Abstract
Glypican 3 (GPC3) belongs to a family of glycosylphosphatidylinositol-anchored, cell-surface heparan sulfate proteoglycans. GPC3 is over-expressed in hepatocellular carcinomas (HCC). Loss-of-function mutations of GPC3 result in the Simpson-Golabi-Behmel syndrome, an X-linked disorder characterized by overgrowth of multiple organs, including liver. Our previous study showed that GPC3 plays a negative regulatory role in hepatocyte proliferation, and this effect may involve CD81, a cell membrane tetraspanin. To further investigate GPC3 in vivo, we engineered transgenic (TG) mice over-expressing GPC3 in liver under the control of the albumin promoter. GPC3 TG mice with hepatocyte-targeted over-expressed GPC3 develop normally compared with their non-transgenic littermates, but have a suppressed rate of hepatocyte proliferation and liver regeneration after partial hepatectomy (PHx). Moreover, gene array analysis revealed a series of changes in the gene expression profiles in TG mice, both in normal mice and during liver regeneration. In unoperated GPC3 TG mice there was over-expression of Runx3 (7.6 fold), C/EBPα (2.5 fold) and GABA A Receptor (2.9 fold) and Wnt7b (2.8 fold). There was down-regulation of IGF BP1 (8.4 fold), Rab2 (5.6 fold), beta Catenin (1.7 fold), TGF beta 1 (3.1 fold), Nodal (1.8 fold) and Yap (1.4 fold). Changes after hepatectomy included decreased expression in several cell cycle related genes.
Conclusion
Our results indicate that in GPC3 transgenic mice, hepatocyte over-expression of GPC3 suppresses hepatocyte proliferation and liver regeneration, and alters gene expression profiles in GPC3 TG mice, in which potential cell cycle related proteins and multiple other pathways are involved and affected.
Keywords: glypican 3, liver regeneration, partial hepatectomy, gene array, cell cycle
GPC3 is a heparan sulfate proteoglycan (HSPG) that is bound to the cell surface through a glycosylphosphatidylinositol (GPI) anchor.1 Expression of GPC3 is reported to be high during embryogenesis and organogenesis.2,3 In adults, GPC3 can only be detected in a limited number of tissues.4 A loss-of-function mutation in the GPC3 gene causes Simpson-Golabi-Behmel syndrome (SGBS), which is characterized by pre- and post-natal overgrowth, increased risk of embryonic tumors during early childhood, and numerous visceral and skeletal anomalies.5,6 The involvement of GPC3 in SGBS was confirmed by Filmus et al. using GPC3-deficient mice (GPC3−/−) which display some of the phenotypic features of SGBS.7 Since the functional loss of GPC3 leads to enlarged organs (including liver) in humans as well as in mice, it is reasonable to speculate that GPC3 may play a negative role in regulation of organ growth. On the other hand, previous studies have shown that GPC3 is highly up-regulated in hepatocellular carcinoma (HCC) and hepatoblastoma, but not in normal liver or tissue adjacent to tumors.8 These findings suggest that GPC3 plays an important and diverse regulatory role during normal liver growth as well as tumorigenesis, but the exact manner in which GPC3 functions is still not well characterized.
Liver is an organ with great regenerative potential.9,10 In the mouse 2/3 partial hepatectomy (PHx) model, specific mouse liver lobes that account for about 2/3 of the total mass are removed and the residual lobes enlarge to make up for the loss of mass. 11 Following PHx, hepatocytes enter into the cell cycle from their habitual quiescent phase and proliferate to restore normal hepatic mass and hepatic functional capacity.10 The whole process lasts about one week in mice, during which the hepatocytes divide first, at about 36–48 hours post-PHx, followed by expansion of non-parenchymal cells (NPC).10
In our previous study, we showed that GPC3 plays an overall growth inhibitory role in hepatocyte proliferation in primary cultures.12 Our previous study also demonstrated that GPC3 mRNA and protein increase in a time frame that coincides with the termination of proliferative activities of either hepatocytes (day 2 after PHx and day 8–12 in culture) or NPCs (day 5–6 after PHx) in rats.12 In vitro studies showed that hepatocyte growth was promoted when GPC3 expression was blocked using antisense Morpholino oligonucleotides. The yeast-two hybrid assay revealed that GPC3 interacts with CD81, a cell membrane tetraspanin,13,14 which was further confirmed by co-immunofluorescence and co-immunoprecipitation studies. CD81 mRNA and protein increase in grossly the same time frame after PHx. The co-localization of GPC3 and CD81 at day 2 and 6 after PHx indicated the potential for an important regulatory interaction between the two proteins.
While the evidence for GPC3 as growth suppressor is derived primarily from loss-of-function models in rodents and humans, the effects of GPC3 as a positive growth suppressor in rodents with transgenic organ targeted over-expression have not been studied. To further study the role and mode of action of GPC3 during liver regeneration, we generated GPC3 transgenic (TG) mice under the control of the mouse albumin promoter/enhancer, in order to over-express GPC3 specifically in mouse hepatocytes. The GPC3 TG mice appeared phenotypically normal and were indistinguishable from their non-transgenic littermates. Further investigation by western blotting and immunofluoresence staining revealed an up-regulation in GPC3 levels in the liver of these TG mice. We found that after 2/3 PHx, liver regeneration and hepatocyte proliferation was suppressed in these TG mice, as shown by assessment of hepatocyte proliferation and liver weight growth curve. Gene array analysis demonstrated an altered gene expression profile in the TG mice compared with their wild-type (WT) littermates. A comparison of gene expression profiles at various time points after PHx revealed a panel of cell cycle related genes and growth arrest genes which were either up- or down-regulated. Western blotting confirmed the changes of key factors in specific pathways. The overall findings suggest that GPC3 is indeed a suppressor of hepatocyte growth and that it may play a role in regulation of the processes resulting in termination of liver regeneration.
Materials and Methods
Results
Generation of Transgenic Mice Over-expressing GPC3 in the Liver
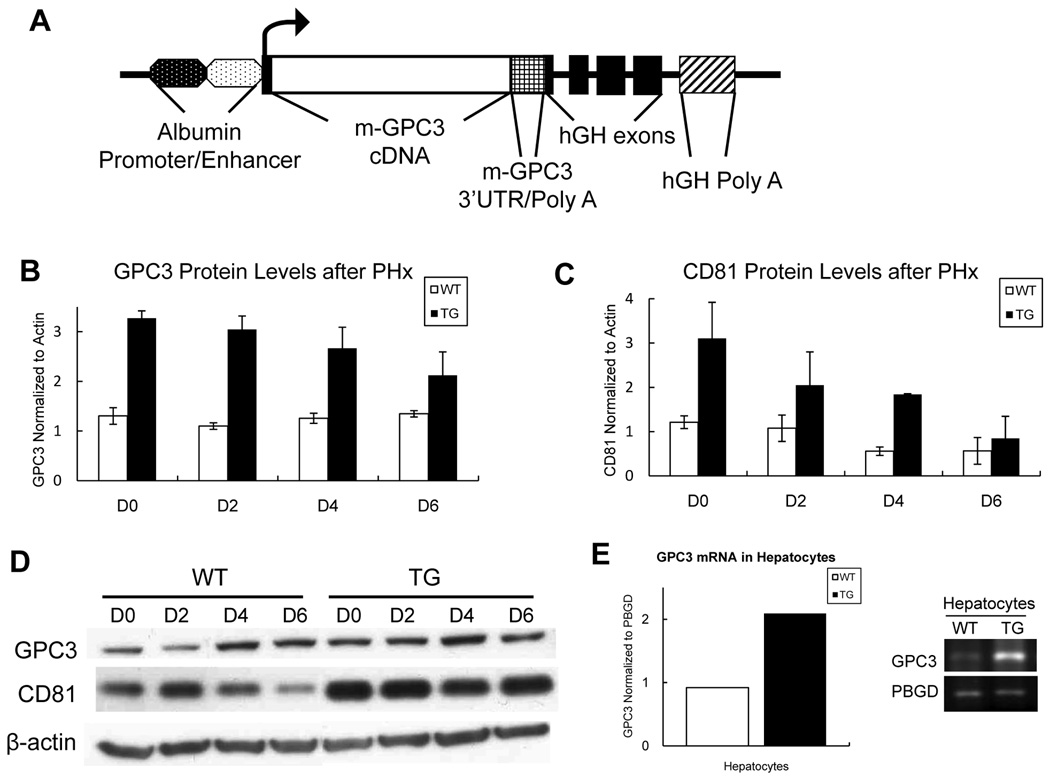
To generate GPC3 transgenic mice, we designed and cloned a transgenic expression construct containing the 1.8 kb coding region of GPC3 cDNA under the transcriptional control of the mouse albumin promoter/enhancer (Fig. 1A). The insertion of GPC3 transgene was confirmed by PCR and Southern blotting in GPC3 TG mice (data not shown). To examine the changes in the protein levels of GPC3, total liver protein extract from a minimum of three mice per time point was tested and analyzed individually (per mouse) using western blotting in TG mice and their WT littermates. As expected, there was increased GPC3 protein levels in TG mice at all times after PHx compared with their non-transgenic littermates (Fig. 1B), with a slight downtrend after PHx (Fig. 1B). Of interest, CD81 protein levels were also significantly up-regulated in GPC3 TG mice in general, but also with a downtrend during liver regeneration process (Fig. 1C). The pooled protein samples from each group were also tested by western blotting, showing the same results (Fig.1D). RT-PCR results quantitated using ImageJ software demonstrated an increase in GPC3 expression in hepatocytes of TG mice (Fig. 1E). Using immunofluorescence, we saw an increase in GPC3 protein on hepatocyte membrane as well as in cytoplasm in TG mice compared with their WT littermates (Fig. 2A).
Fig.1.
The construct utilized for generation of the GPC3 transgene and GPC3 expression. (A) Schematic representation of the GPC3 transgene consisting of the 1.8 kb mouse GPC3 cDNA (open box) with GPC3 3’UTR/Poly A (square mesh box) inserted into the first exon of the human growth hormone gene (black boxes) controlled by the mouse albumin enhancer/promoter (dotted ovals), and possessing a human growth hormone Poly A site (cross box). (B) and (C): Total liver protein was extracted from a minimum of three mice per time point and GPC3 and CD81 protein levels were individually examined by western blotting. The signal intensity of each protein band was quantitated using ImageJ software and was statistically analyzed. Each bar signifies the mean and standard error of measurements from minimally three separate animals. Each measurement involved intensity of GPC3 and CD81 band normalized to β-actin. TG: GPC3 transgenic mice. WT: FVB wild-type littermate mice. (D) Western blots showing GPC3 and CD81 levels in pooled samples during liver regeneration in WT and TG mice. (E) RT-PCR of GPC3 mRNA in hepatocytes in WT and TG mice. The results were normalized to PBGD expression and quantitated using ImageJ software.
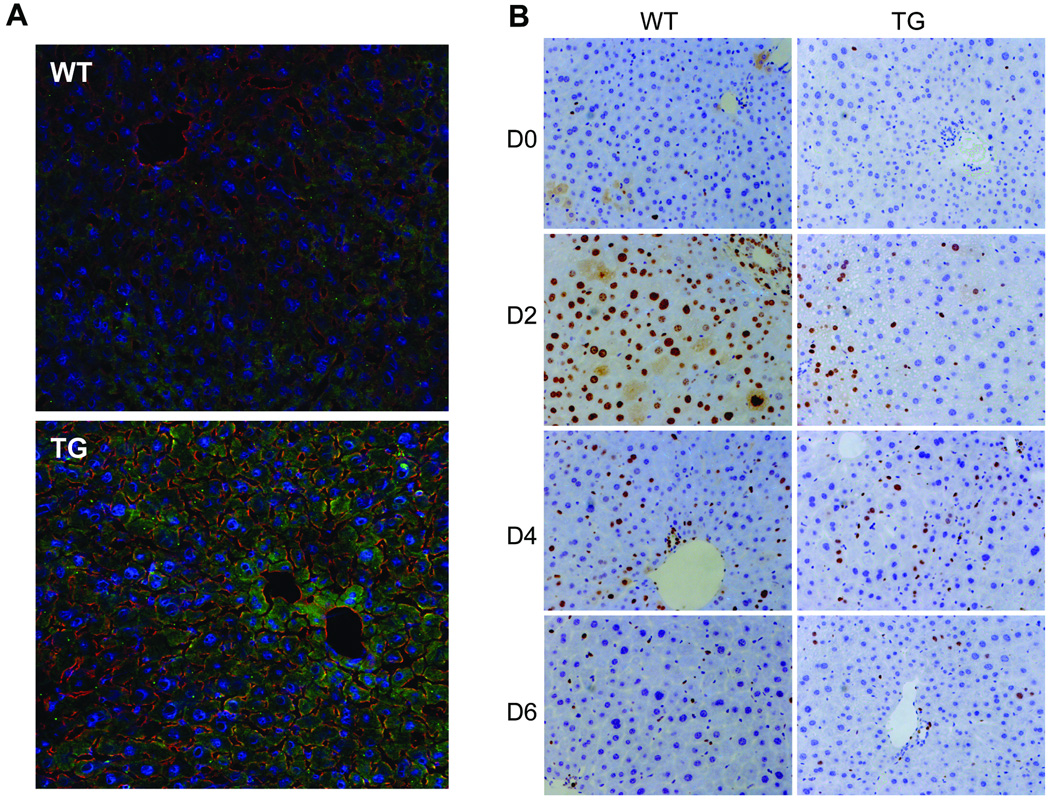
Fig.2.
Immunofluoresence staining of GPC3 and immunohistochemistry staining of Ki67 in TG and WT mice. (A) Olympus Fluoview 500 Confocal Microscope was used to visualize the images of the liver in TG and WT mice. GPC3 (green), CD81 (red) and DRAQ5 nucleus staining (blue) were shown in the merged images with a magnification of 200×. TG mice showed a significant increased level of GPC3. (B) Immunohistochemistry staining and Ki67 (brown), a proliferation marker, in paraffin sections of TG and WT mice livers after PHx. The Ki67 positive rate was suppressed in TG mice at day 2 after PHx compared with WT mice.
Growth Suppression after PHx in GPC3 TG Mice
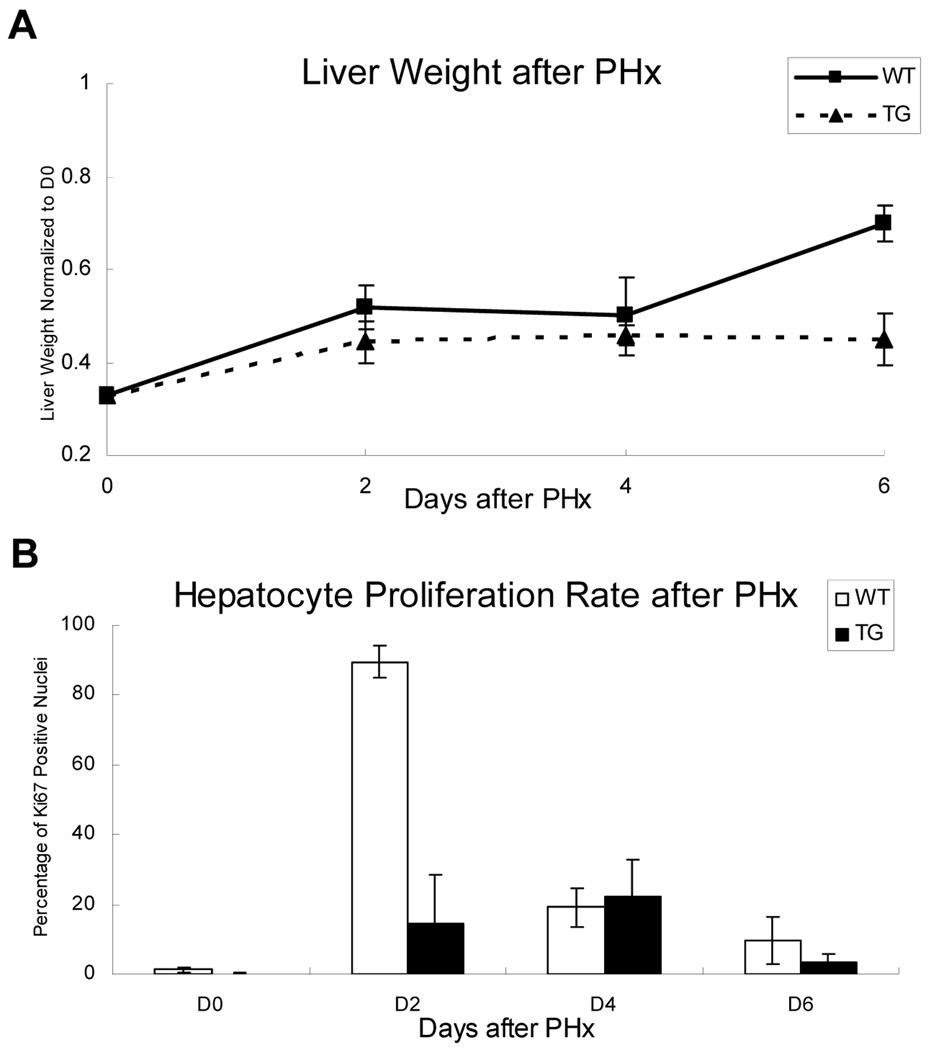
To further study the function and role of GPC3 in vivo, 2/3 PHx were performed in both GPC3 TG and WT male mice (littermates), and liver tissue samples were collected at day 2, 4, 6 after hepatectomy. Liver weight was assessed by normalizing to the day 0 total liver weight in each group. The data are presented in Figure 3A. There was no significant difference of the total (pre-hepatectomy) liver weight at day 0 comparing TG and WT mice (data not shown). At day 6 after PHx, the liver weight of TG mice was significantly lower than WT mice (Fig. 3A). Ki67 immunohistochemistry results (Fig. 2B) showed that in WT mice there was abundant hepatocyte proliferation at 2 days after PHx, demonstrated by the percentage of Ki67 positive nuclei. In GPC3 TG mice, the hepatocyte proliferation was significantly suppressed at day 2 post-PHx, with no significant difference at day 4 and 6 compared with WT (Fig. 3B).
Fig. 3.
Suppression of liver regeneration and hepatocyte proliferation in GPC3 TG mice. (A) Statistic analysis of liver weight of TG and WT mice after PHx, normalized to day 0 total (pre-hepatectomy) liver weight. D0 in the graph represents the remaining liver weight after PHx, which is 1/3 of the total liver weight. There were no significant differences in the pre-hepatectomy liver weights at day 0 between TG and WT mice. (B) Percent of Ki67 positive nuclei in hepatocytes of TG and WT mice after PHx. A significant decrease in Ki67 positive nuclei was observed in TG mice at day 2 after PHx.
Gene Expression Profile Alteration in Unoperated TG Mice
Microarray analysis was used to investigate the expression profile and transcriptional regulation in liver tissues of TG and WT mice under normal conditions (unoperated mice). The Mouse Genome 430 2.0 GeneChip array is a single array analyzing the expression level of over 39,000 transcripts and variants, including over 34,000 well-characterized mouse genes (Affymetrix, CA, USA). Direct comparison between TG mice and WT mice expression pattern revealed 44 genes up-regulated and 58 genes down-regulated in TG with > 1.5 fold change (Supplementary Material, Table S1). Among the most interesting are Runx3 (Runt related transcription factor 3, up by 7.6-fold), Jak3 (Janus kinase 3, up by 6.4-fold), Dbp (D site Albumin promoter binding protein, up by 4-fold),Gabaar (GABA-A receptor, up by 2.9-fold), Wnt7b (Wingless-related MMTV integration site 7B, up by 2.8-fold), Cebpα (CCAAT/enhancer-binding protein alpha, up by 2.5-fold) and Egfr (epidermal growth factor receptor, up by 1.6-fold). Most strongly down-regulated genes are Igfbp1 (insulin-like growth factor binding protein 1, down by 8.4-fold), Rab2 (RAS oncogene family, down by 5.6-fold), Egr1 (Early growth response 1, down by 5.3-fold), Tgfb1 (Transforming growth factor beta type I, down by 3.1-fold), Ndl (Nodal, down by 1.8-fold), Catnb (beta catenin, down by 1.7-fold) and Yap (Yes-associated protein, down by 1.4-fold) in GPC3 TG mice. A complete list of genes showing either up- or down-regulation in expression of TG mice can be found in Supplementary Material, Table S1.
Gene Expression Profile Altered after PHx in TG Mice
A comparison of expression profile between TG mice and WT mice after PHx also indicated dysregulation of many genes in GPC3 TG mice during liver regeneration. A panel of important genes known to be involved in hepatocyte cell cycle and growth arrest was examined and analyzed at 2 days after PHx in TG mice compared with their WT littermates, when there was the biggest suppression of hepatocyte proliferation (Supplementary Material, Table S2). Some cell cycle related genes were observed to be down-regulated in TG mice at day 2 post-PHx, such as Ccna1 (Cyclin A1, by 7.6-fold), Ccnm1 (Cyclin M1, by 5.4-fold), Ccnd1 (Cyclin D1, by 1.4-fold), Ccne2 (Cyclin E2, by 1.7-fold), and Gas1 (Growth arrest specific 1, by 1.7-fold). While the up-regulated genes after PHx in TG mice were: Runx3 (by 8-fold), Gadd45b (Growth arrest and DNA-damage-inducible 45 beta, by 3.9-fold), Fgf14 (Fibroblast growth factor 14, by 2-fold) and Tgfb1 (by 1.6-fold). A series of Cdks (Cyclin-dependent kinases), Ccns (Cyclins) as well as Igfbps (Insulin-like growth factor binding proteins) was also observed to be altered at day 2 after PHx in TG mice. Some other important genes that are related to growth and proliferation with less significant changes in TG mice, if any, were also listed for reference. (Supplementary Material, Table S2)
Growth Related Protein Changes after PHx between TG and WT mice
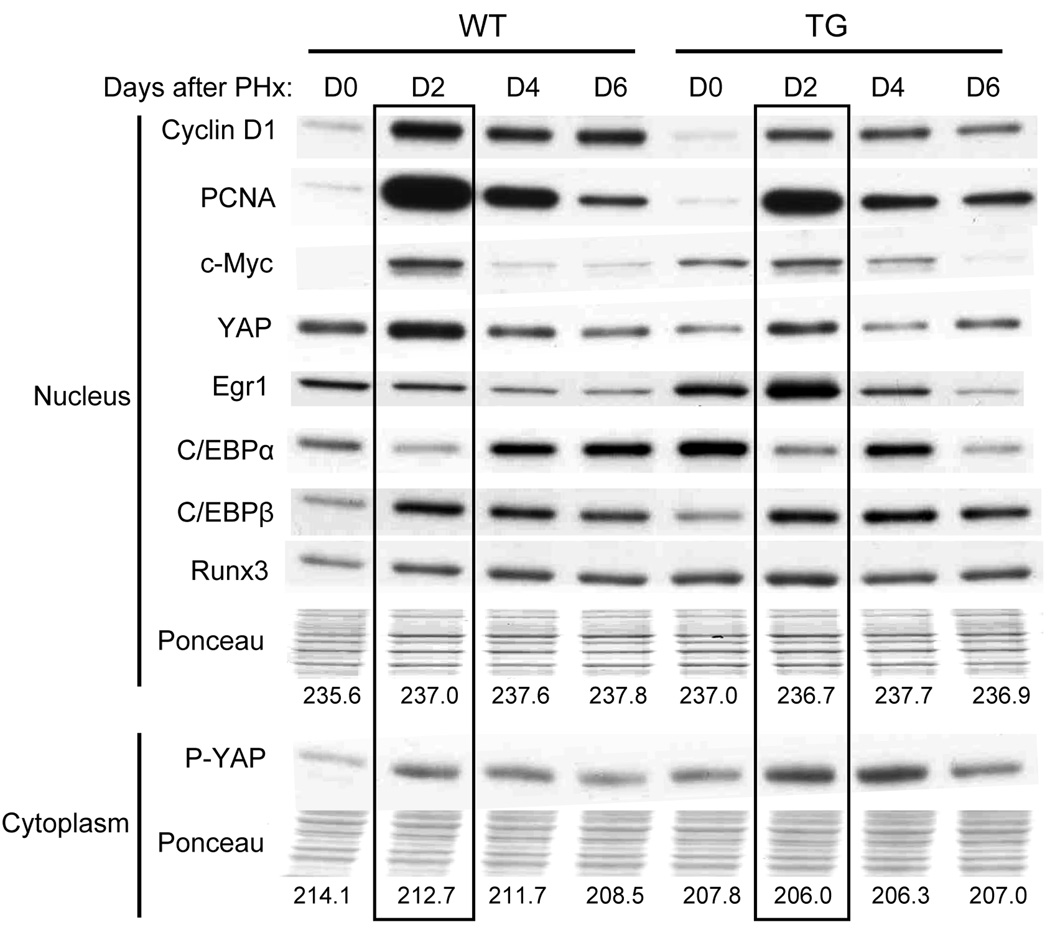
Gene array results (shown above) revealed multiple cell cycle related and growth related genes, the expression of which was altered in GPC3 TG mice after PHx compared to their WT littermates. Western blotting was performed to further investigate the protein levels in some of these genes (Fig. 4). For each time point, the liver samples from a minimum of three mice were pooled and the nucleus and cytoplasmic proteins were separated for analysis by western blotting. Prior to PHx, there was an increase in the protein levels of c-Myc, Egr1, C/EBPα and Runx3 in TG mice compared with WT. Except for Egr1, these findings correlate with the changes seen for these proteins by gene array analysis. After PHx a decrease of levels of some of the proteins in the nucleus was observed at day 2 in GPC3 TG mice, such as Cyclin D1, PCNA, c-Myc and YAP. There was also increased phosphorylated YAP (p-YAP) protein in cytoplasm, which is the inactive form of YAP. C/EBPβ level was increased at day 4 and day 6 after PHx in TG mice. These results coincided with the suppression of liver regeneration and hepatocyte proliferation after PHx.
Fig. 4.
Western blotting results showing changes in protein levels of selected genes compared between GPC3 TG mice and WT mice during liver regeneration. Protein levels in nucleus and cytoplasm of the liver from GPC3 TG and WT mice were examined. Ponceau staining of each protein lane was used as loading control and the average grey value was shown below each column. The protein levels at day 2 after PHx was highlighted in TG and WT mice.
Discussion
Liver regeneration is a complicated process and involves multiple factors and pathways.10 Our previous study suggested that GPC3 plays an inhibitory role during rat liver regeneration and hepatocyte proliferation. The work performed in this study further suggests the effects of GPC3 on hepatocyte growth are primarily inhibitory. Over-expression of GPC3 in hepatocytes is associated with suppression of liver regeneration and concomitant persistent decrease in liver to body weight ratio in the GPC3 transgenic mice. Despite the new evidence that presents growth suppression as one aspect of GPC3 function, the mechanism by which such actions are exerted remains elusive. The overall growth regulatory effects of GPC3 transcend mere growth suppression and may extend to lineage determination, in view of the fact that increase expression of GPC3 is seen in hepatic progenitor cells.15
We have always chosen to use mice of at least 20 weeks of age for our studies related to liver regeneration. Our choice is driven by earlier studies demonstrating that in male mice and rats, hepatocyte nuclei undergo extensive polyploidization, transforming hepatocyte nuclei from 2n to (on average) 8n.16–19 This occurs in the first 15 weeks of age.19 Polyploidization of hepatocyte nuclei and the attendant DNA synthesis interfere with assessment of background DNA synthesis and interpretation of studies associated with liver regeneration. The mice in this study, as with our previous studies,20 were chosen to be minimally at 20 weeks of age.
It is reported that GPC3 is involved in several pathways during development and tumorigenesis, and acts as potential co-receptor to many growth factors.21 To study the pathways that might be altered and affected by over-expression of GPC3 transgene in the liver, we examined and compared the expression profile of TG mice and WT mice during liver regeneration. Our gene array data revealed an alteration of over 100 gene expression in GPC3 TG mice, with GPC3 expression up-regulated by 1.5 fold. Western blotting and immunofluoresence showed an increase of GPC3 protein level in the liver of TG mice. 22,23 Although in TG mice overall higher GPC3 levels were observed compared with WT mice, there was a slight decrease in transgene expression, at 4 and 6 days post-PHx. This might be due to a down-regulation of albumin promoter, which is controlling the GPC3 transgene, in early stage of liver regeneration.23–25 Previous studies showed that a decrease of nuclear factors C/EBPα and Dbp22 and an increase of C/EBPβ23 were involved in the down-regulation of albumin expression during liver regeneration. Our western blotting results confirmed the down-regulation of C/EBPα and up-regulation of C/EBPβ after PHx in GPC3 TG mice (Fig. 4) In gene array analysis, we also observed an 11.5-fold decrease in the expression of Dbp at 2 days after PHx in TG mice (data not shown). The changes of these nuclear factors may lead to down-regulation of the Albumin promoter controlling the GPC3 transgene in TG mice, which may explain the decrease of GPC3 protein levels after PHx.
The suppression of regeneration seen in this study well corresponds with some of the changes noted in the TG mice prior to PHx, using gene array analysis. GABA A receptor is over expressed by 2.9-fold, as shown above. Previous studies have shown that GABA and its receptor have inhibitory effects on liver regeneration.26 Runx3, another over-expressed gene in the TG mice, is associated with suppression of Notch signaling events in the liver.27 NICD, the Notch intracellular domain, is associated with stimulation of hepatocyte proliferation during liver regeneration.28 The role of Wnt7b is not clear. There is minimal expression of Wnt7 in normal mouse liver and, if any, it is expressed in non-parenchymal cells.29 Surprisingly, there is over-expression of EGFR, a major mitogenic receptor for hepatocytes.30 This may reflect a compensatory effect associated with the increase of the growth-inhibitory GPC3 in the liver of the TG mice. We have further validated some of the changes in gene array data using western blotting. There was good correlation between the changes in mRNAs as shown in the gene array and an increase in C/EBPα and Runx3 and a decrease in Yap in GPC3 TG mice at day0 (Fig.4). Although the expression of Egr1 is down-regulated by 5.3-fold, there is enhanced protein level of Egr1 in TG mice, which might be due to an effect related to unknown microRNAs or the protein stability. The enhanced protein level of Egr1 may also correlate with the observed effects on liver regeneration since Egr1 is known to suppress expression of the HGF receptor.31 The changes associated with cell cycle related genes after PHx are easier to correlate with the observed suppression of liver regeneration, especially in relations to cyclin A and D1, well known to be associated with entry of hepatocytes into S-phase of the cycle.32 The observed increase in C/EBPα also relates to the overall suppression of regeneration, since C/EBPα is known to decrease following PHx.33 Equally important is the observed decrease in YAP nuclear levels in TG mice, associated with increase in cytoplasmic levels of p-YAP. YAP is known to correlate with hepatocyte growth and adjustment of liver size.34,35 Phosphorylation of YAP is associated with export from the nucleus to the cytoplasm and suppression of its effects on regulation of growth related genes.35
The results of our current work further extend the evidence that GPC3 has overall growth-suppressive effects on hepatocytes. GPC3 however is greatly elevated in human hepatocellular carcinoma.8 One can rationalize that the increased expression of GPC3 in liver cancer reflects an existing gene expression algorithm by which proliferating hepatocytes regulate their growth, as in liver regeneration. In the case of hepatocellular carcinoma, this approach to growth regulation fails due to intrinsic genomic irregularities that render carcinomas unresponsive to GPC3. GPC3, however, is also reported to be elevated in liver progenitor (oval cell) populations,15 a cell population which is not neoplastic. Despite all accumulated literature on growth regulation by GPC3, its mechanisms of action remain currently elusive. It is associated with CD81, as shown in our previous study. The latter also has growth regulatory effects in specific cell populations.12 We showed in our previous study that there is enhanced association between GPC3 and CD81 after PHx.12 This may result in suppression of CD81 signaling, of which much less is known even in comparison to GPC3. Since not much is known about the pathways involved in termination of liver regeneration, further studies should be performed to fully understand the signaling pathways associated with the effects of GPC3 in normal liver, in order to better conceptualize its high levels of expression in liver neoplasia.
Supplementary Material
Acknowledgments
We thank Ann Orr for valuable assistance with all aspects of histology in this manuscript.
This work was supported by the National Institutes of Health (NIH) grants CA30241 and CA35373 (GKM).
Abbreviations
- GPC3
glypican 3
- HSPG
heparan sulfate proteoglycan
- GPI
glycosylphosphatidylinositol
- SGBS
Simpson-Golabi-Behmel syndrome
- PHx
partial hepatectomy
- HCC
hepatocellular carcinoma
- NPC
non-parenchymal cells
- TG
transgenic
- WT
wild-type
- Poly A
polyadenylation
- PBGD
porphobilinogen deaminase
- YAP
yes-associated protein
- Egr-1
early growth response factor 1
- Dbp
D site Albumin promoter binding protein
- Jak3
Janus kinase 3
- Gabaar
GABA-A receptor
- Wnt7b
wingless-related MMTV integration site 7B
- Egfr
epidermal growth factor receptor
- Igfbp1
insulin-like growth factor binding protein 1
- Rab2
RAS oncogene family
- Tgfb1
transforming growth factor beta type I
- Catnb
beta catenin
- Ndl
Nodal
- Ccna1
Cyclin A1
- Ccnm1
Cyclin M1
- Ccnd1
Cyclin D1
- Ccne2
Cyclin E2
- Gas1
growth arrest specific 1
- Runx3
runt related transcription factor 3
- Gadd45b
growth arrest and DNA-damage-inducible 45 beta
- Fgf14
fibroblast growth factor 14
- C/EBPα
CCAAT-enhancer-binding protein α
- C/EBPβ
CCAAT-enhancer-binding protein β
References
- 1.Fransson LA. Glypicans. International Journal of Biochemistry & Cell Biology. 2003;35:125–129. doi: 10.1016/s1357-2725(02)00095-x. [DOI] [PubMed] [Google Scholar]
- 2.Song HH, Filmus J. The role of glypicans in mammalian development. Biochimica et Biophysica Acta. 2002;1573:241–246. doi: 10.1016/s0304-4165(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 3.Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19r. [DOI] [PubMed] [Google Scholar]
- 4.Iglesias BV, Centeno G, Pascuccelli H, Ward F, Peters MG, Filmus J, Puricelli L, et al. Expression pattern of glypican-3 (GPC3) during human embryonic and fetal development. Histol Histopathol. 2008;23:1333–1340. doi: 10.14670/HH-23.1333. [DOI] [PubMed] [Google Scholar]
- 5.Hughes-Benzie RM, Pilia G, Xuan JY, Hunter AG, Chen E, Golabi M, Hurst JA, et al. Simpson-Golabi-Behmel syndrome: genotype/phenotype analysis of 18 affected males from 7 unrelated families. Am J Med Genet. 1996;66:227–234. doi: 10.1002/(SICI)1096-8628(19961211)66:2<227::AID-AJMG20>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini M, Pilia G, Pantano S, Lucchini F, Uda M, Fumi M, Cao A, et al. Gpc3 expression correlates with the phenotype of the Simpson-Golabi-Behmel syndrome. Dev Dyn. 1998;213:431–439. doi: 10.1002/(SICI)1097-0177(199812)213:4<431::AID-AJA8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 7.Cano-Gauci DF, Song HH, Yang H, McKerlie C, Choo B, Shi W, Pullano R, et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146:255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 10.Michalopoulos GK. Liver regeneration. Journal of Cellular Physiology. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins G, Anderson R. Experimental pathology of liver: restoration of liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 12.Liu B, Paranjpe S, Bowen WC, Bell AW, Luo JH, Yu YP, Mars WM, et al. Investigation of the role of glypican 3 in liver regeneration and hepatocyte proliferation. Am J Pathol. 2009;175:717–724. doi: 10.2353/ajpath.2009.081129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oren R, Takahashi S, Doss C, Levy R, Levy S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol Cell Biol. 1990;10:4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy S, Todd SC, Maecker HT. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annual Review of Immunology. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 15.Grozdanov PN, Yovchev MI, Dabeva MD. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Lab Invest. 2006;86:1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- 16.Ohtsubo K, Nomaguchi TA. A flow cytofluorometric study on age-dependent ploidy class changes in mouse hepatocyte nuclei. Mech Ageing Dev. 1986;36:125–131. doi: 10.1016/0047-6374(86)90013-8. [DOI] [PubMed] [Google Scholar]
- 17.Shima A, Sugahara T. Age-dependent ploidy class changes in mouse hepatocyte nuclei as revealed by Feulgen-DNA cytofluorometry. Exp Gerontol. 1976;11:193–203. doi: 10.1016/0531-5565(76)90033-4. [DOI] [PubMed] [Google Scholar]
- 18.Stocker E, Wullstein HK, Brau G. [Capacity of regeneration in liver epithelia of juvenile, repeated partially hepatectomized rats. Autoradiographic studies after continous infusion of 3H-thymidine (author's transl)] Virchows Arch B Cell Pathol. 1973;14:93–103. [PubMed] [Google Scholar]
- 19.Nadal C, Zajdela F. Hepatic polyploidy in the rat. IV. Experimental changes in the nucleolar volume of liver cells and their mechanisms of regulation. Exp Cell Res. 1967;48:518–528. doi: 10.1016/0014-4827(67)90318-7. [DOI] [PubMed] [Google Scholar]
- 20.Apte U, Gkretsi V, Bowen WC, Mars WM, Luo JH, Donthamsetty S, Orr A, et al. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50:844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filmus J, Capurro M. The role of glypican-3 in the regulation of body size and cancer. Cell Cycle. 2008;7:2787–2790. doi: 10.4161/cc.7.18.6672. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto T, Tsujinaka T, Yano M, Iijima S, Ebisui C, Kan K, Kishibuchi M, et al. Regulation of albumin synthesis after hepatectomy and in the acute inflammation phase of rat liver. The Journal of Nutritional Biochemistry. 1995;6:522–527. [Google Scholar]
- 23.Trautwein C, Rakemann T, Pietrangelo A, Plumpe J, Montosi G, Manns MP. C/EBP-beta/LAP controls down-regulation of albumin gene transcription during liver regeneration. J Biol Chem. 1996;271:22262–22270. doi: 10.1074/jbc.271.36.22262. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto T, Tsujinaka T, Yano M, Ogawa A, Kishibuchi M, Morita S, Shiozaki H, et al. Regulation of albumin mRNA and its promoter-binding nuclear factors under different perioperative nutritional methods in hepatectomized rats. Am J Surg. 1998;175:221–225. doi: 10.1016/s0002-9610(97)00296-1. discussion 225–226. [DOI] [PubMed] [Google Scholar]
- 25.Mueller CR. The down-regulation of albumin transcription during regeneration is due to the loss of HNF-1 and the D-site transcription factors. DNA Cell Biol. 1992;11:559–566. doi: 10.1089/dna.1992.11.559. [DOI] [PubMed] [Google Scholar]
- 26.Minuk GY, Gauthier T. The effect of gamma-aminobutyric acid on hepatic regenerative activity following partial hepatectomy in rats. Gastroenterology. 1993;104:217–221. doi: 10.1016/0016-5085(93)90854-6. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Chen Y, Wu KC, Liu J, Zhao YQ, Pan YL, Du R, et al. RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Exp Cell Res. 2010;316:149–157. doi: 10.1016/j.yexcr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, Michalopoulos GK. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng G, Awan F, Otruba W, Muller P, Apte U, Tan X, Gandhi C, et al. Wnt'er in liver: expression of Wnt and frizzled genes in mouse. Hepatology. 2007;45:195–204. doi: 10.1002/hep.21473. [DOI] [PubMed] [Google Scholar]
- 30.Skarpen E, Oksvold MP, Grosvik H, Widnes C, Huitfeldt HS. Altered regulation of EGF receptor signaling following a partial hepatectomy. J Cell Physiol. 2005;202:707–716. doi: 10.1002/jcp.20171. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Liu Y. Suppression of HGF receptor gene expression by oxidative stress is mediated through the interplay between Sp1 and Egr-1. Am J Physiol Renal Physiol. 2003;284:F1216–F1225. doi: 10.1152/ajprenal.00426.2002. [DOI] [PubMed] [Google Scholar]
- 32.Mullany LK, White P, Hanse EA, Nelsen CJ, Goggin MM, Mullany JE, Anttila CK, et al. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle. 2008;7:2215–2224. doi: 10.4161/cc.7.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenbaum LE, Cressman DE, Haber BA, Taub R. Coexistence of C/EBP alpha, beta, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. Implications for maintenance of the differentiated state during liver growth. J Clin Invest. 1995;96:1351–1365. doi: 10.1172/JCI118170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.