-helix near the C-terminus (residues 97–114) were identified. However, the lack of a PP1:DARPP-32 complex structure prohibits further analysis of this data.
Tertiary contacts in the unbound ensembles
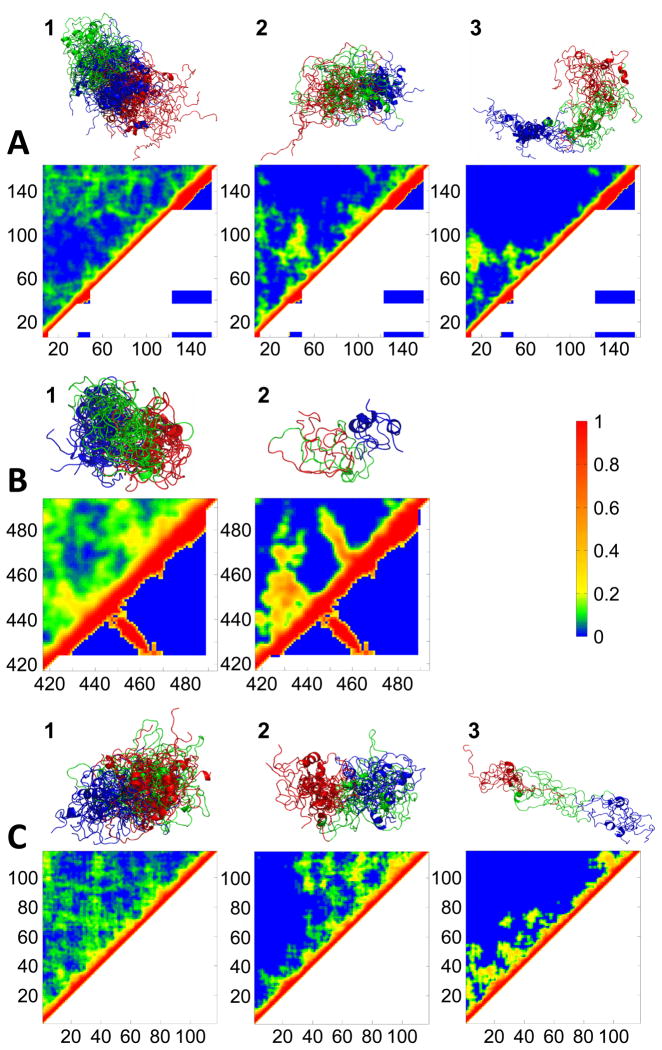
The identification and interpretation of tertiary structure in IDPs is much more complicated than secondary structure. Although contact plots are commonly used to demonstrate tertiary structure in folded and disordered proteins, low populated contacts within a heterogeneous ensemble can be difficult to discern. Thus, to analyze tertiary structure in our ensembles, we used a clustering algorithm to divide the ensembles into clusters of structurally similar conformers (Marsh and Forman-Kay, 2009). The tertiary structure of each cluster is presented in Figure 2 using fractional contact plots (Marsh and Forman-Kay, 2009) which show tertiary contacts within these clusters. Contact plots for the PP1-bound structures of I-2 and spinophilin are shown in the bottom halves of the plots. Populations and structural properties of the clusters are presented in Table S1. For both I-29–164 and DARPP-321–118, a large number of PRE distance restraints were utilized in the ENSEMBLE calculations (Table 1). However, the spinophilin PP1-binding domain is particularly difficult to work with as it is rapidly degraded by proteases. Thus, accurate data from only one spin-label site was available.
Figure 2.
Significantly populated clusters and their fractional contact plots for (a) I-29–164, (b) spinophilin417–494 and (c) DARPP-321–118. Fractional contact plots represent the fractional formation of contacts between pairs of residues, with a contact being defined as any two heavy atoms being within 10 Å of each other. The upper halves of the contact plots show contacts present in the clusters while the lower halves show contacts present in PP1-bound I-2 and spinophilin. White represents regions not present in the PP1-bound structures, either as no electron density was detected or because different construct lengths were used in the studies. Structures are colored as follows: I-2 – residues 6–58 red, 59–111 green, 112–164 blue; spinophilin – residues 417–442 red, 443–468 green, 469–494 blue; DARPP-32 – residues 1–39 red, 40–78 green, 79–118 blue. See also Table S1.
Contact patterns observed in the three major clusters of I-29–164 were analyzed. Cluster 1 is highest populated (44%) and most compact. It is dominated by contacts involving the C-terminal region of I-29–164, particularly residues 120–164. Cluster 2 (17% populated) has significant tertiary contacts between regions around residues 50 and 90. Cluster 3 (14% populated) has contacts between its N-terminus and the region around residue 80. Direct comparison of tertiary structure between I-29–164 clusters and PP1-bound I-2 is difficult as electron density for I-2 was only observed for three isolated segments which lack intramolecular contacts. Nevertheless, although none of these clusters resemble the bound state, it is interesting that I-2 regions that directly interact with PP1 in the crystal structure (residues 11–16, 42–54 and 128–167) participate in a substantial fraction of the contacts detected in unbound I-2.
Upon binding to PP1, spinophilin417–494 undergoes a folding-upon-binding transition and forms secondary and tertiary structure elements including two β-strands that extend a large βsheet in PP1 and an α-helix. Spinophilin417–494 cluster 1 (68% populated) has a large number of tertiary contacts, particularly near the N- and C-termini; none of these contacts resemble the topology of PP1-bound spinophilin. Cluster 2 (18% populated), on the other hand, has tertiary interactions that clearly resemble the β-strand contacts of the PP1-bound state, seen as contacts perpendicular to the diagonal in the contact plot. Although this cluster also contains contacts not seen in the bound state, this suggests that free spinophilin417–494 has a minor population in which contacts resembling the bound state are beginning to form. Notably, these contacts are close to the position of the spin label, providing confidence in their accuracy.
Three clusters are identified for DARPP-321–118. Cluster 1 (50% populated) is very compact. Cluster 2 (29% populated) is dominated by contacts involving the C-terminal region. Finally, cluster 3 (12% populated) is extended and has only very few tertiary contacts. Although these results suggest interesting tertiary structure in unbound DARPP-321–118, further analysis is limited due to the lack of a PP1:DARPP-32 complex structure.
Notably, all three IDPs have at least one cluster with tertiary structures involving the N- and/or C-termini. This is interesting, given the observation that contacts involving the termini should be more likely due to the excluded volume of the polymer chain (Chan and Dill, 1989). This was also previously noted in the unfolded state of the drkN SH3 domain (Marsh and Forman-Kay, 2009). Thus, it seems likely that contacts involving the termini are a common structural feature of IDPs.
Calculation of an ensemble model of the partially folded PP1:I-2 complex
As noted earlier, electron density for I-2 in the PP1:I-2 complex was only detected for ~25% of the protein and, thus, I-2 remains largely disordered, even upon binding to PP1 (Hurley et al., 2007). The 2D [1H,15N] HSQC/TROSY spectra of unbound 15N-labeled I-2 and 15N,2H-labeled I-2 in complex with unlabeled PP1 are shown in Figure 3. Chemical shift changes between unbound I-2 and PP1-bound I-2 were limited to residues that are present in the PP1:I-2 crystal structure. Signals of regions with no electron density in the crystal structure retained the characteristic traits of disordered proteins, including very narrow line-widths typical for fast reorientation in solution. This strongly suggests that regions not observed in the PP1:I-2 crystal structure are highly similar in the free and bound states of I-2. Therefore, we used ENSEMBLE to determine a model of the full PP1:I-2 complex using the assumption that experimental restraints from the free state of I-2 could be used to describe the disordered regions of PP1- bound I-2.
Figure 3.
PP1:I-2 NMR study. (a) 2D [1H,15N] HSQC/TROSY spectra of unbound 15N-labeled I-29–164 (black) and 15N,2H-labeled I-29–164 in complex with unlabeled PP1 (red). Spectra were aligned by overlaying the peaks belonging to the Nε1 of Trp46. (b) Chemical shift changes in I-29–164 upon addition of PP1 are small and are primarily localized to regions observed in the crystal structure of the bound complex. The chemical shift differences (Δδ) between peaks in unbound and bound spectra were calculated as .
The resulting PP1:I-2 complex model is shown in Figure 4A. In Figure 4B, the most extended conformation from the model is shown to fit into the PP1:I-2 crystallographic unit cell, providing substantial support for this model. If I-2 adopted a more compact conformation upon PP1 binding, our extended model would be unlikely to fit into the unit cell. The most remarkable feature of this complex is the long loop connecting residues 55–127 (Figure 4A). Although this loop is somewhat heterogeneous, with varying conformations between the different ensemble conformers, a significant fraction of structures adopt a distinct loop structure. This loop contains residue Thr72 which is essential for the biological function of the PP1:I-2 complex, as phosphorylation of Thr72 by GSK-3β reactivates the PP1:I-2 complex (Jurgensen et al., 1984). Interestingly, this loop also contains a transiently populated α-helix (residues 97–105, ~35% populated). Finally, although most conformers show this loop region extending away from PP1, several conformers show the loop closer to PP1, indicating that there may be transient interactions between this loop and PP1which potentially adding stability to the overall complex.
Figure 4.
The PP1:I-2 complex. (a) PP1 is shown in a grey surface representation while I-2 is shown in various colors: the region observed in the PP1:I-2 crystal structure is shown in light blue while the three disordered regions of I-2, residues 6–10, 17–41 and 55–127, are colored orange, green and red, respectively. Thr72 is shown in black. (b) Fitting of the PP1:I-2 complex model into the crystallographic unit cell. The most extended conformer of the PP1:I-2 complex model was overlaid with the PP1:I-2 crystal structure, and unit cell information from the crystal structure was applied to the complex model. Symmetry mates were then added to determine whether the PP1:I-2 model fits into the crystallographic unit cell. (c) Fractional contact plots for the unbound I-2 ensembles (top left) and the PP1-bound I-2 ensemble model (bottom right). The color scale is the same as in Figure 2.
The PP1:I-2 complex: assessment using SAXS measurements
To experimentally validate the PP1:I-2 complex ensemble model we utilized small angle X-ray scattering (SAXS) measurements, which report on the overall size and shape of a protein in solution. Using the Guinier approximation of four independent SAXS samples a radius of gyration (Rg) of 29.8 ± 0.4 Å was calculated for PP1:I-2 (Figure 5A, all measurement statistics are reported in Table 3). This is in excellent agreement with the average Rg (28.5 ± 1.6 Å) for the nineteen calculated ensemble conformations of PP1:I-2. Furthermore, theoretical scattering curves were generated for each ensemble model, which fit the experimental scattering data well with an average χ of 2.1 ± 0.9 (Figure 5B). Finally, a molecular envelope of the PP1:I-2 complex was calculated using GASBOR (Svergun et al., 2001) by averaging sixteen independent ab initio models (Svergun et al., 2001). These models represent the scattering data well with an overall fit of χ = 1.22 ± 0.02 (Figure S8). The models were generated with a maximum length of 100 Å as determined by the analysis of the pair-distance distribution function (Figure S8). The individually calculated ab initio models were averaged and the resulting envelope was aligned with the nineteen ensemble models of PP1:I-2. The envelope and ensemble models are visually and statistically similar, aligning with an average normalized spatial discrepancy (NSD) of 1.50 ± 0.14 (Figure 5C) (Kozin and Svergun, 2001). The calculated PP1:I-2 envelope represents the largest population of the ensemble models with the distinct loop structure of I-2 extended away from the center of PP1. Although the minor populations deviate slightly from the overall envelope, this is likely due to the averaging of SAXS data in solution. Thus the SAXS data supports the correctness of the overall ensemble model of the PP1:I-2 complex.
Figure 5.
SAXS data and analysis for the PP1:I-2 complex. (a) The Guinier region of the scattering curve for 0.8 mg/ml and 0.5 mg/ml PP1:I-2. (b) Average theoretical scattering curve for the ensemble models (red line) fit to the experimental scattering data for PP1:I-2 (squares). (c) Overlay of the SAXS envelope (surface representation, blue) with the PP1:I-2 ensemble models. PP1 is shown as a cartoon representation in black and I-2 is shown as a cartoon representation in red. See also Figure S8.
Table 3.
Statistics for SAXS analysis of the PP1:I-2 complex. NSDa is the discrepancy between the ab initio models. NSDe is the discrepancy between the envelope and the ensemble models.
| PP1:Inhibitor-2 | |
|---|---|
| Guinier approximation | |
| Rg (Å) | 29.8 ± 0.4 |
| P(r) function calculation | |
| q-range (Å−1) | 0.015–0.300 |
| Rg (Å) | 30.2 |
| Dmax (Å) | 100 |
| Structure modeling | |
| χ | 1.22 ± 0.02 |
| NSDa | 1.13 ± 0.03 |
| NSDe | 1.50 ± 0.14 |
Discussion
Structural properties of the unbound ensembles and their implications for PP1 binding
Two questions are fundamental for the molecular understanding of IDP interactions: 1) Are the conformational propensities of unbound states similar to their bound structures? 2) What is the role of transient secondary and tertiary structure for specific interactions with their targets? In the “conformational selection” model of binding, the IDP adopts the bound-state conformation in its free state at a limited population (Weber, 1972). Only these pre-populated binding-competent conformations will interact and thus shift the equilibrium towards the bound state. In contrast, in the “induced fit” model, the bound conformation only becomes energetically accessible in the presence of the binding partner (Koshland, 1958). Current evidence suggests that both induced fit and conformational selection mechanisms are important for IDP function (Sugase et al., 2007). Certainly the interaction of any IDP undergoing a large folding-upon-binding transition must involve an induced-fit component, as the probability of all elements of the bound-state structure occurring simultaneously with the correct 3D geometry seems low. However, this does not exclude conformational selection, as individual elements of secondary or tertiary structure might be fractionally populated and selected for during the binding process.
The most remarkable feature of unbound I-29–164 is the presence of a highly populated α-helix (residues 130–142) that is also observed in the PP1:I-2 complex. This α-helix is vital for PP1 inhibition, as it folds over and blocks the PP1 active site. The high population of this helix suggests it is likely to be critical for the association with PP1. A C-terminal adjacent helix (residues 153–159) is ~10% populated in the unbound state and primarily folds upon binding to PP1, possibly mediated by the binding of the first I-2 helix. Additional secondary structural elements observed in the I-2-bound state, including an α-helix (residues 47–54), are lesser populated in the unbound state and are thus induced upon binding. In addition, the transient intramolecular contacts of unbound I-29–164 show no significant resemblance to I-2 in the complex, although many contacts are in regions that directly interact with PP1. Therefore, the initial binding of the C-terminal helix may initiate the release of many of these intramolecular contacts in unbound I-2. In particular, the region surrounding Trp46, which includes the RVxF motif, may become more accessible, thus permitting complex formation.
Spinophilin417–494 contains a ~30% populated α-helix in the unbound state near the C-terminus that becomes fully formed upon PP1-binding. Furthermore, a region corresponding to the second β-strand (residues 456–461) shows a significant peak in β-strand-like Ramachandran angles, suggesting it transiently populates conformations that are optimal for PP1 binding. Finally, there is evidence for low populated tertiary contacts occurring in spinophilin417–494 that resemble its structure in the complex. Thus, many of the intramolecular structural features observed in PP1-bound spinophilin are also detected in unbound spinophilin, suggesting that conformational selection is likely important for PP1 binding.
Although no structure of the PP1:DARPP-32 complex has been reported, it is likely that transient structure present in the free state of DARPP-32 is important for this interaction. In particular, the fractionally formed α-helices probably play an important role in PP1 binding, just as α-helices identified in the ensembles of both I-2 and spinophilin correspond closely to their PP1-bound crystal structures. The ~30% populated α-helix lies between the RVxF motif of DARPP-32 (KKIQF, residues 7–11) and Thr34. Phosphorylation of Thr34 by PKA activates DARPP-32 as a PP1 inhibitor (Hemmings et al., 1984). Just C-terminal to this transient α-helix is a stretch of basic residues (RRRRPT, residues 29–34), which complement the acidic groove of PP1. The acidic groove, one of three predicted substrate binding grooves on the PP1 surface, connects the RVxF motif binding site and the active site of PP1. Thus, binding of the helix and the C-terminal positively charged residues is likely important in the positioning of Thr34 in the active site of PP1 for inhibition.
Biological implications of the PP1:I-2 complex model
The ensemble model of the PP1:I-2 complex provides molecular insight into the dynamic regions of I-2 that are not observed in the crystal structure, but which fulfill essential biological functions. The most notable structural feature of the complex is the 73 amino acid loop that connects I-2 residues 55 to 127. This loop contains Thr72, a GSK-3β phosphorylation site, which is important for the formation of the reactivated ATPMg-dependent PP1 phosphatase complex (i.e. phosphorylation of Thr72 reverses the inhibition of PP1 by I-2 without release of I-2 from PP1) (Jurgensen et al., 1984). Phosphorylation of Thr72 on I-2, when bound to PP1, has been shown to increase the fluorescence anisotropy of a label covalently linked to a cysteine engineered into residue 129 of I-2 (Picking et al., 1991). This suggests a conformational change in PP1-bound I-2 upon phosphorylation at Thr72, corresponding change in unbound I-2 was not seen. In agreement with this data, we observe contacts between residues near 50–70 and 130 in PP1-bound I-2 (Figure 4C). These interactions only exist in the PP1-bound form of I-2 and are likely required for the structural rearrangement induced by phosphorylation.
The flexibility of this loop likely guides the intricate regulation of PP1 by I-2 for at least three reasons. First, phosphorylation of Ser86 on I-2 has been shown to prime I-2 for GSK-3β phosphorylation at Thr72 (Park et al., 1994). However, the thirteen residue linker between these two residues is much larger than the typical three residue linker between phosphorylation sites for GSK-3β recognition (Park and DePaoli-Roach, 1994). Interestingly, in conformers that adopt the extended loop structure of our PP1:I-2 model, Ser86 and Thr72 are in close proximity (average distance of 16.4 Å ± 5.8 Å). This is in contrast to the second set of conformers, where the loop is folded back onto PP1. In this set, the average distance between these two residues is 30.6 Å ± 8.5 Å. Thus, the positioning of the loop allows for regulation of the phosphorylation state of Thr72 on I-2, and thus regulation of the activation state of PP1. Second, Thr72 must be accessible for phosphorylation by GSK-3β as well as for subsequent dephosphorylation by PP1 in order to reactivate the complex (Jurgensen et al., 1984). In the inactivated PP1:I-2 complex, Tyr147 of I-2 is placed in the active site of PP1 for inhibition (Hurley et al., 2007). Therefore, both Tyr147 and Thr72 must be able to reach the active site of PP1, which is easily accomplished with the long, flexible loop of PP1-bound I-2. Finally, the extended loop structure of PP1-bound I-2 creates an additional protein interaction surface on I-2 that is extended away from PP1. This loop contains a transient α-helical region from residues 97–105 (~35% populated). Given the tendency of transient helices in IDPs to serve as binding sites, it seems quite likely that this α-helix is important for I-2 function. Its accessible position would facilitate interactions of the PP1:I2 holoenzyme with other proteins. Alternatively, this α-helix might facilitate the binding of I-2 to other proteins, as I-2 has previously been shown to activate Aurora-A kinase (Satinover et al., 2004) and regulate the activity of the prolyl isomerase Pin1 (Li et al., 2008).
A structural continuum from folded to disordered states
There is increasing evidence that proteins do not exist in either discrete random coil or folded states – rather, they exist in a continuum of possible states ranging from folded to disordered. The ensembles calculated in this study convincingly demonstrate this continuum: disordered states can have varying amounts of secondary and tertiary structure, ranging from highly disordered to the almost fully formed α-helix in I-2. In addition, it is now understood that complete folding-upon-binding is only one possible scenario for IDPs. An IDP can also retain substantial flexibility upon complex formation (Mittag et al., 2009; Permyakov et al., 2003; Sigalov et al., 2008; Sigalov et al., 2007; Tompa and Fuxreiter, 2008). The model of the PP1:I-2 complex presents a unique structural picture of a protein that undergoes partial folding but remains largely disordered upon complex formation. In fact, dynamic complexes such as PP1:I-2 are likely far more common than might be expected based upon current experimental evidence, given that X-ray crystallography is heavily biased against disorder and flexibility. Structurally characterizing these highly dynamic complexes is not simple, but the presented approach of combining various experimental measurements with previously known structural information should be applicable to numerous systems. Thus, these combined experimental and computational ensemble-modeling techniques will likely prove to be extremely valuable for improving our understanding of the structural properties and the biological functions of these dynamic complexes.
Experimental Procedures
15N-labeled and 2H,15N-labeled I-29–164 and PP1 were produced as described (Dancheck et al., 2008; Kelker et al., 2009). For the purification of the PP1:I-2 complex, cells were lysed by high-pressure homogenization (Avestin C-3 Emulsiflex). The soluble protein fraction was loaded onto a Ni-NTA column (Invitrogen). Purified I-29–164 was incubated on the PP1-bound Ni-NTA beads, and the complex was eluted with imidazole. The eluate was injected onto a Superdex 75 26/60 size exclusion column (GE Healthcare) equilibrated with 20 mM Tris (pH 7.5), 50 mM NaCl, 0.5 mM TCEP. Fractions containing the pure protein complex, as confirmed by SDS-PAGE, were pooled and concentrated to 60 μM for subsequent NMR measurements. The 2D [1H,15N]-HSQC spectrum of unbound I-29–164 was measured at 298 K on a Bruker Avance II 500 MHz spectrometer; the 2D [1H,15N]-TROSY spectrum of the PP1:I-2 complex was measured at 298 K on a Bruker Avance II 800 MHz spectrometer. Both spectrometers were equipped with TCI HCN z-gradient cryoprobes. The NMR spectra were processed with Topspin 1.3 (Bruker) and analyzed with the CARA software package (www.nmr.ch).
Ensemble models of intrinsically disordered I-29–164, spinophilin417–494 and DARPP-321–118 were calculated with ENSEMBLE using a nearly identical protocol to that previously described for the drkN SH3 domain unfolded state (Marsh and Forman-Kay, 2009) and the experimental restraints given in Table 1. The model of the PP1:I-2 complex was calculated using ENSEMBLE with modifications to the previous protocol. The PP1 structure was kept identical to the crystallographic model and all refinement was performed on I-29–164. The regions of I-2 observed in the crystal structure were restrained to be nearly identical to the crystal structure (< 2 Å CαCα RMSD). Chemical shift and 15N R2 restraints from unbound I-2 were used for the disordered regions not observed in the bound complex. All unbound state PRE restraints were used except those between pairs of residues both observed in the bound crystal structure. The PRE restraints were weighted according to the chemical shift changes that were observed upon binding PP1, whereby residues that had large chemical shift changes had their PRE restraints significantly underweighted. In addition, the maximum energy penalty from any single restraint was limited; this prevented individual poorly fit restraints from dominating the ENSEMBLE calculations. One of the final unbound I-29–164 ensembles was used as the starting ensemble and, rather than using the simplest-ensemble approach, the number of structures was kept constant at 19. The calculation proceeded until the chemical shift and 15N R2 restraints were entirely fit and then continued to fit the PRE restraints as optimal as possible. This step-wise procedure was chosen as it was expected that the unbound-state PRE restraints of I-2 would have a lower fit in the bound form of I-2.
The PP1:I-2 complex sample for SAXS data collection was produced within 24 hours of data acquisition and stored at 4°C. Prior to scattering experiments the sample was filtered through a 0.02 μm filter (Whatman). Synchrotron X-ray scattering data were collected at the National Synchrotron Light Source (NSLS) beamline X9. Small angle x-ray scattering (SAXS) data were collected using a MarCCD 165 located at 3.4 m distance from the sample. Wide angle x-ray scattering (WAXS) data were collected simultaneously with SAXS data using a Photonic Science CCD located at 0.47 m from the sample. 20 μl of sample was continuously pushed through a 1 mm diameter capillary for either 180 or 360 s of total measurement time and exposed to a 400 × 200 microns X-ray beam. Scattering data for the complex was collected at 0.5 mg/ml and 0.8 mg/ml PP1:I-2 concentrations. Normalization for beam intensity, buffer subtraction and merging of the data from both detectors were carried out using PRIMUS (Konarev et al., 2003). A Guinier approximation, I(q) = I(0)exp(-q2Rg2/3), where a plot of I(q) and q2 is linear for q<1.3/Rg, was performed on four independent scattering trials and averaged to determine the radius of gyration (Guinier, 1939). HYDROPRO was used to calculate the Rg for the individual ensemble models for comparison (Garcia De La Torre et al., 2000). GNOM was used to determine the pair distribution function [P(r)] and maximum particle dimension (Dmax) (Svergun, 1992). Sixteen independent ab initio models were calculated using GASBOR (Svergun et al., 2001). The models were aligned and averaged using DAMAVER (Volkov and Svergun, 2003). The averaged ab initio envelope was aligned with nineteen ensemble models for the PP1:I-2 complex using SUPCOMB (Kozin and Svergun, 2001). The normalized spatial discrepancy (NSD) values were averaged and used to represent the overall fit of the ENSEMBLE model to the ab initio envelope. Theoretical scattering curves were calculated for each of the ensemble structures using CRYSOL (Svergun et al., 1995). The scattering curves and χ values were averaged to represent the overall fit of the theoretical scattering curve to the experimental data.
Supplementary Material
Acknowledgments
This material is based upon work supported by a Natural Sciences and Engineering Research Council of Canada fellowship to J.A.M and a National Science Foundation Graduate Research Fellowship to B.D. The authors thank Dr. Lin Yang for his support at beamline X9. The project described was supported by Grant R01NS056128 from the National Institute of Neurological Disorders and Stroke to W.P and funding from the Canadian Institutes for Health Research to J.D.F.-K. 800 MHz NMR data were recorded at Brandeis University (NIH S10-RR017269). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. W.P. is the Manning Assistant Professor for Medical Science at Brown University.
Footnotes
Supplementary information is available on the Structure website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH, Gleeson JG. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129:579–591. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. Bioessays. 2002;24:371–381. doi: 10.1002/bies.10069. [DOI] [PubMed] [Google Scholar]
- Chan HS, Dill KA. Interchain loops in polymers: effects of excluded volume. J Chem Phys. 1989;90:492–509. [Google Scholar]
- Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308:1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Csizmok V, Bokor M, Banki P, Klement E, Medzihradszky KF, Friedrich P, Tompa K, Tompa P. Primary contact sites in intrinsically unstructured proteins: the case of calpastatin and microtubule-associated protein 2. Biochemistry. 2005;44:3955–3964. doi: 10.1021/bi047817f. [DOI] [PubMed] [Google Scholar]
- Dancheck B, Nairn AC, Peti W. Detailed structural characterization of unbound protein phosphatase 1 inhibitors. Biochemistry. 2008;47:12346–12356. doi: 10.1021/bi801308y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Feldman HJ, Hogue CW. A fast method to sample real protein conformational space. Proteins. 2000;39:112–131. [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A. La diffraction des rayons X aus tres petits angles: application a l’etude de phenomenes ultramicroscopiques. Ann Phys. 1939;12:161–237. [Google Scholar]
- Hemmings HC, Jr, Greengard P, Tung HY, Cohen P. DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- Hsieh-Wilson LC, Allen PB, Watanabe T, Nairn AC, Greengard P. Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry. 1999;38:4365–4373. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Jurgensen S, Shacter E, Huang CY, Chock PB, Yang SD, Vandenheede JR, Merlevede W. On the mechanism of activation of the ATP X Mg(II)-dependent phosphoprotein phosphatase by kinase FA. J Biol Chem. 1984;259:5864–5870. [PubMed] [Google Scholar]
- Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009;385:11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- Li M, Satinover DL, Brautigan DL. Phosphorylation and functions of inhibitor-2 family of proteins. Biochemistry. 2007;46:2380–2389. doi: 10.1021/bi602369m. [DOI] [PubMed] [Google Scholar]
- Li M, Stukenberg PT, Brautigan DL. Binding of phosphatase inhibitor-2 to prolyl isomerase Pin1 modifies specificity for mitotic phosphoproteins. Biochemistry. 2008;47:292–300. doi: 10.1021/bi701819k. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Forman-Kay JD. Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J Mol Biol. 2009;391:359–374. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Neale C, Jack FE, Choy WY, Lee AY, Crowhurst KA, Forman-Kay JD. Improved structural characterizations of the drkN SH3 domain unfolded state suggest a compact ensemble with native-like and non-native structure. J Mol Biol. 2007;367:1494–1510. doi: 10.1016/j.jmb.2007.01.038. [DOI] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2009;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Morishita W, Connor JH, Xia H, Quinlan EM, Shenolikar S, Malenka RC. Regulation of synaptic strength by protein phosphatase 1. Neuron. 2001;32:1133–1148. doi: 10.1016/s0896-6273(01)00554-2. [DOI] [PubMed] [Google Scholar]
- Nairn AC, Svenningsson P, Nishi A, Fisone G, Girault JA, Greengard P. The role of DARPP-32 in the actions of drugs of abuse. Neuropharmacology. 2004;47(Suppl 1):14–23. doi: 10.1016/j.neuropharm.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Park IK, DePaoli-Roach AA. Domains of phosphatase inhibitor-2 involved in the control of the ATP-Mg-dependent protein phosphatase. J Biol Chem. 1994;269:28919–28928. [PubMed] [Google Scholar]
- Park IK, Roach P, Bondor J, Fox SP, DePaoli-Roach AA. Molecular mechanism of the synergistic phosphorylation of phosphatase inhibitor-2. Cloning, expression, and site-directed mutagenesis of inhibitor-2. J Biol Chem. 1994;269:944–954. [PubMed] [Google Scholar]
- Permyakov SE, Millett IS, Doniach S, Permyakov EA, Uversky VN. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- Picking WD, Kudlicki W, Kramer G, Hardesty B, Vandenheede JR, Merlevede W, Park IK, DePaoli-Roach A. Fluorescence studies on the interaction of inhibitor 2 and okadaic acid with the catalytic subunit of type 1 phosphoprotein phosphatases. Biochemistry. 1991;30:10280–10287. doi: 10.1021/bi00106a028. [DOI] [PubMed] [Google Scholar]
- Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs Protein Phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrouilhe D, di Tommaso A, Metaye T, Ladeveze V. Spinophilin: from partners to functions. Biochimie. 2006;88:1099–1113. doi: 10.1016/j.biochi.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Satinover DL, Leach CA, Stukenberg PT, Brautigan DL. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc Natl Acad Sci U S A. 2004;101:8625–8630. doi: 10.1073/pnas.0402966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalov AB, Kim WM, Saline M, Stern LJ. The intrinsically disordered cytoplasmic domain of the T cell receptor zeta chain binds to the nef protein of simian immunodeficiency virus without a disorder-to-order transition. Biochemistry. 2008;47:12942–12944. doi: 10.1021/bi801602p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect transform methods using perceptual criteria. J Appl Crystallogr. 1992;26:495–503. [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002a;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002b;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- Wang W, Stukenberg PT, Brautigan DL. Phosphatase inhibitor-2 balances protein phosphatase 1 and aurora B kinase for chromosome segregation and cytokinesis in human retinal epithelial cells. Mol Biol Cell. 2008;19:4852–4862. doi: 10.1091/mbc.E08-05-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Huang HB, Horiuchi A, da Cruze Silva EF, Hsieh-Wilson L, Allen PB, Shenolikar S, Greengard P, Nairn AC. Protein phosphatase 1 regulation by inhibitors and targeting subunits. Proc Natl Acad Sci U S A. 2001;98:3080–3085. doi: 10.1073/pnas.051003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.