Abstract
In humans, peroxisomes harbor a complex set of enzymes acting on various lipophilic carboxylic acids, organized in two basic pathways, α-oxidation and β-oxidation; the latter pathway can also handle ω-oxidized compounds. Some oxidation products are crucial to human health (primary bile acids and polyunsaturated FAs), whereas other substrates have to be degraded in order to avoid neuropathology at a later age (very long-chain FAs and xenobiotic phytanic acid and pristanic acid). Whereas total absence of peroxisomes is lethal, single peroxisomal protein deficiencies can present with a mild or severe phenotype and are more informative to understand the pathogenic factors. The currently known single protein deficiencies equal about one-fourth of the number of proteins involved in peroxisomal FA metabolism. The biochemical properties of these proteins are highlighted, followed by an overview of the known diseases.
Keywords: adrenoleukodystrophy, ATP binding cassette-transporter, carnitine, cholestanoic acid, chondrodysplasia, dicarboxylic acids, docosahexaenoic acid, lignoceric acid, phytanic acid, phytol, Refsum, Zellweger
The report by Goldfischer et al. (1) in 1973 on the lack of peroxisomes in liver and kidney of patients with the cerebro-hepatorenal syndrome of Zellweger was the first one associating peroxisomes with an inherited lethal human disorder. At that time, peroxisomes were thought to be remnants of a respiratory ancestor organelle, and mitochondria, also affected in this disorder, were thought to contribute primarily to the pathology. In the meantime, knowledge about the metabolic role of peroxisomes in humans has increased substantially. With regard to FAs, in the context of this review defined in a broader sense as lipophilic carboxylic acids, peroxisomes are required for the degradation of very long-chain (VLC) FAs (VLCFA), pristanic acid, and various other carboxylic acids and play an essential or important role in the formation of primary bile acids and PUFA (via β-oxidation). The α-oxidative degradation of phytanic acid and α-hydroxylated FAs turned out to be confined to peroxisomes as well. In addition to Zellweger syndrome (ZS) and related disorders, other diseases have been linked to peroxisomes, notably X-linked adrenoleukodystrophy (X-ALD) and Refsum disease (Table 1). To understand some aspects of the peroxisomal α- and β-oxidation pathways in humans, the main topic of this review, a brief introduction on the biogenesis of peroxisomes is given, followed by some discussion about their membrane properties. This will also be useful to comprehend the underlying causes of certain peroxisomal diseases, with new entities being added in recent years. Fundamental knowledge about peroxisomal metabolism is mainly based on studies in rodents. However, these findings cannot always be extrapolated to humans, sometimes even not from rats to mice. As far as known, the situation in humans has been described in this text, or possible differences are indicated. Unless otherwise mentioned, genes/enzymes will be named according to recommendations by HUGO.
TABLE 1.
Human diseases linked to impaired peroxisomal α- and/or β-oxidation
PEROXISOME BIOGENESIS
Based on recent proteomic studies (2–4) it is estimated that mammalian peroxisomes contain about 80–90 proteins. These are involved in reactive oxygen, glyoxylate, polyamine, purine, amino acid, and lipid metabolism. Enzymes belonging to the latter pathways are highly expressed in liver, but the peroxisomal content can differ according to tissue and cell type (Table 2). More than 80% of the organellar proteins are matrix enzymes. These are synthesized on free polyribosomes in the cytosol and posttranslationally imported in import-competent (pre)peroxisomes (5, 6) (Fig. 1). For their import, matrix proteins rely on targeting sequences, named peroxisome targeting signal (PTS) 1 and PTS2. PTS1 consists of a C-terminal dodecamer ending with SKL or a related sequence (S/A/V/P - K/R/H/Q/N - L/I/M-COOH) in mammals) (7); its binding protein is called peroxin 5 (PEX5). The latter is a cytosolic, tetratricopeptide repeat-containing protein that docks at the membrane of import competent vesicles, a process controlled by PEX14. In humans, a short and long PEX5 isoform exist, both derived from PEX5 by differential splicing. PTS2 is a nonapeptide located near the N terminal of proteins, with a consensus motif NH2-X(1-30)-R-L-XXXXX-H-L for mammals. It is recognized by another cytosolic receptor protein, PEX7, which is characterized by a WD40 motif. The PTS2-PEX7 complex also docks at PEX14 but indirectly via interaction with PEX5L, the longer PEX5 isoform (8). In mammals, PTS2 proteins are processed after import, with the N-terminal PTS2 being cleaved off by TYSND1, a peroxisomal cysteine endopeptidase. Somewhat mysteriously, only three matrix proteins employ the PTS2-PEX7 import system. Not all PTS1 variants are equally effective, and proteins containing a less potent PTS1 are partially cytosolic or face competition with proteins containing a strong PTS1, like SKL. This explains differences between species, or sometimes between cell types, with regard to the cytosolic/peroxisomal distribution of matrix proteins. A few proteins are known to contain, in addition to PTS1, a mitochondrial targeting sequence at the N terminus. Apparently, these signals and their respective import machinery have a comparable strength, and such enzymes display a bimodal distribution.
TABLE 2.
List of peroxisomal proteins involved in α- and β-oxidation in humans, chromosome mapping, tissue expression, and targeting
| Approved Gene Symbol | Approved Gene Name | Aliases | Accession No. | Locus | Tissue Expression | Targeting Sequence | Remarksb |
|---|---|---|---|---|---|---|---|
| ABCD1 | ATP-binding cassette, subfamily D (ALD), member 1 | AMN, ALDP, ALD, ABC42 | NM_000033 | Xq28 | G (high in cervix, pancreas, skin; nerves; low in liver) | mPTS | |
| ABCD2 | ATP-binding cassette, subfamily D (ALD), member 2 | ALDL1, ALDR, ABC39, ALDL1, ALDRP, hALDR | NM_005164.3 | 12q11-q12 | R (brain, liver) | mPTS | Low expression compared with ABCD1 |
| ABCD3 | ATP-binding cassette, subfamily D (ALD), member 3 | PMP70, ABC43; PXMP1 | NM_002858 | 1p22-p21 | U | mPTS | Not expressed in adipose |
| ACAA1 | Acetyl-CoA acyltransferase 1 | Peroxisomal thiolase, THIOL | NM_001607 | 3p22-p21.3 | U (high in liver, spleen; intestines) | PTS2 | Likely splice forms |
| ACAD11 | Acyl-CoA dehydrogenase family, member 11 | FLJ12592; MGC150619 | NM_032169 | 3q22.1 | U (high in kidney, thymus) | TAKI | |
| ACNAT1 | Acyl-CoA amino acid N-acyltransferase | AL359893 | 9q22.3 | RSKL | Pseudogene of BAAT | ||
| ACSL1 | Acyl-CoA synthetase long-chain family member 1 | LACS2, LACS, ACS1, LACS1, FACL1 | NM_001995 | 4q35 | U (high in adipose tissue, mammary gland, adrenals, liver) | TIKV | Active on C16:0, Phyt, Pris |
| ACSL4 | Acyl-CoA synthetase long-chain family member 4 | FACL4, ACS4, LACS4 | NM_004458 | Xq22.3-q23 | U | Membrane associated ? | Peripheral membrane protein in rat |
| SLC27A2 | Solute carrier family 27 (FA transporter), member 2 | ACSVL1FATP2, hFACVL1, VLACS, VLCS | NM_003645 | 15q21.2 | G (high in kidney, liver, placenta) | TLKL ? | Also ER associated active on Phyt, Pris, C24:0, THCA |
| SLC27A4 | Solute carrier family 27 (FA transporter), member 4 | ACSVL5FATP4 | NM_005094.2 | 9q34.13 | G (high in skin) | EEKL ?(peripheral) | Also mitochondria, MAM, ER in fibroblasts |
| ACOX1 | Acyl-CoA oxidase 1a acyl-CoA oxidase 1 b | ACOX, SCOX, PALMCOX, palmitoyl-CoA oxidase | NM_004035 NM_007292 | 17q25.1 | G | QSKL QSKL | Two splice forms with different exon 3 |
| ACOX2 | Acyl-CoA oxidase 2 | BCOX, BRCOX, THCCox, BRCACOX, branched chain acyl-CoA oxidase | NM_003500 | 3p14.3 | R (liver, nerves) | RSKL | |
| ACOX3 | Acyl-CoA oxidase 3 | Pristanoyl-CoA oxidase | NM_003501 | 4p15.3 | G (prostate, adrenals) | KSKL | Present as protein ? a shorter C-truncated splice form exists (NM_001101667) |
| ACOXL | Acyl-CoA oxidase-like | ACOX4 | NM_018308 | 2q13 | R (lung) | absent | Major transcript truncated; a splice form ending in GAKL might be targeted |
| ACOT2 | Acyl-CoA thioesterase 2 | Mte1, PTE2 | NM_006821 | 14q24.3 | G (liver>kidney) | PSKV | Likely only mitochondrial; active on medium and long-chain esters |
| ACOT4 | Acyl-CoA thioesterase 4 | PTE-Ib; PTE1B; PTE2B | NM_152331 | 14q24.3 | R (liver<kidney; present in adipose tissue) | VPKL | Combined activities of mouse ACOT3,4,5; active on succinyl-, medium, and long-chain CoA-esters |
| ACOT6 | Acyl-CoA thioesterase 6 | Pristanoyl-phytanoyl-CoA thioesterase | NM_001037162 | 14q24.3 | R (testis, connective tissue) | HSKI ? | Compared with mouse counterpart, lacking first exon with startcodon; not targeted in fibroblasts |
| ACOT8 | Acyl-CoA thioesterase 8 | hACTE-III, PTE-2, PTE1, TE2, het, HNAACTE | NM_005469.2 | 20q12-q13.1 | U | ESKL | Active on diC5 to diC12-, choloyl-, prostaglandin-, pristanoyl-CoA; interacts with NEF |
| ACOT12 | Acyl-CoA thioesterase 12 | Cach, THEAL, STARD15 | NM_130767 | 5q14.1 | R (liver specific) | VSTF ?? | |
| ALDH3A2 | Aldehyde dehydrogenase 3 family, member A2 | SLS, ALDH10, FALDH | NM_000382.2 | 17p11.2 | U | SKQR ? | Major splice variant FALDH-V (membrane bound); ER-localized isoform FALDH-N corresponds to NM_001031806.1 |
| AMACR | α-Methylacyl-CoA racemase | Epimerase | NM_014324 | 5p13.2-q11.1 | U | KASLa | Different splice forms |
| BAAT | Bile acid CoA-amino acid N-acyltransferase | BAT | NM_001701.3 | 9q22.3 | R (liver specific) | TSQL | Might be partially cytoplasmic |
| CAT | Catalase | NM_001752 | 11p13 | U | KANL | ||
| CRAT | Carnitine acetyltransferase | CAT1 | NM_000755.3 | 9q34.1 | G (skeletal muscle> heart, liver and pancreas> brain, placenta, lung, kidney). | RAKL | By alternatively splicing, an isoform with an N-terminal mitochondrial transit peptide |
| CROT | Carnitine O-octanoyltransferase | COT | NM_021151 | 7q21.1 | G (low in heart, adipose tissue) | STHL | Malonyl-CoA inhibitory |
| DECR2 | 2,4-Dienoyl CoA reductase 2, peroxisomal | PDCR, SDR17C1 | NM_020664 | 16p13.3 | G/R | SAKL | |
| DHRS4 | Dehydrogenase/reductase (SDR family) member 4 | SCAD-SRL, SDR-SRL, SDR25C1 | NM_021004 | 14q11.2 | U/G | PSRL | Reduction of 3-oxosteroids; oxidation of 3β-hydroxysteroids; lower activity on retinal/retinol than pig or mouse DHRS4 |
| ECH1 | Enoyl CoA hydratase 1, peroxisomal | Δ3,5-Δ,4-dienoyl-CoA isomerase, HPXEL | NM_001398.2 | 19q13.1 | U (high in adipose tissue, muscle, heart) | FSKLa | |
| FAR1 | Fatty acyl CoA reductase 1 | SDR10E1 | NM_032228 | 11p15.2 | G | TMRY ? | Partial colocalization |
| FAR2 | Fatty acyl CoA reductase 2 | SDR10E2 | NM_018099 | 12p11.23 | R (sebaceous glands) | TLKV ? | Partial colocalization |
| HACL1 | 2-Hydroxyacyl-CoA lyase 1 | 2-HPCL | NM_012260 | 3p24.3 | U | RSNM | |
| HAO2 | Hydroxyacid oxidase 2 (long-chain) | HAOX2, GIG16 | NM_001005783 | 1p13.3-p13.1 | R (liver, kidney) | FSRL | |
| HSDL2 | Hydroxysteroid dehydrogenase like 2 | SDR13C1 | NM_032303 | 9q32 | U (kidney, liver, testis, ovary, pancreas) | NARL | C-terminal SCP2 domain; likely targeted to mitochondria as well; substrate? |
| IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | IDH, IDP, IDCD, IDPC, PICD, IDH1 | NM_005896.2 | 2q33.3 | U | GAKL | Mainly cytosolic |
| EHHADH | Enoyl-CoA, hydratase/3-hydroxyacyl-CoA dehydrogenase | MFP1 MFE-1, L-BP, ECHD | NM_001966 | 3q26.3-q28 | G(high in liver>kidney) | SSKL | Evidence for a N-truncated isoform (NM_001166415) |
| 17HSD4 | Hydroxysteroid 17β-dehydrogenase 4 | MFP2 MFE-2, DBP, SDR8C1 | NM_000414 | 5q21 | U (high in liver, adipose, spleen, brain) | YAKL | |
| NUDT7 | Nudix (nucleoside diphosphate linked moiety X)-type motif 7 | NM_001 105663.1 | 16q23.1 | R(spleen, prostate) | TSRL | Active on acyl-CoA, CoA | |
| NUDT12 | Nudix (nucleoside diphosphate linked moiety X)-type motif 12 | NM_031438 | 5q21.2 | G (high in liver, kidney) | NPNL | Active on NAD(P)H | |
| PECI | Peroxisomal Δ3,Δ2-enoyl-CoA isomerase | ACBD2, DRS1, HCA88 | NM_006117 | 6p24.3 | U (high in liver, low brain, muscle) | KSKL | |
| PECR | Peroxisomal trans-2-enoyl-CoA reductase | TERP, SDR29C1 | NM_018441 | 2q35 | G(high in liver, kidney) | KAKL | |
| PHYH | Phytanoyl-CoA 2-hydroxylase | PAHX, PHYH1 | NM_006214 | 10p13 | U | PTS2 | |
| PXMP2 | PMP2, 22 kDa | PMP22 | NM_018663.1 | 12q24.33 | G | mPTS | |
| SCP2 | SCPx | SCPx | NM_002979 | 1p32 | U | NAKL | Evidence for C-truncated splice form (NM_001007098.1) |
| SCP2 | SCP2 | SCP2; NLTP; NSL-TP | NM_002979 | 1p32 | U | NAKL | Three variants |
| SLC25A17 | Solute carrier family 25 (mitochondrial carrier; PMP, 34 kDa), member 17 | PMP34 | NM_006358 | 22q13.2 | U (high in kidney, liver) | mPTS | |
| SLC22A21 | Solute carrier family 25 member 21 | OCNT3 | ?? | mPTS | Mouse specific ? | ||
| TYSND1 | Trypsin domain containing 1 | NM_173555.2 | 10q22.1 | R (bone marrow>lung>liver) | RSKL | Evidence for a truncated spliceform (NM_001040273.1) |
Entries are arranged alphabetically according to their gene symbol (bold) or the preferred abbreviation used in the main text (alias or name in bold). Expression, based on published tissue distribution, mRNA analysis, and/or in silico expression data (http://www.ncbi.nlm.nih.gov/unigene), is defined as ubiquitous (U, in all tissues), general (G, in most tissues), or restricted (R). The targeting sequence is defined as mPTS (for integral membrane proteins), PTS2, or PTS1. For the latter, the last four amino acids are given, because weaker tripeptides are often preceeded by a positive charged amino acid (printed bold). If the peptide sequence if followed by a question mark, the PTS1 functionality is doubtful.
Same protein is targeted to mitochondria via N-terminal mitochondrial sequence.
NEF, negative factor of immunodeficiency virus; Phyt, phytanic acid; Pris, pristanic acid.
Fig. 1.
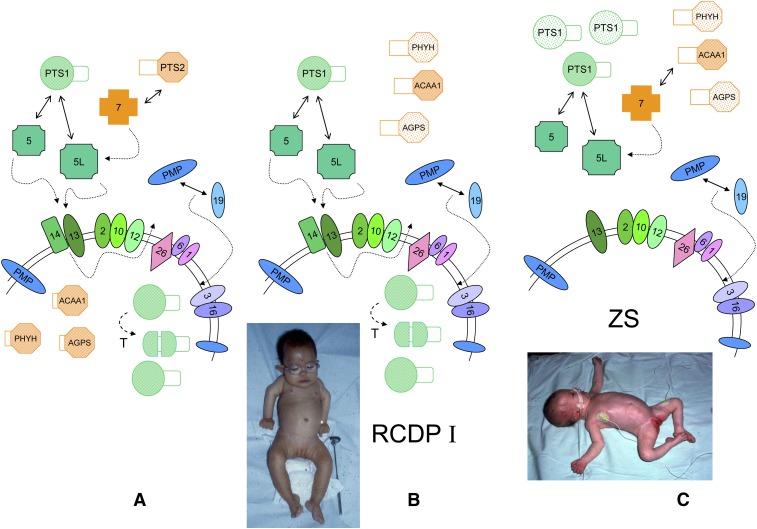
Peroxisome biogenesis in humans. A: A simplified scheme for protein import in mammalian peroxisomes under normal conditions and involved peroxins, indicated by their PEX number and arranged according to known interactions, is shown. The complex of PTS1-matrix proteins (in green) and one of the PEX5 isoforms, formed in the cytoplasm, will dock at the membrane, followed by translocation involving the RING peroxins PEX2, 10, and 12 (dashed arrows). After cargo release, PEX5 is recycled to the cytoplasm with the help of PEX1, 6, and 26 (dashed arrow). The role of mono- and polyubiquitination of PEX5 in this process is not displayed. Import of PTS2-matrix proteins (in orange), captured by PEX7, is mediated by the long PEX5 isoform. After import, some PTS1 proteins are proteolytically processed (T), and the targeting signal of PTS2-proteins is removed (ACAA1, thiolase; PHYH, AGPS). Newly synthesized PMPs (in blue) can interact with PEX19 in the cytoplasm (or with the membrane associated PEX19), followed by docking and membrane insertion. Peroxins playing a role in PMP targeting are depicted in different shades of blue. In the absence of a functional PEX7, PTS2 proteins are not imported and processed and are degraded in the cytoplasm (faintly colored), but the matrix is still filled with PTS1 proteins (B). This condition results in RCDP type I. Patients with RCDP type I have rhizomelia and congenital cataracts (notice the spectacles in this 19-month-old infant). When peroxins involved in PTS1 import are missing, e.g., PEX14 (C), peroxisomes are empty (ghosts) causing the severe ZS. Both PTS1 and PTS2 proteins remain cytoplasmic and unprocessed and can be degraded. The picture shows a very hypotonic baby with head malformation, low ear implantation, and hepatomegaly. In cases where PEX19 or other peroxins involved in PMP import are mutated (not shown), all peroxisomal proteins remain in the cytoplasm or are degraded, and no ghosts can be observed. Pictures courtesy of Dr. L. Van Maldergem, Centre de Génétique Humaine, Université de Liège, Belgium (B) and Dr. J. Jaeken, UZ-Leuven, Belgium (C) with informed consent of the parents.
With regard to peroxisomal integral membrane proteins (PMP), also posttranslationally imported, the targeting sequences, called mPTS, are less well delineated (9). Various PMPs interact with PEX19, a predominantly cytosolic protein containing a C-terminal prenylation site, but some PEX19 is found on the peroxisomal membrane. In the absence of PEX19 and other proteins involved in peroxisomal membrane biogenesis (PEX3, PEX16), cells lack peroxisomes and no membrane remnants are found (Fig. 1). In the absence of PEX5, or downstream binding partners, cells still have peroxisomes, although empty and without matrix content (ghosts) (Fig. 1). Indeed, due to the role of PEX5L in PTS2-import, both PTS1 and PTS2 proteins are mistargeted. Depending on their stability, the nonimported proteins can sometimes be detected in the cytosol, some are even active (e.g., catalase, alanine glyoxylate aminotransferase), but PTS2-proteins and some PTS1-proteins (see “Acyl-CoA oxidases” and “2-Enoyl-CoA hydratases and 3-hydroxyacyl-CoA dehydrogenases”) are not processed.
Mutations in PEX5 and in genes coding for downstream peroxins or peroxins involved in membrane biogenesis cause rather similar phenotypes, currently named peroxisome biogenesis disorders (PBDs), the prototype being the ZS, an autosomal recessive lethal disorder (ZS; OMIM #214100; Table 1.Fig. 1) (10). ZS patients present with severe hypotonia, craniofacial malformations, liver and kidney problems, seizures, retinopathy and neuron migration defects, and die usually within the first year of life. Other clinical descriptions, with a milder phenotype but still fatal, include neonatal adrenoleukodystrophy (OMIM #202370) and infantile Refsum disease (OMIM #266510) (10) (Table 1). The biochemical abnormalities reflect the absence of functional peroxisomes: accumulation of unusual bile acid intermediates, of VLCFA, of pristanic acid, and of phytanic acid; lack of plasmalogen; shortage of docosahexaenoic acid (DHA; C22:6n-3). ZS patients and patients with milder clinical symptoms (collectively named the Zellweger spectrum) can be classified into 12 complementation groups depending on the underlying PEX mutation. Mutations in PEX7 result in a separate phenotype, rhizomelic chondrodysplasia punctata type 1 (RCDP; OMIM #215100). Characteristics for this rare autosomal recessive disorder are the symmetrical shortening of the upper limbs, calcific stippling, a typical dysmorphic face, and growth retardation (Fig. 1). Most patients present with cataract, and ichthyosis is quite common. Compared with ZS, RCDP type 1 patients live longer, some surviving into the second decade. In this syndrome, PTS2 enzymes, which catalyze reactions in three major peroxisomal pathways (plasmalogen synthesis, α-oxidation, and β-oxidation) are mislocalized. As a consequence, plasmalogens are low and phytanic acid accumulates; PTS1 enzymes are, however, targeted and functional (Fig. 1). Clinically, patients with type 1 RDCP cannot be distinguished from patients with a defect in one of the peroxisomal enzymes of the plasmalogen synthesis, glyceronephosphate O-acyltransferase (GNPAT) deficiency or RCDP type 2 (OMIM #222765) and alkylglycerone-phosphate synthase (AGPS) deficiency or RCDP type 3 (OMIM #600121).
Besides these biogenesis disorders, various other disorders have been diagnosed due to mutations in a single peroxisomal protein (Table 1). Many of these display perturbations in the oxidation of lipophilic carboxylic acids. These pathways will be discussed further. In Table 2, the targeting sequences of all known enzymes/proteins involved in peroxisomal α- and β-oxidation are given together with gene acronyms, chromosome mapping, and other relevant information.
PEROXISOMAL MEMBRANE PROPERTIES
The pioneering studies of de Duve and coworkers (11) revealed particular aspects of the membrane permeability of isolated peroxisomes: lack of osmotic behavior causing sedimentation to high density in small solute gradients and lack of latency for matrix enzymes1. In the 1980s, the permeability of rat liver peroxisomes was biochemically and electrophysiologically investigated (12), resulting in the concept of a nonselective pore-forming protein; a likely candidate was PXMP2, better known as PMP22, a rather abundant PMP (subunit size of 22 kDa). Following the discovery of some peroxisomal membrane transporters, such as members of the ATP binding cassette (ABC) transporter and the mitochondrial solute transporter families, the pore hypothesis was disfavored. Recent reinvestigations of the permeability, however, led to similar “leaky” conclusions (13). Based on studies in a knockout mouse (14), PXMP2 is responsible for the permeability, forming a homotrimeric general diffusion pore (estimated diameter 1.4 nm) with weak cation selectivity and no voltage dependence. This leads to the following picture: the PXMP2 pore allows access of small, uncharged or weak anionic solutes (<300 Da) with an exclusion limit of ∼600 Da; entry or exit of bulky polar molecules (e.g., CoA, ATP) is restricted and is or has to be mediated by (specific) carriers/transporters, or, alternatively, exit could occur after hydrolysis to smaller fragments. Presently, known transporters include three ABC-transporters (ABCD1, ABCD2, ABCD3) (discussed further in “ABCD-transporters and β-oxidation”) and two solute carriers (SLC25A17 or PMP34 and SLC22A21 or OCTN3) (see “Auxiliary enzymes and proteins related to α-oxidation” and “Auxiliary enzymes and new candidates”). Based on indirect data, a few other transporters are postulated (15, 16), but so far without molecular or genetic evidence. Hence, compared with the inner mitochondrial membrane, mammalian peroxisomes contain only a limited number of solute transporters. Moreover, proteomics on purified peroxisomal membranes (4, 17) failed to reveal additional transporters. With regard to degradation of bulky cofactors, nudix hydrolases (18) acting on NAD(P) H and CoA have been shown to be present in peroxisomes. For more insight/discussion on the membrane properties of peroxisomes, the reader is referred to recent reviews (15, 16). This brief description of the enigmatic properties of these organelles, however, should be sufficient to understand the difficulties when studying the peroxisomal uptake/transport of amphipatic molecules such as FAs and acyl-CoAs or related processes such as activation and to propose models.
PEROXISOMAL α-OXIDATION
The prime substrate in humans for α-oxidation, being defined as the degradation of carboxylic acids by removal of one carbon atom, is phytanic acid (3,7,11,15-tetramethylhexadecanoic acid) (Fig. 2). More recently, long-chain 2-hydroxylated FAs (2OH-FA) were proven to be α-oxidized (19), and an α-oxidative mechanism is supposedly responsible for the presence/formation of odd-numbered long-chain FAs in brain (20).
Fig. 2.
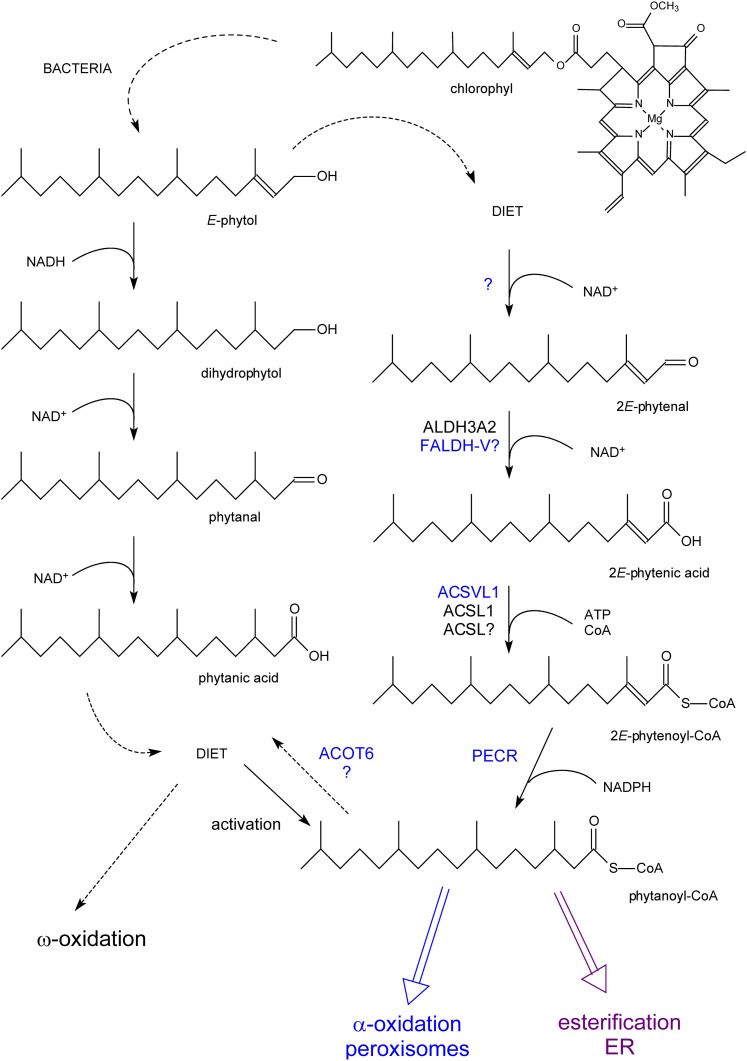
Formation of phytanic acid from phytol and its metabolism. Phytol, derived from chlorophyll, can be converted to phytanic acid by rumen bacteria (left side) and taken up via the diet or to phytanoyl-CoA in mammals with phytenoyl-CoA as intermediate (right side). Phytanoyl-CoA can be incorporated in lipids (esterification), shortened by α-oxidized, or hydrolyzed back to phytanic acid. In cases where α-oxidation is impaired, phytanic acid will be degraded starting from the ω-end (ω-oxidation; see supplementary Fig. II). Enzymes printed in blue are associated with peroxisomes. Reactions for which the responsible enzyme have not yet been clarified are marked by a question mark.
Phytanic acid (racemic at position 3; Fig. 2) is present in our diet, mostly in dairy products, meat, and certain fishes, with an estimated daily intake of 1 mg/kg body weight for an adult. This methylbranched FA originates from phytol, which, when esterified to chlorophyll, serves to anchor this pigment in the thylakoid membranes of chloroplasts. Humans and mice cannot hydrolyze esterified phytol, but in ruminants, phytol is liberated from ingested plants and further oxidized to phytanic acid by the bacterial flora (Fig. 2). Apparently, the capacity of ruminants to degrade phytanic acid is not sufficient, and it is stored in neutral and phospholipids of tissues and milk (21). In addition to direct intake, humans can convert free dietary phytol to phytanic acid. The enzymatic steps involved are (partly) peroxisomal (22) (Fig. 2). Once ingested and after entering the circulation (via lymph and lipoproteins), phytanic acid is taken up by most tissues, but especially by heart, muscle, and liver. Kidney and adipose tissue also store phytanic acid; uptake in brain, however, is small (based on animal studies) (23).
Enzymology of peroxisomal α-oxidation
Until 1993, phytanic acid degradation was described as an oxidative decarboxylation, generating pristanic acid and CO2 (23), with some hints for 2-hydroxyphytanic acid being an intermediate. Starting with the discovery of formate being produced from phytanic acid by Poulos et al. (24), the pathway was revised considerably, with the major findings being the discovery of an α-hydroxylase (25) and a lyase (26) (for review, see 27–29).
Within the cell, phytanic acid (3S or 3R isomer, both present in our diet) is activated to its CoA-ester, which can be either directed to degradation or esterified into triglycerides and phospholipids (30; Fig. 2; supplementary Fig. I). At the subcellular level, the activation can occur in mitochondria, endoplasmic reticulum (ER), and peroxisomes. Based on activity measurements in transfected cells, ACSL12 (33) and ACSVL1 (SLC27A2)3 (37), a long-chain and VLC acyl-CoA synthetase, respectively, can be responsible for this reaction. One generally assumes that phytanoyl-CoA is formed outside peroxisomes. Following its transport into the peroxisomes, the CoA-ester will be hydroxylated at the second carbon by phytanoyl-CoA α-hydroxylase (PHYH) (Fig. 3). This enzyme belongs to the family of dioxygenases and uses Fe2+, 2-oxoglutarate, and O2 as cofactors. It mediates a two-electron oxidation reaction whereby 2-oxoglutarate is converted into CO2 and succinate. One of the oxygen atoms of O2 is incorporated into succinate, the other in the acyl-CoA ester. In vitro, substrate inhibition is seen with phytanoyl-CoA (and long-chain 3-methylacyl-CoA) already starting around 10 µM. This can be counteracted by albumin (38). Others have claimed that the enzyme prefers substrate bound to sterol carrier protein 2 (SCP2) (39), which indeed has a good affinity for phytanoyl-CoA (40).
Fig. 3.
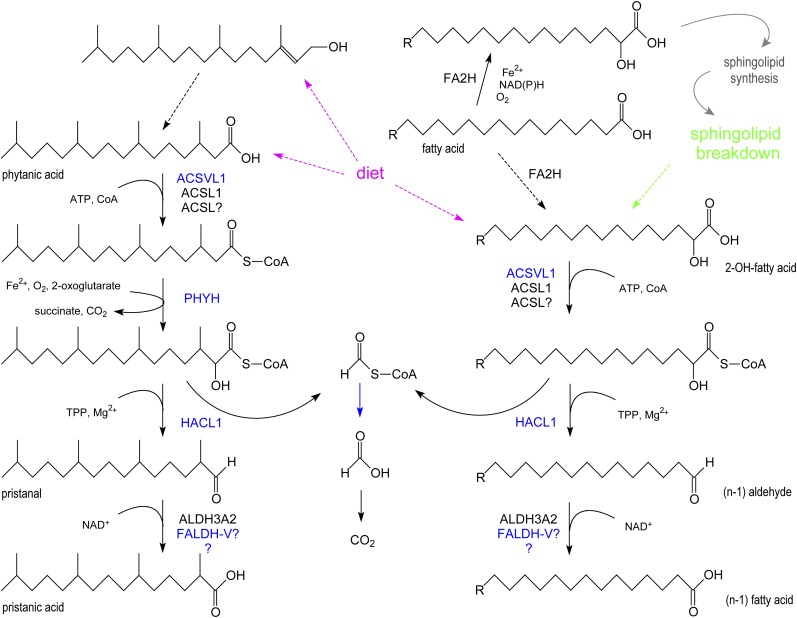
Peroxisomal α-oxidation. At the left side, the revised α-oxidation pathway for phytanic acid is shown. Both phytanic acid and its precursor, phytol, are dietary lipids (magenta dashed arrows). At the right side, the degradation of 2-hydroxy FAs is depicted. 2-Hydroxy FAs are derived from the diet (magenta dashed arrow), formed by lysosomal breakdown of sphingolipids (green dashed arrow), or generated by hydroxylation of FAs in the ER by FA2H (dashed black arrow). The contribution of the latter step seems to be small, and the formed 2OH-FA, after activation, is mainly used for the N-acylation of sphingoid bases resulting in the formation of ceramide species containing 2OH-FA, which are incorporated in sphingolipids. In blue, peroxisomal enzymes/reactions.
PHYH hydroxylates not only phytanoyl-CoA but also other 3-methyl-branched acyl-CoA esters with a chain length of 10–18 carbons. A bulky group at the ω-end (e.g., 5-phenyl-3-methylpentanoyl-CoA) is tolerated, but free 3-methyl FAs are not substrates. Both the length and the position of the branch are important; extending the branch to a 3-ethyl group lowers the activity considerably and 2-methyl- and 4-methylbranched acyl-CoAs are not hydroxylated (38). With an excess of enzyme, a low activity with straight chain acyl-CoAs can be seen (41). Rather strangely, both 3S- and 3R-methyl isomers are recognized but are selectively transformed into the 2R-hydroxy,3S-methyl- and 2S-hydroxy,3R-methyl-, respectively (42). Consequently, both naturally occurring isomers of phytanic acid can be degraded (supplementary Fig. I).
Human PHYH is synthesized as a 38.6 kDa protein and contains a PTS2 at residues 9–17. After import, mediated via PEX7, it is processed to a 35 kDa protein. In the primary amino acid sequence, an oxygenase motif (HXD/E…H) that binds iron is present. By site-directed mutagenesis, the role of H175QD was established, H264 being a likely candidate for the second histidine. A motif for binding 2-oxoglutarate (RXS) is not present, but R275 was identified as the binder of the 5-carboxygroup of 2-oxoglutarate (43).
The formed 2-hydroxyphytanoyl-CoA is cleaved into pristanal and formyl-CoA by 2-hydroxyphytanoyl-CoA lyase (26), now renamed 2-hydroxyacyl-CoA lyase (HACL1) (Fig. 3). This reaction requires thiamine-pyrophosphate (TPP) and Mg2+ as cofactors and is optimal around pH 7.5–8.0 and in the presence of albumin. HACL1 is a tetrameric protein, composed of 60 kDa subunits ending in a noncanonical PTS1, SNM, in humans. A TPP-binding motif (P-X8-GD-S/G-X24-27-NN; prosite PDOC00166), typical for decarboxylases/transketolases, is present in the C-terminal half of the protein. The metal cofactor Mg2+, coordinated to the pyrophosphate group of TPP, is likely bound by D455 and N482 (of human HACL1). The importance of D455 and S456 was confirmed by site directed mutagenesis (P. Fraccascia, M. Casteels, and P.P. Van Veldhoven, unpublished observations). Generally, decarboxylases act on 2-oxocarboxylic acids, generating CO2 and aldehyde. HACL1 (and the closely related oxalyl-CoA decarboxylases) appear to be an exception, whereby a nucleophilic attack is presumed to take place at the carbonyl of the thioester linkage. Whereas HACL1 is mainly peroxisomal in rat liver, subcellular studies on human liver revealed some cleavage activity in the microsomal fraction, which is associated with ER vesicles (V. Foulon, P.P. Van Veldhoven, and M. Casteels, unpublished observations).
Pristanal, formed by HACL1, is transformed into pristanic acid. It is unclear which aldehyde dehydrogenase was/is responsible. The ER-bound fatty aldehyde dehydrogenase ALDH3A2 (previously FALDH or ALDH10), the deficiency of which causes Sjögren-Larsson syndrome (SLS) (44), was postulated to be involved (45). This enzyme is active on pristanal, and the activity is reduced in SLS cells. Subcellular fractionation studies revealed, however, that pristanal (or a synthetic analog) can be oxidized in peroxisomes (and mitochondria, although the bulk activity is microsomal). Recent data suggest that alternative splicing of ALDH3A2 could explain the peroxisomal activity (46). Whereras ALDH3A2 is targeted to the ER, a truncated splice form, named FALDH-V, is directed to peroxisomes (although no apparent PTS is present). However, FALDH-V seems to be a membrane bound enzyme, whereas the peroxisomal pristanal dehydrogenation is catalyzed by a matrix enzyme. Hence, the link with SLS remains debatable: phytanic acid is not accumulating in this disorder (45) and α-oxidation in SLS fibroblasts is normal (47). Pristanal likely can be converted by more than one peroxisomal enzyme and/or can leave the peroxisomal compartment and reenter as pristanic acid after oxidation at another site. Pristanic acid, characterized by a methyl branch at position 2, is a substrate for peroxisomal β-oxidation (see “Substrates for β-oxidation”; supplementary Fig. I).
Regarding the fate of formyl-CoA, the other lyase product, in vitro data suggest that a major4 portion is enzymatically hydrolyzed to formate and CoA in the matrix (Fig. 3). Formate is further oxidized to CO2 in the cytosol, mainly via the folate-dependent one carbon pathway.
Hence, although the endproducts of phytanic acid breakdown were correctly formulated by the pioneers of α-oxidation research, the intermediary metabolites were not and appear be more complex than anticipated.
Substrates for α-oxidation
Although not documented completely, other 3-methyl (isoprenoid) FAs, or indirectly their alcohol/aldehyde precursors, are postulated to use the above described α-oxidation pathway. In fact, this was proven for synthetic phytanic acid analogs like 3-methylhexadecanoic acid (30), 3,14,14-trimethylpentadecanoic acid (49), and the shorter 3,6-dimethyloctanoic acid (50). Notably, various PET-tracers for myocardial imaging (51) are 3-methylbranched FAs5 and are, although slowly, metabolized. Further candidates include metabolites of farnesol, geranylgeraniol, and dolichols. Similarly to the phytol to phytenic/phytanic acid conversion (see “Auxiliary enzymes and proteins related to α-oxidation”), farnesol and geranylgeraniol can be oxidized to 2-enoic/2,3-dihydro acids in mammalian cells (52, 53). Attempts to study the degradation of dolichols are rather limited, although the presence of dolichoic acid was documented several years ago in bovine thyroid (54) and more recently in the substantia nigra in humans (55). When MDCK cells were supplied with [1-14C]dolichoic acid, labeled formate was produced, supporting an α-oxidative mechanism, although apparently not operational in HepG2 cells (56).
Investigation of the substrate specificity of HACL1 revealed that peroxisomes are involved in the degradation of 2OH-FA as documented for long-chain 2-hydroxyoctadecanoic acid (2OH-C18:0) (19) and VLC cerebronic acid (2OH-C24:0) (M. Sniekers, M. Casteels, and P.P. Van Veldhoven, unpublished observations). In contrast to phytanic acid, 2OH-FA are endobiotics, rather abundantly present in cerebrosides and sulfatides of brain and kidney and ceramides of skin. They were postulated to play a role in myelin formation and stability. This seems to be confirmed by recent findings in mice (57) and patients (58) lacking FA α-hydroxylase (FA2H), an ER enzyme responsible for the formation of 2OH-FA. These patients suffer from spastic paraparesis and dystonia caused by degeneration of the white matter (leukodystrophy).
Given extracellularly, the majority of 2OH-FA appear to be degraded, with few molecules being incorporated into structural lipids (19, 59, 60). The steps involved in the degradation of 2OH-FA are an activation, uptake of the CoA-ester in peroxisomes (if not formed in the matrix), and cleavage by HACL1 into formyl-CoA and an (n-1) straight-chain aldehyde (Fig. 3). The latter, similarly to pristanal, is oxidized to a (n-1) FA. Activation of 2OH-FA can be catalyzed by different acyl-CoA synthetases (M. Sniekers, M. Casteels, and P.P. Van Veldhoven, unpublished observations), such as ACSL6, ACSVL1, and ACSVL4. 2OH-FA formed by lysosomal hydrolysis of sphingolipids likely will undergo the same degradative route, whereas hydroxylation of FAs by FA2H and incorporation into sphingolipids are thought to be well-connected processes without release of the intermediary 2OH-FA (61) (Fig. 3).
An alternative pathway for degradation of 2OH-FAs is their oxidation to 2-oxo-FAs by HAO2, a peroxisomal long-chain L-hydroxyacid oxidase. HAO2 prefers medium-chain 2OH-FA, but to be recognized, these must possess the L-configuration, and HAO2 has a limited tissue distribution (mainly kidney). Studies with thiamine depleted cells/animals and the use of thiamine antagonists6 indicate that this probably constitutes a minor pathway and pinpoint HACL1 as the main player (59).
The presence of straight-chain odd-numbered FAs (C23:0, C25:0) in brain is well known. Older in vivo experiments indicated that these are derived from one carbon longer FAs, with 2-OH FAs being an intermediate (62). This fits perfectly with the revised peroxisomal α-oxidation pathway for 2OH-FA as depicted in Fig. 3.
Auxiliary enzymes and proteins related to α-oxidation
As shown in Fig. 2, phytol can be converted to phytanic acid by mammals. Interestingly, this is a (partial) peroxisomal process (22) that differs from the conversion occurring in ruminant bacteria. In mammals, phytol is oxidized to phytenal, followed by dehydrogenation to 2E-phytenic acid. The latter is converted in ER or peroxisomes to a CoA-ester, which is reduced to phytanoyl-CoA by PECR, a peroxisomal long chain 2-enoyl-CoA reductase (Fig. 2). Interestingly, one phytol isomer is preferred in mammals, resulting in formation of 2E-phytenic acid and 2E-phytenoyl-CoA. The phytol/phytenic acid conversion is dependent on (microsomal) ALDH3A2 (63). Because ALDH3A2 generates a peroxisomal splice variant (see above), this process might take place in peroxisomes.
To what extent HSDL2, a member of the short-chain dehydrogenase/reductase family (hydroxysteroid dehydrogenase subfamily) present in peroxisomes (64), can contribute to phytol metabolism is not known. Its substrate has not yet been determined, but the presence of an SCP2 domain is indicative of a role in lipid metabolism. Another member of this family, DHRS4 (originally named carbonyl reductase), is also reported to be a peroxisomal enzyme. It acts as a retinal reductase/retinol dehydrogenase (65), but activity toward phytol/phytenal has not been tested.
Another gene product that can modulate α-oxidation is SCP2, which stimulates in vitro PHYH activity. This could be a nonspecific effect of SCP2, well known for its lipid binding properties (66). The ligand binding cavity of SCP2 can accommodate multiple lipids, including long-chain acyl-CoAs, which are high affinity ligands. Affinity of SCP2 for phytanoyl-CoA (Kd 255 nM) is, however, higher than for other long-chain acyl-CoA-esters or for the acid (40). Hence, SCP2, which contains a PTS1 and is present in the peroxisomal matrix (but also in the cytosol), might be required for proper delivery of substrate to PHYH. In agreement with this idea, SCP2-deficient mice fed a phytol-enriched diet develop a cardiac phenotype (67). However, in these mice, the thiolase required for the degradation of pristanic acid (SCPx; see “3-Oxoacyl-CoA thiolases”) is also absent. On the other hand, a selective accumulation of phytanic acid, but not of pristanic acid, was found in the myocardial phospholipids and might be causative for the high rate of sudden cardiac arrest.
In mice, a peroxisomal acyl-CoA thioesterase (ACOT6) has been described with specificity toward phytanoyl-CoA (and pristanoyl-CoA), with a high expression (based on mRNA level) in white adipose tissue, followed by kidney and brown adipose tissue (68). The ACOT6 gene is present in humans, but expression seems restricted to testis. The physiological function of ACOT6 is unknown, and its CoA ester hydrolysis would be counterproductive with regard to degradation of phytanic acid (or pristanic acid) (Fig. 2). Westin et al. (68) speculate that hydrolysis would mediate exit of phytanic acid (or pristanic acid) from peroxisomes in case of limiting α-oxidation (or β-oxidation) capacity, thereby allowing their esterification into triglycerides or transport to a more catabolically active organ. At least for mice, this would be consistent with the rather selective expression of ACOT6 in adipose tissue.
An extensive discussion about the import of cofactors for α-oxidation does not seem justified given the enigmatic permeability of the peroxisomal membrane described above (see “PEROXISOMAL MEMBRANE PROPERTIES”). Concerning 2-oxoglutarate, it was postulated that isocitrate dehydrogenase (IDH1), ending in a PTS1 and to some extent localized in peroxisomes, would provide this cosubstrate of PHYH. If so, α-oxidation would be dependent on isocitrate, NADP+, and IDH1. Alternatively, based on liposomal reconstitution, the presence of a peroxisomal 2-oxoglutarate/isocitrate transporter was suggested (16). On the other hand, given the properties and exclusion limit of the pore forming PMXP2 (14), the matrix concentration of these small cofactors will reflect the cytoplasmic concentration and carriers are not crucial.
Isolated peroxisomes contain a pool of TPP, most likely bound to HACL1 (69). This cofactor appears to be imported together with HACL1 and hence a specific transporter is not required ((P. Fraccascia, M. Casteels, and P.P. Van Veldhoven, unpublished observations).
To what extent ATP, CoA, or acyl-CoA transporters are involved in α-oxidation depends largely on the locations of synthetases acting on phytanic/phytenic acid and 2OH-FA. This is a complicated issue, because the number of involved acyl-CoA synthetases, their topology, and their contribution are rather unclear. Substrate spectra can overlap, often phytanic acid (and pristanic acid) is not included in specificity studies, and modern tools like expression of tagged proteins can lead to wrong conclusions, because the localization of overexpressed synthetases differs from that of the endogenous protein (36). Various reports claimed that peroxisomal phytanoyl-CoA synthetase activity is facing the cytoplasm, and ACSL1 was thought to be responsible for this reaction (33). Lewin et al. (70), however, argued that ACSL4, based on the use of selective antibodies, is the only long-chain acyl-CoA synthetase (ACSL type) present in (rat liver) peroxisomes. ACSL4 is peripherally associated with the mitochondrial-associated membranes and the peroxisomal membrane. It likely faces the cytoplasm and prefers PUFA, but its activity toward phytanic acid has not been tested. ACSVL1, associated mainly with ER but also with peroxisomes, can also act on phytanic acid (in addition to pristanic acid and various other straight-chain FAs). Its catalytic site is oriented toward the matrix (37). Finally, in mouse skin fibroblasts, ACSVL5 [FA transport protein (FATP) 4] is associated with multiple organelles, including peroxisomes (71), but activity toward phytanic acid was neither reported nor disproved.
So, the best conclusion is that phytanic acid activation can occur on both sides of the peroxisomal membrane. For 2OH-FA, similar conclusions can be drawn. If it proceeds on the outside, the involvement of an ABC-transporter (see “ABCD-transporters and β-oxidation”) can be postulated. If activated within the matrix, CoA and ATP are obviously essential cofactors. CoA, although being present in peroxisomes (72), is not available for this reaction, and a transporter is not known. SLC25A17, a member of the mitochondrial solute transporters (better known as PMP34) is described as a peroxisomal ATP transporter (73, 74). The phytol-induced pathology seen in Slc25a17−/− mice, with accumulation of phytanic and pristanic acid (E. Van Ael, M. Baes, and P.P. Van Veldhoven, unpublished observations), is at least consistent with an intra-peroxisomal need for ATP.
In addition to α-oxidation, phytanic acid can be ω-oxidized via the cytochrome P450 dependent pathway. More specifically, different members of the CYP4 family can ω-hydroxylate phytanic acid in humans (75, 76) (supplementary Fig. II). After further oxidation, breakdown of the resulting ω-carboxyphytanic acid, starting now from the ω-carboxylgroup via β-oxidation, will produce 3-methyladipic acid (3-MAA) as the endproduct (supplementary Fig. II), which is excreted in the urine. Given the presence of a methylgroup in the ω-1 position of ω-carboxyphytanic acid, its shortening is postulated to be carried out by peroxisomes (see “Substrates for β-oxidation”).
Human diseases linked to α-oxidation
In PBDs in which both PTS1 and PTS2 import is impaired (1), the key enzymes of α-oxidation, PHYH and HACL1, are mistargeted, resulting in the accumulation of phytanic acid (10). The mistargeted hydroxylase appears to be degraded in ZS samples, but the lyase remains partly active (77). In RCDP type-1, import of PHYH is affected due to mutations of PEX7 (Table 1). Because RCDP type 1 patients generally live longer than Zellweger patients, phytanic acid elevations, which are age dependent, are easier to document.
PHYH deficiency results in Refsum's disease (OMIM #266500), an autosomal recessive disorder, also known as heredopathia atactica polyneuritiformis (28, 29). In affected individuals, phytanic acid slowly accumulates in tissue lipids, and levels in plasma, based on total lipid analysis, will increase to >200 µM, even up to 5 mM (controls <10 µM). Other α-oxidation substrates (see “Substrates for α-oxidation”) also accumulate; e.g., dihydrogeranylgeranoic acid has been detected in urine and plasma of Refsum patients (78, 79). In late adulthood, the first clinical symptoms appear, notably retinitis pigmentosa and night blindness. Anosmia is a constant feature, and peripheral neuropathy, cerebellar ataxia, and nerve deafness are common abnormalities in later life. If untreated (dietary restriction, plasmapheresis), cardiac arrhythmia can lead to death. A shortened metacarpal or metatarsal is present in some Refsum patients at birth.
About 20 mutations have now been described for PHYH. These can be grouped into several classes: mutations that affect the targeting (P29S), the active sites for binding Fe2+ and/or oxoglutarate (D177G, R275Q, R275W), the overall conformation (W193F, I199F, F257S), or those that result in truncations, frame shift, and deletions (80, 81). In case the mutation affects the 2-oxoglutarate binding site (R275Q, R275W), the activity can be restored by the use of alternative 2-oxoacids (chemical cosubstrate rescue), at least in vitro (82).
In Refsum patients, the above-described ω-oxidation salvage pathway (see “Auxiliary enzymes and proteins related to α-oxidation”; supplementary Fig. II) has been documented (28). Based on the excretion of 3-MAA (and its immediate precursor, 2,6-dimethyloctanedioic acid), it is activated about 2-fold, but apparently this is not sufficient to avoid pathology. Nevertheless, this pathway is of interest, because it can be pharmacologically upregulated and could offer a therapeutic treatment for Refsum disease (28). Currently, therapy is limited to dietary manipulation and plasmapheresis.
Genetic analysis showed that not all Refsum patients carry mutations in the PHYH gene (locus 10p13). Depending on the cohort of patients studied, about 20–50% (83, 84) are due to defects at other loci. The first of these to be established by family linkage studies involves chromosome 6q22-24, which includes PEX7. In the affected family members, impaired import of PHYH (and other PTS2 proteins) was indeed demonstrated, but apparently they display a milder phenotype than classical RCDP type 1 patients. In fact, one of the first patients diagnosed by Refsum belongs to this group (85). Mutations in PEX7 are present, but these allow for some residual functional activity (83). The other loci have not been mapped yet, but 1p32 (SCP2), 22q13.2 (SLC25A17), and 3p24.3 (HACL1) are likely candidates, and maybe 12q24.33 (PXMP2).
For HACL1, no deficiency has been described yet. Because not only phytanic acid but also 2OH-FA will accumulate, patients might present with a more severe phenotype than in Refsum disease. Given TPP is a cofactor for HACL1, thiamine (vitamin B1) supply was postulated to modulate α-oxidation. Indeed, breakdown of 3-methylbranched FAs and 2OH-FA are impaired in cells cultured in presence of thiamine antagonists or in hepatocytes of oxythiamine-treated rats (59). Furthermore, in thiamine responsive megaloblastic anemia (OMIM #249270), a disease caused by mutations in SLC19A2 coding for a thiamine plasma membrane importer, α-oxidation is more sensitive to thiamine depletion (59). This raises the possibility that α-oxidation is affected when dietary thiamine is restricted, such as in beri-beri and Wernicke-Korsakoff syndrome. Thiamine defiency combined with phytol or phytanic acid supplementation is even toxic to rats (59). So far, the effects of thiamine deficiency were only linked to energy processes, never to (brain) lipid abnormalities.
Accumulation of phytanic acid not only is seen in Refsum disease (and PBDs) but is in fact found in all diseases in which the breakdown of its metabolite, pristanic acid, is reduced, most likely due to product inhibition or competition (see “PEROXISOMAL β-OXIDATION”). In some of these patients, clinical symptoms typical for Refsum disease were reported, such as sensory motor neuropathy and adult onset retinitis pigmentosa in patients with 2-methylacyl-CoA racemase deficiency (86) (see “Diseases linked to β-oxidation”), likely caused by the accumulation of pristanic acid. On the other hand, such symptoms are not due per se to phytanic acid accumulation. A Refsum-like disorder has been described, linked to 20p11.21-q12, in which phytanic and pristanic acid levels are normal (87).
Cellular and molecular pathology of phytanic acid
Toxicity of phytanic acid was initially explained by structural homology to other isoprenoids (farnesol, tocopherols), especially to retinoic acid, given the retinopathy. Hence, it could interfere with isoprenoid metabolism and protein prenylation, but few supportive data exist. Another factor is the disturbance of the bilayer packing upon incorporation of phytanic acid in phospholipids, causing changes in proton permeability and likely affecting phospholipid signaling. Phytanic acid accumulation in the peripheral nerves is more pronounced than in the central nerves; phosphatidylcholine of the sciatic nerve of a 57-year-old female Refsum patient was reported to contain 24% phytanic acid (88). This could underlie the peripheral neuropathy and demyelinisation of the peripheral nerves in Refsum disease.
More recently, mitochondria are proposed as the main target of phytanic acid. Some cell types (ciliary ganglion cells, hippocampal astrocytes) (89) do not tolerate well phytanic acid, and in mice lacking PHYH, loss of Purkinje cells and spermatogonia was seen after phytol supplementation (90). In cultured astrocytes, phytanic acid would uncouple complex I, resulting in reactive oxygen species production, cytosolic calcium release, and apoptosis (89, 91, 92); in human skin fibroblasts, a protonophoric action of phytanic acid was documented. Some of the cellular findings can be mimicked with isolated mitochondria exposed to phytanic acid (rather high concentrations of 50–100 µM). To what extent the mitochondrial problems contribute to the pathology of Refsum disease is not known. Intracellular levels of free phytanic acid are normally kept low by binding to fatty acid binding protein (FABP) and SCP2, both abundantly present (0.2–0.4 mM) (93), and a large portion will be converted into phytanoyl-CoA. Effects of phytanoyl-CoA on intra-mitochondrial processes (94) are questionable, because the CoA-ester is not recognized by the carnitine-palmitoyl transferase/translocation machinery (95).
Besides these toxic effects, phytanic acid (but also pristanic acid and phytol) influences transcription by activating nuclear receptors RXRα and PPARα and possibly PPARγ (96). These receptors are not only important regulators of lipoprotein and lipid metabolism but are also linked to carcinogenesis.
PEROXISOMAL β-OXIDATION
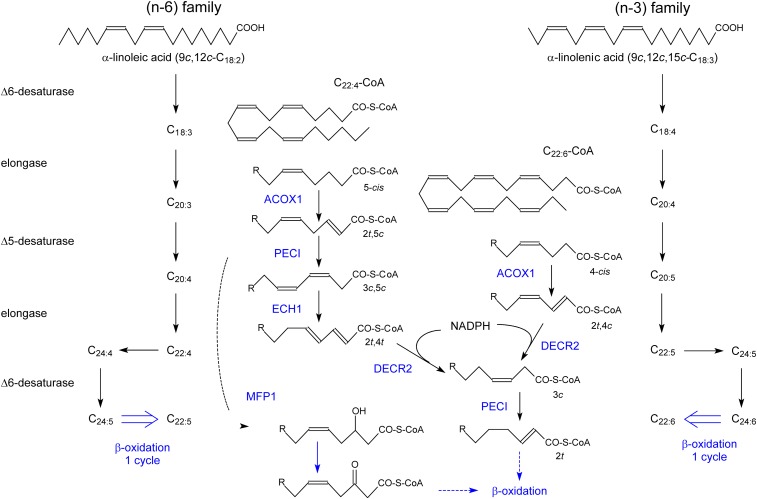
Shortly after the description of a FA β-oxidation pathway in rat liver peroxisomes in 1976 (97), the presence of a similar system in human liver was established (98). The role of this β-oxidation system was initially a controversial issue, partly because it was based on the use of palmitic or oleic acid, but slowly a specialized function emerged and a specific set of substrates were delineated. Based on biochemical work in rodents and clinical data in patients, following substrates are shortened in peroxisomes, either by one (bile acid intermediates) or more cycles, such as VLCFA, pristanic acid, PUFA, eicosanoids, epoxy FAs, xenobiotics, and various ω-oxidized metabolites derived from saturated long-chain FAs, leukotrienes, PUFA, and likely phytanic acid. With regard to PUFA, peroxisomes not only degrade these compounds but are also involved in their formation (see “PUFAs”). Given the role of PUFA in brain-related processes such as memory and behavior in humans, this particular peroxisomal function seems to be undervalued. As a common property, substrates for peroxisomal β-oxidation play no significant role in energy production. Often, these compounds are poor or no substrates for the mitochondrial carnitine palmitoyltransferase (99, 100), which is controlling the entry of long-chain FAs into mitochondria. Furthermore, the presence of a methyl branch at the second carbon relative to the carboxy-thio CoA-ester will determine by which β-oxidation enzymes the particular compound will be degraded (27, 101).
Enzymology of peroxisomal β-oxidation
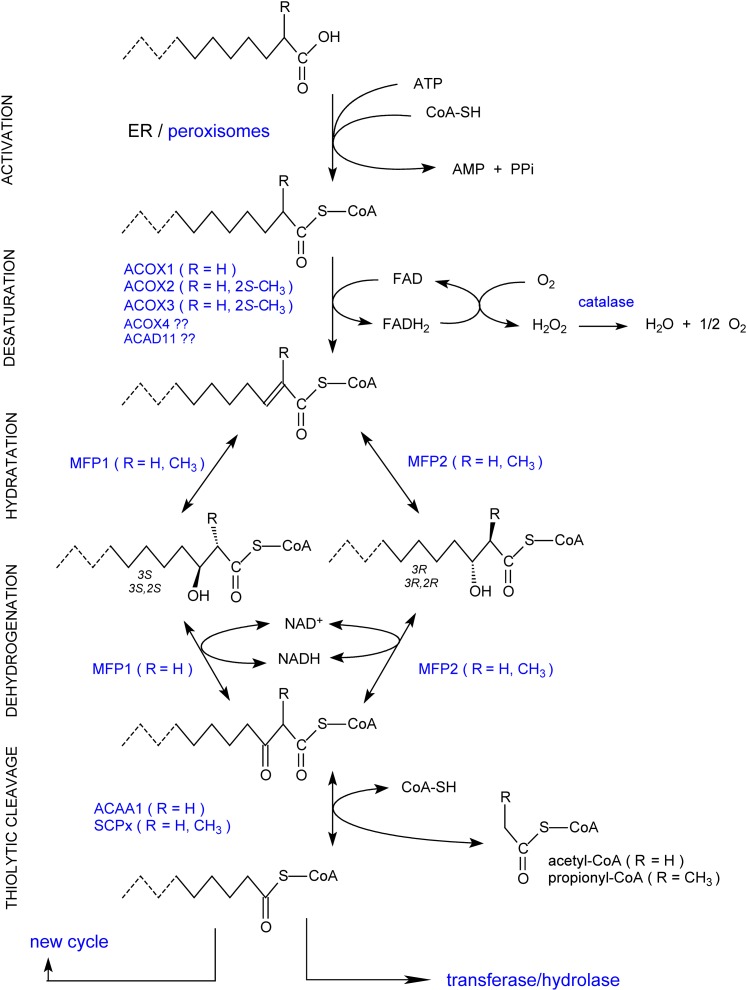
The four basic β-oxidation steps for an acyl-CoA ester, desaturation, hydration of the formed 2E-enoyl-CoA, dehydrogenation of 3-hydroxyacyl-CoA, and thiolytic cleavage of 3-oxoacyl-CoA, are catalyzed by three enzymes in human peroxisomes: an acyl-CoA oxidase, a multifunctional protein (or multifunctional enzyme), and a thiolase (Fig. 4).
Fig. 4.
Overview of peroxisomal β-oxidation reactions. The basic steps of the peroxisomal β-oxidation sequence are shown. Depending on the chain length, substituents, especially the presence of a 2-methyl group, and structure of the acyl-chain of the CoA esters, the involved enzymes will differ. The shortened acyl-CoA can reenter the β-oxidation cycle or undergo other conversions. For the influence of the 2-methyl group configuration, see also Figure 5.
Acyl-CoA oxidases.
In the desaturation step, three acyl-CoA oxidases have been well characterized, differing in substrate spectrum, properties, and tissue distribution7. In an FAD-dependent manner, they introduce a 2-trans bond in the acyl-CoA ester; reoxidation of the produced FADH2 requires molecular oxygen, resulting in the production of H2O2 (Fig. 4). ACOX1 (previously named palmitoyl-CoA oxidase) can only desaturate straight-chain substrates8 and is expressed in most tissues. In vitro highest activity is seen with medium-chain acyl-CoAs, and modifications at the ω-end of the alkyl-chain are tolerated (CoA-esters of dicarboxylic acid, xenobiotics, prostaglandins). Due to a splicing event, the ACOX1 gene encodes two isoforms (a or I and b or II), differing in only a few amino acids encoded by two alternatively used exons. Although this splicing is preserved in mammals and even in lower vertebrates such as fish (102), the role of the isoforms is not entirely clear; tissue distribution and substrate spectrum were reported to be very similar. Recent in vitro data indicate that only (human) isoform b can desaturate VLC acyl-CoAs (103). This would fit with the crystal structure of (rat) ACOXIb revealing a hydrophobic crevice that can bind fatty acyl chains of up to 23 carbon atoms (104).
ACOX2 (originally named trihydroxycholestanoyl-CoA oxidase in rodents and branched-chain acyl-CoA oxidase in humans) acts on 2-methylacyl-CoAs and in humans takes care of cholestanoyl-CoA and pristanoyl-CoA. Additionally, it can also desaturate straight-chain acyl-CoAs. In humans, in contrast to rodents, ACOX2 is ubiquitously expressed. Recognition of 2-methyl-acyl-CoAs by ACOX2 is stereo-specific, because only 2S-methylacyl-CoAs (or 25S for the CoA-esters of bile acid intermediates) are desaturated. As a consequence, an extra enzyme is required to degrade 2R-methylacyl-CoAs (Fig. 5). A third oxidase, ACOX3 (originally named pristanoyl-CoA oxidase), is highly expressed in rat liver but apparently not so in humans and mice. This oxidase recognizes different acyl-CoAs, with or without a 2-methyl branch, but a bulky or constrained group at the ω-end is not tolerated (105). ACOX3, similar to ACOX2, recognizes the 2S-methyl configuration (106). Although no convincing evidence has been reported in human samples for ACOX3 at the protein level, the ACOX3 gene clearly is actively transcribed in humans and modulation of mRNA levels have been described (e.g. in prostate cancer) (107) (see “Perspectives”). Finally, databases suggest the presence of a fourth ACOX in mammals, ACOX4 (or ACOXL), which is most similar to ACOX3, with a lung-specific expression. In humans, ACOX4 mRNA, however, encodes a truncated, likely inactive protein (P.P. Van Veldhoven and E. De Schryver, unpublished observations).
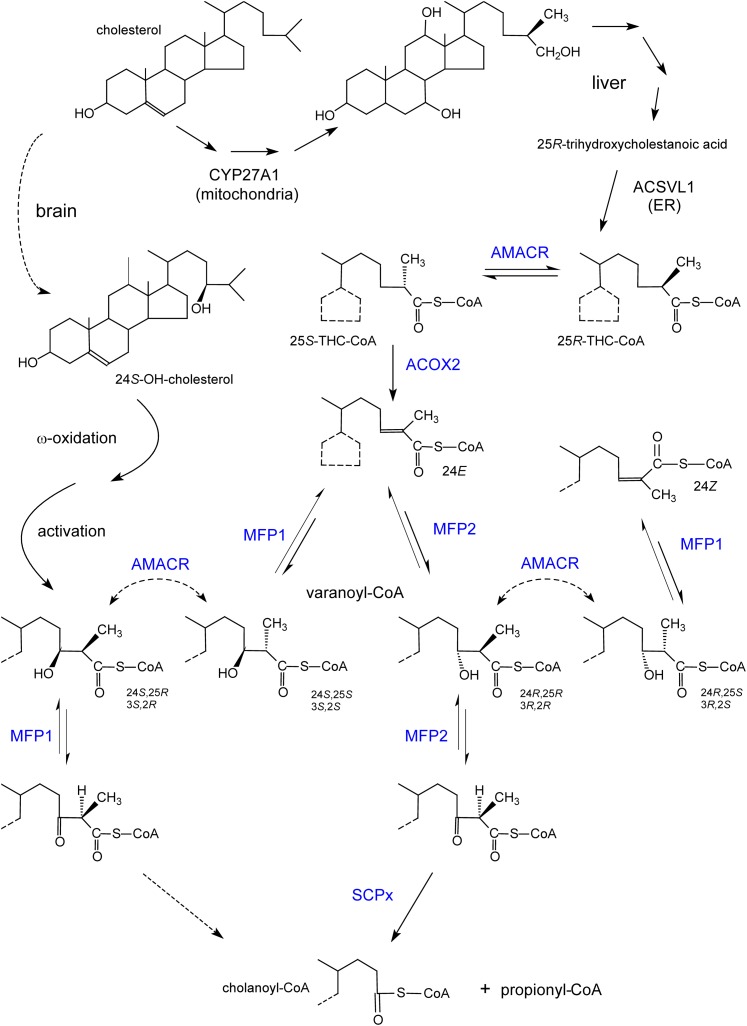
Fig. 5.
Formation of bile acids and stereochemistry of the involved enzymes. Shown at the top, starting from cholesterol through different steps (multiple arrows), mainly 25R-bile acid intermediates (e.g., trihydroxycholestanoic acid) are formed in humans due to the stereospecific reaction of 27-hydroxylase (CYP27A1). After activation in the ER, the CoA-ester is transported into peroxisomes and undergoes a chiral inversion to a 25S-isomer, which is desaturated by ACOX2. The enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase domains of MFP1 and MFP2 can produce or act on only one of the four possible 24-hydroxy-25-methyl-trihydroxycholestanoyl-CoA (also called varanoyl-CoA) stereoisomers. In contrast to the physiological pathway via MFP2 (with 24R-hydroxy-25R-methyl-cholestanoyl-CoA as intermediate), cholanoyl-CoA formation via MFP1 requires the assistance of racemase. A similar reasoning can be applied to pristanic acid (3R-hydroxy-2R-methyl-pristanoyl-CoA as the normal intermediate). The conversion of the 24R,25R-intermediate into 24Z-cholestenoyl-CoA, apparently a dead end product, via AMACR followed by a dehydratation by MFP1, has only been demonstrated in vitro. Shown at the left side, the ω-oxidized and activated product of the brain-derived cerebrosterol (24S-OH-cholesterol), given the 24S-hydroxy configuration, might be converted to primary bile acids via MFP1. Enzymes depicted in blue are peroxisomal.
Mammalian ACOXs display a high degree of amino acid similarity and belong to the superfamily of acyl-CoA dehydrogenases/acyl-CoA oxidases (108). In mammals, all ACOXs have a subunit of about 75 kDa (calculated) and employ FAD as cofactor, the latter being stably (ACOX1) or loosely (ACOX2) bound. ACOX1 and ACOX2 are dimeric and rat ACOX3 is octameric. A conserved glutamate (around amino acid 420–430) constitutes the catalytic site involved in the α-proton abstraction from the substrate whereby the β-hydride is transferred to the enzyme-bound FAD. A FAD binding motif (CGGHGY) is situated in front of this glutamate. Near the N terminal, a conserved stretch of six amino acids (K-W/F-W-I/V/P-G-G/N/D) is present, proposed as an ACOX motif.
After import, ACOX1 is proteolytically processed into the so-called B (50 kDa) and C (20 kDa) subunits that remain together as a heterodimer; the cleavage occurs with both isoforms and is catalyzed by the peroxisomal TYSND1 protease (109). The other oxidases are not cleaved. To measure the different ACOX enzymes, selective assays and/or substrates are required (110), but so far only ACOX1 deficiencies have been diagnosed (see “ACOX1 deficiency”).
2-Enoyl-CoA hydratases and 3-hydroxyacyl-CoA dehydrogenases.
Regarding the multifunctional proteins, the first one discovered, multifunctional protein 1 (MFP1)9, (multifunctional enzyme type 1, originally called bifunctional protein) catalyzes the hydration of 2E-enoyl-CoA into 3S-hydroxyacyl-CoA (3-L-hydroxyacyl-CoA) and dehydrogenation of 3S-hydroxyacyl-CoA (Fig. 4) but also displays Δ3,Δ2-enoyl-CoA isomerase activity. When given 2Z-enoyl-CoA, 3R-hydroxyacyl-CoAs are formed. MFP1 can act on 2-methyl-2E-enoyl-CoA, but the formed 3S-hydroxy-2S-methylacyl-CoA is not a substrate for its dehydrogenase domain (Figs. 4, 5). The second discovered multifunctional protein, MFP210 (D-bifunctional protein; multifunctional enzyme type II) transforms 2E-enoyl-CoA into 3R-hydroxyacyl-CoA (3-D-hydroxyacyl-CoA) and dehydrogenates this product (hence, referred to as D-specific multifunctional enzyme). In case of a 2-methylbranch, the formed 3R-hydroxy-2S-methyl stereoisomer is recognized by the dehydrogenase domain (Figs. 4, 5). The described stereoselectivities of MFP1 and MFP2 are maintained in the reversed reactions. Based on these facts, both MFP1 and MFP2 can act on straight-chain compounds (MFP1 with a preference for shorter chain substrates in vitro), whereas MFP2 is involved in the degradation of compounds containing a 2-methylbranch such as pristanic acid and bile acid intermediates.
Although both MFPs have a subunit size of about 80 kDa, the different stereospecificity suggests already that MFP1 and MFP2 are not homologous. Indeed, their primary structure and domain organization are quite different. MFP1 has an N-terminal 2-enoyl-CoA hydratase domain (amino acids 1–190), which also harbors the enoyl-CoA isomerase activity. Active sites are two acidic amino acids (E104 and E124). Via a stretch of about 90 amino acids, the hydratase domain is linked to the dehydrogenase domain. The latter one consists of two subdomains, one (amino acids 281–474) containing a Rossman fold binding the dinucleotide with a G304XGXXG motif, the other one (amino acids 480–583) displaying homology to the dimerization domain of mitochondrial acyl-CoA dehydrogenases. The last domain carries the PTS1 and might be involved in protein folding and stabilizing the two active domains (111).
N terminally, (human) MFP2 contains a short-chain alcohol dehydrogenase domain (amino acids 1–254) with a Rossmann fold for binding the NAD(H)-cofactor (G16XXXGXG) and a conserved catalytic tetrad (D39-S151-Y164-K168), whereby Y164 is the catalytic site. Centrally, one finds an enoyl-CoA hydratase domain (amino acids 336–603), with E366 and D510 being involved in the catalysis. At the C terminus, a SCP2-like domain (amino acids 612–731) is present ending in a PTS1 (112).
Unlike monomeric MFP1, MFP2 is a dimeric protein, which is partly processed after import by TYSND1 (109) into two dimers corresponding to the dehydrogenase (subunit size of 35 kDa) hydratase plus SCP2-like domains (subunit size of 45 kDa), respectively. Both separate domains are active. The extent of cleavage differs between tissues and species, but processing is not required for activity and MFP2 is active in the absence of peroxisomes.
Regarding the tissue distribution, MFP1 is most abundant in liver and kidney. For MFP2, the highest mRNA levels are found in liver, heart, hippocampus, prostate, and testis of humans (for an extensive review on MFP2, see 112). So far, only MFP2 deficiency has been reported in humans (see “MFP2 deficiency”).
3-Oxoacyl-CoA thiolases.
The thiolytic cleavage is done by either acetyl-CoA acyltransferase (peroxisomal 3-oxoacyl-CoA thiolase; ACAA1)11 or by SCP X (SCPx) (Fig. 4). The latter, a fusion of a thiolase domain and a C terminally SCP2 domain, is derived from the largest transcript of the SCP2 gene; a smaller transcript, with an independently regulated more distal promotor, encodes SCP2. Due to splicing, even more transcripts can be formed, and both SCPx and SCP2 can undergo post-translational processing (113). For SCPx, the proteolysis takes place within the peroxisomal matrix (and for SCP2 at least to some extent). Of the two thiolases present, only SCPx catalyzes the cleavage of 3-oxo-2-methylacyl-CoA (114). Hence, SCPx is required to degrade bile acid intermediates and pristanic acid, and during the cleavage reaction of these substrates, propionyl-CoA is produced.
All enzymes described above, except ACAA1, contain a PTS1. The N-terminal PTS2 of ACAA1 (41 kDa) is cleaved off by TYSND1 after import. In its mature form, this thiolase is a dimeric protein of 40 kDa subunits and is found in most tissues. Important catalytic residues12 are C123, H377, and C408; C123 is positioned close to the 3-oxo-group of the substrate to form an intermediate.
Isolated SCPx is dimeric, being either homo- or heteromeric combinations of 58 and 46 kDa subunits. The processed 46 kDa subunit lacks the SCP2 domain, but all three forms are active. A conserved cysteine (C94 of human SCPx) is proposed as catalytic site. The protein is most abundant in liver, colon, adrenal, and kidney. In adult mice, there is a striking sexual difference in the amount of hepatic SCPx (10-fold less in females) (115, 116). It is not known whether a similar gender effect is present in humans. Regarding disorders related to peroxisomal thiolases, so far only SCPx deficiency has been reported (see “SCPX deficiency”).
2-Methylacyl-CoA racemase.
To degrade compounds containing a 2R-methylbranch, e.g., bile acid intermediates and pristanic acid, the above described enzymes are not sufficient and an extra enzyme is required. In humans, cholesterol is converted via the acidic pathway, involving ω-hydroxylation/oxidation by sterol 27-hydroxylase (CYP27A1), into 3α,7α,12α-trihydroxy-25R-cholestanoic acid (THCA) or 3α,7α-dihydroxy-25R-cholestanoic acid (DHCA). After activation, at the level of the ER, the CoA-ester is transported to and into peroxisomes (Fig. 5). The R-configuration of the methyl branch at carbon 25 (α-positioned with regard to the 26-carboxy group) has to be converted to 25S before the CoA-ester can be desaturated by ACOX2. The racemisation of the methylgroup, which can proceed in both directions, is catalyzed by 2-methylacyl-CoA racemase [α-methylacyl-CoA racemase (AMACR)] (117). Pristanic acid (formed from 3R/S-phytanic acid) is racemic at position 2, while the 6- and 12-methyl groups possess an R-configuration. To get rid of 2R-pristanic acid here, racemisation at the level of the CoA-ester is needed and catalyzed by the same racemase (Fig. 5; supplementary Fig. I). AMACR is a monomeric protein of 43 kDa, present both in peroxisomes and mitochondria. This bimodal distribution, seen in all mammals investigated, is controlled by two targeting sequences, PTS1 at the C-end and a mitochondrial targeting sequence at the N terminal. Mitochondrial acyl-CoA dehydrogenases, similar to acyl-CoA oxidases, are stereospecific13, and mitochondrial AMACR can help to form the proper isomer of short-chain 2-methylbranched acyl-CoA (see “Pristanic acid”). Apparently, no cofactor or cosubstrate is needed for the AMACR reaction, which proceeds via deprotonation and reprotonation of the α-carbon (and an enol/keto tautomerisation of the α-carbanion). Thioesterification of the substrate thereby serves to increase the acidity of the α-carbon hydrogen. Based on the crystal structure of AMACR from Mycobacterium (118), amino acids R87, H122, D123, D152, and E237 of human AMACR are important for catalysis. The histidine base (H122) would abstract a proton from the α-carbon of the substrate in the keto-form and the acidic aspartate (D152) would donate a proton to the enolic form of the bound substrate.
The in vitro finding that AMACR acts on 3-hydroxy-2-methyl-acyl-CoAs (e.g., 24S,25S-varanoyl-CoA into the 24S,25R-stereoisomer), although with a low efficiency compared with acyl-CoA esters with no 3-hydroxy group (119), suggests that there might be a salvage pathway by which 2-methylbranched substrates can be β-oxidized without the involvement of MFP2 but by a sequence of hydration by MFP1, racemisation by AMACR, and dehydrogenation by MFP1 (Fig. 5).
Although outside the scope of this review, it is of interest to note that AMACR controls the action of certain pain relievers (Ibuprofen, Nurofen, Advil, Nuprin). These 2-arylpropionates, an important subclass of nonsteroidal antiinflammatory agents, can indeed be considered as 2-methylbranched FAs. Moreover, only the 2S-isomer is pharmacologically active, but generally the formulation is racemic given the observed epimerisation upon intake. The actual process consists of an activation, followed by chiral inversion of the CoA-ester, and hydrolysis. The responsible enzyme was called 2-arylpropionyl-CoA epimerase (120); however, it is identical to AMACR. In which cell compartment 2-arylpropionates are racemiced in vivo, mitochondria or peroxisomes or both, is not known. Cholestatic liver disease has been seen after Ibuprofen overdose (121) and could be due to deregulation of peroxisomal bile acid β-oxidation. The latter is manifestly seen in AMACR deficiency (see “2-Methyl acyl-CoA racemase deficiency”).
Enzyme complexes.
After the discovery of the enzymes acting on 2-methylbranched substrates, the idea emerged of separate β-oxidation systems with substrate channeling, one for straight chain, another system for branched compounds (122). This hypothesis was strongly supported by the fact that the enzymes not able to act on branched chains are induced severalfold in rodents by peroxisome proliferators, in contrast to other enzymes recognizing branched acyl-CoAs that are not or only modestly increased. As more single enzyme deficiency patients were (correctly) diagnosed (see “ Diseases linked to β-oxidation”) and more knockout mouse models became available (123, 124), a more complicated picture emerged. Whereas some substrates are selectively used, others can be employed by two or more enzymes, as discussed in the next section. Furthermore, given the tissue-dependent content and function of peroxisomes, some caution is warranted when extrapolating from cellular studies on fibroblasts or hepatocytes to other cells or to the situation in the intact organism. Whether or not the above-named enzymes are organized in larger units or complexes allowing substrate channeling is not well studied. Based on the ability to measure both intermediates and end products, enzymes acting on C27-bile acid intermediates (in rat) would not form a tight complex (125). Interestingly, by removing the 25-methylgroup (hence a straight-chain FA attached to a steroid nucleus), other (one or more) enzymes are involved and only end products can be demonstrated. SCP2 can associate with ACOX1, MFP1, and ACAA1 (the interaction with ACOX1 being the strongest) (126), and catalase is reported to associate with MFP1 (127). Kurochkin et al. (109) speculate that the processing of β-oxidation enzymes, notably ACOX1, MFP2, ACAA1, SCP2, and SCPx, by TYSND1 might provoke conformational changes facilitating such complex formation.
Substrates for β-oxidation
In order of their discovery, physiological substrates for peroxisomal β-oxidation include bile acid intermediates, VLCFA, dicarboxylic FAs, xenobiotics, epoxy-FAs, PUFA, prostaglandines, pristanic acid, leukotrienes, thromboxanes, VLC-PUFA, and dicarboxylic PUFA (Fig. 4). Depending on their structure, these substrates can undergo one or more oxidation cycles. The fate of the shortened substrates is diverse; some will be shuttled to mitochondria where they will undergo more cycles (pristanic acid metabolites), a few might be intraperoxisomally converted (long-chain acyl-CoA) or be used for phospholipid synthesis in the ER (PUFA-CoA), whereas others are exported to the bile (primary bile acids) or via blood and kidneys excreted (Fig. 6).
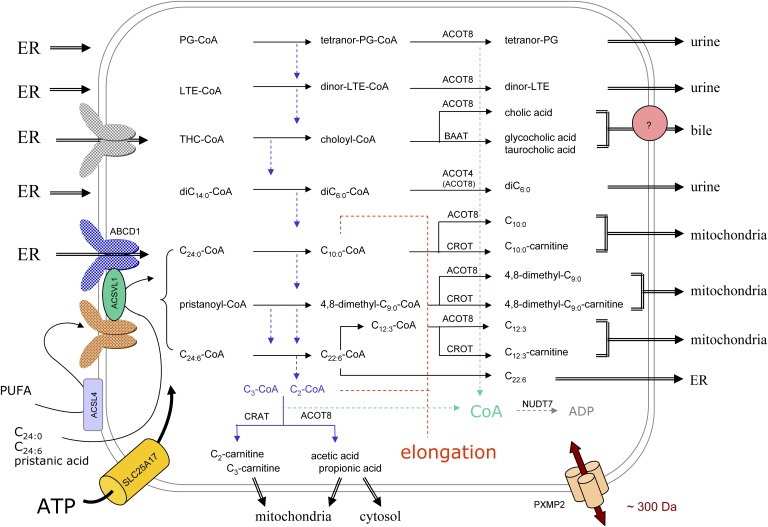
Fig. 6.
Organization of peroxisomal β-oxidation in relation to cellular metabolism. In this scheme, the subcellular origin of the major peroxisomal substrates (left side) and the fate of their degradation products (right side) are depicted. Due to the tissue-specific expression, not all depicted reactions will take place in peroxisomes of a particular cell type. The majority of substrates are activated in the ER (left side) and ABCD transporters, drawn in different colors, likely play a role in the uptake of the CoA-esters. Some FAs can be activated at the peroxisomal membrane or in the matrix, the required ATP entering via SLC25A17, but it is not known how these FAs cross the membrane. Depending on the presence of a 2-methyl group in the substrate, acetyl- or propionyl-CoA is produced (blue dashed arrows) that are normally shuttled to the mitochondria, as carnitine ester or free acid, but can be used for elongation (red dashed lines). The shortened substrates are generally processed in such manner that CoA is formed intra-peroxisomally, apparently a common theme during β-oxidation (green dashed arrows). This CoA can be reused by synthetases or thiolases or degraded by NUDT7. The β-oxidation end products can leave the organelle by passive diffusion, via the PXMP2 pore, or if larger than the pore diameter, via transporters. For bile acid conjugates (433–515 Da), there is biochemical evidence for a transporter (pink circle). Small solutes such as those involved in the conjugation or transferase reactions (glycine, taurine, carnitine) likely enter peroxisomes via the PXMP2 pore.
The acetyl- and propionyl-CoA units are transferred to the mitochondria as carnitine esters or can be released as free acids, which might also enter the mitochondria. The responsible enzymes are a peroxisomal carnitine acetyl-CoA transferase, encoded by CRAT, and an acetyl-CoA thioesterase14 encoded by ACOT8 (see 129 for a review on thioesterases and transferases). Via differential splicing, CRAT gives rise to two mRNAs coding for mitochondrial CRAT (70 kDa) and peroxisomal CRAT, both monomeric proteins. The latter lacks the mitochondrial targeting signal15. CRAT preferably acts on short-chain acyl-CoA (C2-C4) and is especially expressed in muscle and heart. ACOT8 (36 kDa monomeric protein) is considered a “broad range” thioesterase, expressed in most tissues and hydrolyzing short- (acetyl-CoA), medium-, and long-chain CoA-esters as well as esters of prostaglandins and bile acids. Recently, it was suggested that peroxisomally formed acetyl-CoA could be hydrolyzed in rodents by ACOT12, also known as the extra-mitochondrial acetyl-CoA thioesterase (131). ACOT12 was indeed detected during a proteomic analysis of purified kidney peroxisomes, confirming older activity measurements. Whether its carboxy-end, KSVL, considered as a nonconsensus PTS1, is functional has not been established. Human ACOT12, expressed primarily in liver, ends, however, in VSTF, unlikely to be a targeting signal. Depending on the expression of the transferase and thioesterases, the acetate/acetyl-carnitine ratio will be tissue dependent (131). In rat hepatocytes, peroxisomal β-oxidation of dicarboxylic (132) and PUFAs (133) produce mainly acetate. In heart, peroxisomal acetyl-CoA is claimed to be a source for mitochondrial malonyl-CoA (134).
A third pathway for acetyl-CoA, whose physiological significance seems underestimated, is elongation. Whereas data obtained with broken systems or organelles might be difficult to interpret, it is rather convincing that after intravenous injection of [1-14C]lignoceric acid in rats, label is recovered in the 1-C16:1/C18:1-alkenylgroup of the hepatic plasmalogens but not with [1-14C]palmitic acid (135). All peroxisomal β-oxidation enzymes catalyze the reversed reactions, except for ACOXs, but due to the presence of a 2-enoyl-CoA reductase (PECR) (see also “Auxiliary enzymes and proteins related to α-oxidation” and Fig. 2), it is indeed possible to create an elongation pathway. Based on its optimal substrate (136), elongation would start from octanoyl-CoA. In isolated peroxisomes, dodecanoyl-CoA was reported to be the best primer for FA elongation (137).
Regardless of which path prevails, all three assure that CoA remains in the peroxisomal compartment (see Fig. 6). This might be important, because hepatic peroxisomal β-oxidation, in contrast to mitochondrial peroxisomal β-oxidation, is diminished in rats kept on a pantothenate (a precursor of CoA)-deficient diet (138). The shortened substrates also can be subjected to enzymatic conversions whereby CoA is released (Fig. 6). In the subsequent sections, the degradation of some important substrates is described in more detail.
Bile acid intermediates.
Primary bile acids are formed in liver from the longer bile acid intermediates with 27 carbons (cholestanoic acids) and excreted into bile as conjugated bile acids. The latter facilitate the efficient excretion of lipophilic toxins/cholesterol and intestinal absorption of fat-soluble vitamins. Formation of C24-bile acids (cholanic acids) is an exclusive peroxisomal process (139). THCA, derived from cholesterol (Fig. 5), is activated at the cytosolic side of the ER, supposedly by ACSVL1 (140), which is mainly expressed in liver and kidney. The CoA-ester is transported to and taken up by peroxisomes and transformed into choloyl-CoA with one β-oxidation cycle (Fig. 6). In a similar way, DHCA is converted to chenodeoxycholoyl-CoA. Despite this important function, mice lacking ACSVL1 (141) display no obvious abnormalities, implicating additional enzymes acting on cholestanoic acids. ACSVL6, a liver-specific ER enzyme (140), has some activity toward THCA in vitro. The contribution of ACSVL6 seems minor, however, given the normal levels of primary bile acids in knockout mice (142). The majority of these bile acids were unconjugated, confirming primary bile acids as preferential substrates for ACSVL6.
The β-oxidation enzymes needed are ACOX2, MFP2, and SCPx. Prior to desaturation by ACOX2, a chiral change of the 25R-methylgroup by AMACR is needed (Fig. 5). At the end of one cycle, the formed primary bile acid CoA-ester can be hydrolyzed or conjugated (Fig. 6). The hydrolysis has not been studied in detail, but recombinant ACOT8, expressed in liver and other tissues, can act on choloyl-CoA (143). The exact subcellular localization of the bile acid conjugating enzyme(s) has been a matter of debate for some time: depending on the experimental approach and the species investigated, mainly cytosolic or partially peroxisomal and ER bound. In humans, both taurine and glycine conjugation are carried out be one enzyme, bile acid-CoA:amino acid N-acyltransferase (BAAT), which ends in SQL, a less common PTS1-variant. Based on immunocytochemistry of human liver and expression of fusion proteins in hepatocytes, human BAAT is targeted to peroxisomes (144). Upon homogenization of liver, it is partly released from the organelles, explaining its presence in the cytosolic fraction. Apparently, its PTS1 has a weak affinity for PEX5; upon expression in other cell types (fibroblasts, CHO) or upon coexpression in hepatocytes together with another protein with a strong PTS1, BAAT was predominantly cytosolic. Because BAAT is a liver-specific enzyme, the findings in hepatocytes are physiologically relevant and explain some discrepancies in the literature. On the other hand, if no conjugating activity is present in the cytoplasm, peroxisomes are imperative for the reconjugation of secondary bile acids (deoxycholic acid, lithocholic acid) that were deconjugated by the intestinal flora and reabsorbed in the ileum, reentering the liver via portal blood. Because secondary (and primary) bile acids are activated at the ER by ACSVL6 (140, 142, 145), the BAAT localization indicates that peroxisomes are capable of taking up the secondary cholanoyl-CoA-esters.
Detailed analysis of the bile acid composition in patients and mice with MFP2 deficiency revealed, as expected, the presence of C27-bile acids (24E-cholestenoic acids and their 24-hydroxy metabolites). Cholanic acids, although reduced, were still present, suggesting an alternative, perhaps upregulated, pathway. In addition to the 25-hydroxylation pathway, two other scenarios can be described, both involving MFP1 (Fig. 5): 1) 24E-cholestenoyl-CoA can be hydrated by MFP1 to 24S-hydroxy,25S-cholestanoyl-CoA, which is converted by AMACR to the 24S-hydroxy,25R-isomer, which is a substrate for the dehydrogenase of MFP1; or 2) 24S-hydroxycholesterol (cerebrosterol), a cholesterol metabolite excreted from the brain and converted into primary bile acids in humans (146), might enter the β-oxidation as 24S-hydroxy-cholestanoyl-CoA (likely 25R), an isomer that can be dehydrogenated by MFP1, not by MFP2. The bile acid content and composition of bile of adult MFP1-deficient mice is, however, comparable to that of wild-type mice (147), indicative that cerebrosterol-dependent bile acid formation is minor, in agreement with the low 27-hydroxylase activity on cerebrosterol in humans (137). In mice lacking both MFP1 and MFP2, the picture was different; hydroxylated cholestanoic acids16 were the major bile acids, and C24-bile acids were much lower than in MFP2 knockout samples (147). Because ACOX2 was also affected in the double knockout mice, no definitive conclusions can be drawn.
Pristanic acid.
As described in the section on α-oxidation (see “Enzymology of peroxisomal α-oxidation”), pristanic acid can be generated from phytanic acid within peroxisomes (Fig. 2). Given the localization and topology of ACSVL1 (37), this pristanic acid can be activated inside the organelle. On the other hand, pristanic acid derived from dietary sources or if partly formed outside peroxisomes, can be activated at different sites, followed by uptake of the CoA-ester into peroxisomes (or by esterification into glycerolipids at the ER). Recombinant ACSL1 can act on pristanic acid and might be responsible for the mitochondrial and ER activity (33). The latter can also be due to ACSVL1. Whether pristanic acid can be activated at the outside of the peroxisomal membrane, an enduring assumption, is less clear. ACSL1 might not be present in rat liver peroxisomes (70), and ACSL4, peripherally associated with peroxisomes (70), has a more restricted spectrum for PUFAs, but pristanic acid has not been tested as substrate. How pristanoyl-CoA, formed in the cytosol, is taken up is not clear, but a transporter could be involved (see “ABCD-transporters and β-oxidation”; Fig. 6).
ACOX2, MFP2, and SCPx are required for the β-degradation of pristanoyl-CoA. As shown in supplementary Fig. I, both 2R and 2S-pristanoyl-CoA can be formed in human tissues. The 2S-isomer can be directly desaturated; the 2R-isomer is first converted to 2S by AMACR. The product of the first cycle, 4R,8R,12-trimethyltridecanoyl-CoA, undergoes a second cycle, but it has not been established whether the same set of enzymes is involved. Before the third cycle can start, AMACR is again needed to racemise 2R,6R,10-trimethylundecanoyl-CoA (supplementary Fig. I). The product of this cycle, 4R,8-dimethylnonanoyl-CoA, is transported to mitochondria, likely as a carnitine ester (based on fibroblast studies). Peroxisomes do indeed contain an acyl-carnitine transferase (CROT), which displays activity on medium-chain acyl-CoAs, including branched ones like 4,8-dimethylnonanoyl-CoA (148). The carnitine esters enter michondria via the classical carnitine/acylcarnitine carrier and are retroconverted to CoA-esters via carnitine palmitoyl-transferase 2. After one cycle, mitochondrial AMACR comes into play to racemize 2R,6-dimethylheptanoyl-CoA, and β-oxidation halts at the level of isobutyryl-CoA (supplementary Fig. I). Alternatively, 4R,8-dimethylnonanoyl-CoA could leave the peroxisomal compartment as a free acid after hydrolysis by ACOT8 (149). As discussed above for acetyl/propionyl-CoA, CoA is left behind in the peroxisomal matrix with either pathway.
As mentioned in the section on α-oxidation, ω-oxidation is a salvage pathway for phytanic acid breakdown. The formed dicarboxy-compound is degraded from the ω-end, likely in a similar fashion as pristanic acid (supplementary Fig.II). Depending on the stereospecificity of the involved ω-hydroxylase, the ω-1 methyl group will be R or S. Hence, the first cycle of the ω-degradation of dicarboxyphytanic acid might be racemase dependent.
VLCFAs.
VLCFAs, either endogenously synthesized by elongation or from dietary sources, are solely degraded by peroxisomes. In general, the activation of VLCFA is ascribed to ER and peroxisomes. This is based on the initial studies on lignoceric acid (C24:0) degradation performed in rat liver or human skin fibroblasts, but this does not represent the full picture. The rat liver lignoceroyl-CoA synthetase (150) belongs to the SLC27A family, and, in fact, all six members of this family, which also includes FATPs, display synthetase activity toward C24:0 but differ in tissue expression, substrate spectrum, and subcellular localization (34). Furthermore, the brain-specific ACSBG1 (bubblegum, lipidosin) can activate VLCFAs (151), but ACSBG2, expressed in the brainstem and testis, is not active on C24:0 (152). Hence, one cannot predict from in vitro or in situ data the role of these synthetases in an organism. Even the various currently available synthetase knockout mice do not offer proper answers with regard to peroxisomal VLCFA β-oxidation. ACSVL1 is present in peroxisomes, oriented toward the matrix, and at the ER-membrane. Although VLCFA oxidation was reduced in liver of ACSVL1-deficient mice, plasma and tissue VLCFA levels were normal and no obvious phenotype was noticed (141). In brain, skin, and intestines of mice lacking ACSVL5 (FATP4), lignoceroyl-CoA synthetase activity was 8–10-fold lower (153). Tissues in which ACSVL5 is not expressed (liver, lung) were not affected. The reduced activation in Acsvl5−/− murine skin fibroblasts affected the esterification of C24:0 into structural lipids but also peroxisomal C24:0 oxidation (both reduced by <50%) (71). In murine fibroblasts, ACSVL5 was shown to be associated with different intracellular membranes, including peroxisomes. Peroxisomes are, however, devoid of ACSVL6, a liver-specific ER-associated enzyme involved in elongation and lipid synthesis.
After activation, the CoA-ester is taken up (see “ABCD-transporters and β-oxidation”) and desaturated. In humans, as evidenced by work with deficient fibroblasts and the clinical abnormalities in patients, ACOX2, MFP2, and both ACAA1 and SCPx are involved in the degradation of VLCFA. Few data regarding the number of cycles have been reported, and likely during the shortening, other β-oxidation enzymes with higher affinity for shorter chain substrates (e.g., ACOX1, MFP1) might take over. Based on various facts, one assumes that degradation halts at the level of hexanoyl/octanoyl-CoA. These include the high Km of ACOX2 (and ACOX1) for short-chain acyl-CoAs, chain length of accumulating acyl-CoA intermediates in isolated peroxisomes given palmitate, estimated cycles for palmitate in rat hepatocytes, and the chain length specificity of CROT. The latter converts the generated medium-chain acyl-CoAs to carnitine-esters that can leave the peroxisomes and be degraded further in mitochondria. Another export route for medium-chain acyl-CoAs involves the action of thioesterases ACOT8 or ACOT4. The relative expression of CROT and ACOTs will influence the balance between these two exit routes. In adipose tissues, CROT is absent, whereas ACOT4 is highly expressed. If these enzymes are low or not expressed, β-oxidation will be more complete in such tissue (e.g., heart) (129).
However, one should keep in mind that the generated long- or medium-chain acyl-CoA esters can be used for anabolic pathways within peroxisomes. The first one is elongation, mentioned earlier (see “ Substrates for β-oxidation”). The NADPH-dependent peroxisomal 2-enoyl-CoA reductase (PECR) controlling this pathway has a high expression in liver and kidney and has a proper substrate spectrum (C6:1- to C16:1-CoA, optimum with 2-decenoyl-CoA) (136). Palmitoyl-CoA can be employed by peroxisomal glyceronephosphate O-acyltransferase to acylate dihydroxyacetone-phosphate to 1-acyl-dihydroxyacetone-phosphate which can be transformed by alkylglycerone phosphate synthase (AGPS) into 1-alkyl-dihydroxyacetone-phosphate, the obligate precursor of etherlipids/plasmalogens. The long-chain alcohols used by AGPS are derived from long-chain acyl-CoAs (or aldehydes). Upon liver fractionation, the NADPH-dependent acyl-CoA reductase activity has a bimodal distribution, peroxisomal and microsomal. Based on homology to a plant enzyme, two human acyl-CoA reductases, FAR1 and FAR2, were cloned (154). The first one prefers acyl-CoAs with (un)saturated FAs of 16–18 carbons; the second is selective for palmitoyl- and stearoyl-CoA. Though not presenting a defined PTS1, both murine reductases were partially localized to peroxisomes (154). Also, the human counterparts, which are mainly expressed in sebaceous glands and other tissues active in etherlipid synthesis, do not end in a typical PTS1.
Finally, Reilly et al. (155) reported recently that VLC acyl-CoAs can be conjugated to taurine-esters in hepatic peroxisomes of mice. The responsible matrix enzyme, named acyl-CoA:amino acid N-acyltransferase (ACNAT1), has been cloned and is homologous to the bile acid conjugating enzyme BAAT. Whereas the mouse genome likely codes for a second related peroxisomal protein, ACNAT2, it is not clear whether one of these enzymes is expressed in humans. Likely, the murine Acnat1 gene evolved into a BAAT pseudogene in humans.
PUFAs.
Polyenoic acids with 20–22 carbon atoms and 3–6 double bonds (PUFA) are formed from essential FAs by elongation and reduction involving different desaturases (Fig. 7). Due to absence of Δ4-desaturase in mammals, another route is required to form C22:6n-3 from its C22:5n-3 precursor. This pathway consists of an elongation, a Δ6-desaturation, and a single β-oxidation cycle (156) (Fig. 7). This partial shortening, also utilized in the formation of related PUFA (C22:5n-6, C22:4n-9), is referred to as retroconversion. Reduced DHA levels have been reported in Zellweger patients, especially in brain and retina, but also in liver, kidney, and plasma (157). In Pex5−/− mice, brain lipids also contain less DHA, but plasma levels are normal (158).
Fig. 7.
Formation and degradation of PUFA. Starting from the essential FAs oleic acid (not shown), linoleic acid (left), and linolenic acid (right), the various (n-9)-, (n-6)- and (n-3)-PUFA are formed in sequential steps (at the level of CoA-ester). The formation of 22:4(n-9) (not shown), 22:5(n-6), and 22:6(n-3) involves an elongation, a Δ6-desaturase, followed by one β-oxidation cycle in the peroxisomes. In the middle, the degradation of FAs with a 5-cis double bond (left) or a 4-cis double bond (right), either MUFAs or PUFAs such as arachidonoyl-CoA or DHA-CoA, is depicted. Enzymes/reactions in blue are associated with peroxisomes.
For the retroconversion, based on studies in human skin fibroblasts, the enzymes involved are predominantly ACOX1 and MFP2, and both SCPx and ACAA1 (159). This is partly confirmed by mouse models. In Acox1−/− mice, an accumulation of abnormal VLC-PUFA was observed in the hepatic lipids (C24-30 with five and six double bonds), although DHA levels were normal (160). Similarly, in mice lacking MPF2, lipid accumulation is observed in testis, cerebellum, and retina, and abnormal very long PUFA were seen in the FA profiles of testis (161). This suggests that peroxisomes (via ACOX1 and MFP2) constitute a checkpoint on the elongation during C22:6 and C22:5 synthesis; in their absence, an uncontrolled runaway process ensues.
Similar to degradation of saturated and monounsaturated long-chain FAs, arachidonic acid (C20:4) and DHA can be degraded by both peroxisomes and mitochondria. The relative contributions of the organelles are cell and species specific. In human and murine skin fibroblasts, it is a peroxisomal process whereby MFP2 plays a more important role than MFP1 (162). Due to the position of the double bonds in these PUFA, extra enzymes are required to proceed beyond the initial desaturation step. These include a peroxisomal 2,4-dienoyl-CoA reductase (DECR2), Δ3,Δ2-enoyl-CoA isomerase (PECI), and Δ3,5,Δ2,4-dienoyl-CoA isomerase (ECH1) (Fig. 7). Additionally, an enoyl-CoA isomerase activity is associated with MFP1. This set of enzymes is sufficient to degrade MUFAs or PUFAs with a cis-bond at even or odd position or a trans bond at odd-numbered carbons (Fig. 7).
Considering DHA degradation, the CoA-ester is oxidized by ACOX1 to 2trans,4,7,10,13,16,19-docosaheptaenoyl-CoA, which is transformed by DECR2 to 3cis,7,10,13,16,19-docosahexaenoyl-CoA (163) and to 2trans,7,10,13,16,19-docosahexaenoyl-CoA by PECI. After one β-oxidation cycle, the shortened product contains a 5-cis bond and will follow the same path as arachidonoyl-CoA (Fig. 7). In the absence of peroxisomes, shortening of (dietary) PUFA is impaired and uncontrolled elongation can take place. VLC-PUFA (up to C38:6) were described previously in Zellweger brain and are incorporated into the sn-1 position of phosphatidylcholine (164).
Hence, regarding PUFA, peroxisomes have a dual function: synthesis and degradation. How the organelle regulates these opposite processes is not clear. In vitro DECR2, or more correctly the content of its cofactor NADPH, controls degradation.
Medium- and long-chain α,ω-dicarboxylic acids.
In conditions of FA overload (diabetes, fatty liver disease) or intoxication by agents that block mitochondrial β-oxidation (hypoglycin, valproate), ω-oxidized long-chain FAs are formed, mainly in liver and kidney. This process is initiated by CYP4 enzymes acting on the terminal methylgroup. In addition to long-chain FAs, various other carboxylates can be ω-hydroxylated, and depending on the chain length, substituents, and double bonds, different CYP4, belonging to the F and A subfamilies, are involved (165, 166). The ω-hydroxy-FAs are further oxidized to dicarboxylic acids, which are excreted with or without prior shortening via β-oxidation. Depending on chain length and substituents, degradation can occur in mitochondria and/or peroxisomes. Long- and medium-chain dicarboxylic acids are activated in the ER and mainly degraded by peroxisomes. The responsible synthetase is not known. Recently, ω-oxidation of VLCFA was reported in human microsomes, with ω-hydroxylation being catalyzed by CYP4F2 and CYP4F3B (167). Whether this pathway is operational in vivo is not known. The longest dicarboxylic saturated FAs reported in ZS patients are hexa- and pentadecanedioic acid in urine (168) and octadecanedioyl-carnitine in plasma (169).
Based on studies with single enzyme-deficient fibroblasts, combined with clinical data (170), the enzymes degrading hexadecanedioic acid are ACOX1, both MFP1 and MFP2, and SCPx (and maybe ACAA1). In mouse hepatocytes, MFP1 contributes more than MFP2 (171). Based on the presence of ACOT8 and its activity on medium-chain dicarboxylyl-CoA (glutaryl- to dodecanedioyl-CoA) (172), long-chain dicarboxylyl-CoAs are thought to undergo 2–4 cycles. The shortest product formed by isolated rat liver peroxisomes was adipate (173). The fact that mouse peroxisomes contain another thioesterase, ACOT4, which is highly specific for succinyl-CoA, suggests that degradation can go even further. In humans, ACOT4 does not, however, display such specificity and hydrolyzes a variety of other CoA-esters17. The shortened dicarboxylic acids are exported and excreted in the urine, with both even and odd numbers of atoms.
DHA and other PUFA can also undergo ω-oxidation. In this case, the transformation serves to generate bioactive compounds causing vasodilatation and activation of PPARα and -γ (175). These dicarboxy-PUFAs are exclusively degraded by peroxisomes whereby MFP1 plays an important, but not essential, role (176).
Eicosanoids and docosanoids.
Eicosanoids are a group of biologically active compounds derived from (n-6) or (n-3) C20-FAs, with arachidonic acid as the major precursor. Depending on the enzymatic route followed, one distinguishes prostaglandins, prostacyclins, and thromboxanes (generated via cyclooxygenases; also called prostanoids), leukotrienes, lipoxins, and oxylipins (via lipoxygenases), and epoxy- and ω/ω-1-hydroxy-derivatives (via cyt P450 epoxygenases) (see also previous section) (177). Common to these compounds is their local synthesis, local action (para- or autocrine), short half-life, and minor plasma concentrations (<1 nM). After attenuation, often by a hydroxylation or a reduction of a double bond (177), the inactivated eicosanoids undergo further degradation, including ω- and/or β-oxidation, and are finally excreted. Based on analysis of the urinary metabolites in healthy individuals, these compounds undergo one to two β-oxidation cycles. The following prostanoids have been shown to depend on peroxisomes for their shortening: PGF2α (178), 8-iso-PGF2α (179), and TB2 (180, 181). Also for epoxy-eicosatrienoic acids (EET) and monohydroxy-eicosatrienoic acids (12-HETE,15-HETE) (182), leukotrienes (LTB4) (183, 184), and cysteinyl-leukotrienes (LTE4) (183, 185), there is substantial evidence for their peroxisomal degradation. The latter are degraded starting from the ω-end (as dicarboxy compounds; see “Medium- and long-chain α,ω-dicarboxylic acids”). With regard to the involved enzymes, prostaglandins are desaturated by ACOX1 (105); likewise, ACOX1 is needed to degrade 11,12-epoxy-eicosatrienoic acid (11,12-EET) (186). Based on the urinary metabolites in patients, MFP2 is involved in LTB4 and LTE4 shortening, whereas ACAA1 can be excluded (187).
Likely, docosanoids that constitute another class of bioactive lipids involved in antiinflammatory and immunoregulatory processes but derived from C22-PUFAs (188) are subject to a similar degradation in peroxisomes.
Auxiliary enzymes and new candidates
In addition to the auxiliary enzymes already mentioned, a few others remain to be discussed. Screening for PTS1-proteins in humans (V. Mahieu and P.P. Van Veldhoven, unpublished observations; AJ608287) and proteomics on rat liver peroxisomes (2) revealed a multi-domain protein, ACAD11. The C-terminal half of this 87 kDa protein displays similarity to acyl-CoA dehydrogenases (hence its name) and ends in a functional PTS1. Although the function of the other domain, carrying an aminoglycoside phosphotransferase signature, cannot be linked to known peroxisomal functions, similar proteins are found in lower vertebrates, plants, and yeast, all containing a PTS1. Apparently, ACAD11 (and ACAD10, which has a similar domain structure) is encoded by one of the three ancestral acyl-CoA dehydrogenase genes (189); therefore, ACAD11, which is expressed in brain, liver, and kidney, might catalyze a rather essential reaction in β-oxidation (Fig. 4).
SCP2, similar to its role in α-oxidation (see “Auxiliary enzymes and proteins related to α-oxidation”), is thought to assist in substrate delivery. Acyl-CoA esters bind readily to SCP2, the highest affinities being measured with VLC acyl- and 2-enoyl-CoA (190). Hence, given their physical interaction with SCP2, ACOX1 and MFP1 are the prime candidates to be regulated by SCP2. This idea is, however, contradicted by the fact that VLCFA levels are normal in a mouse lacking SCP2/SCPX (39).
As mentioned in the section on membrane properties (section 2), peroxisomes contain two members of the Nudix family. Nudix enzymes are found in all organisms and hydrolyze a wide range of organic pyrophosphates (18). Human NUDT12 converts NADH to NMNH and AMP and NADPH to NMNH and 2′,5′-ADP. It also hydrolyses NAD(P)+ but has a preference for the reduced cofactors (Km NADH = 11 µM; Km NAD+ = 190 µM). Given this specificity, one speculates that NUDT12 regulates the concentration of peroxisomal nicotinamide nucleotide cofactors or is needed to allow exit of the cofactor. How NUDT12 is regulated is unknown. The other hydrolase, mouse NUDT7, cleaves CoA and oxidized CoA into 3′,5′-ADP and the corresponding 4′-phosphopantetheine. More detailed investigations showed that NUDT7 acts also acyl-CoAs, both medium- and long-chain acyl-CoAs, even choloyl-CoA and pristanoyl-CoA (191, 192). The physiological role of NUDT7 is unclear. The existence of acyl-phosphopantetheine esters has never been reported and whether they serve a special role is unknown. Theoretically, NUDT7 could play a role in the export of acyl-CoAs formed by β-oxidation, but its presence causes two problems: its action has to be controlled to avoid hydrolysis before shortening and it will compete with carnitine transferases and thioesterases.
Different from α-oxidation, hydrogen peroxide is produced during β-oxidation (Fig. 4). Hence, catalase is added to this list of auxiliary enzymes.
To what extent peroxisomal β-oxidation is dependent on transporters is uncertain (see “ PEROXISOMAL MEMBRANE PROPERTIES”). The main enzymes are dependent on FAD, NAD+, and CoA; NADPH is required for the breakdown of unsaturated FAs; and carnitine, taurine, or glycine is required for the product conversions. As a general rule, β-oxidation substrates are activated outside peroxisomes, implying the uptake of the CoA-ester; consequently, intraperoxisomal ATP would not be essential (Fig. 6). In accord, bile acids and VLCFA are normal in a mouse lacking PMP34 (E. Van Ael, M. Baes, and P.P. Van Veldhoven, unpublished observations), considered to be an ATP transporter. Murine SLC22A21, a member of the carnitine solute carrier family, is claimed to be peroxisomal (193); in humans, based on in silico data, this member might not be present. Because most conversions of the β-oxidation products release CoA in the matrix (Fig. 6), this seems to serve a purpose such as recycling by the thiolases.
ABCD-transporters and β-oxidation
Imbedded in the peroxisomal membrane are ABC-transporters, members of a superfamily of proteins that bind ATP and transport various molecules across biomembranes (194, 195). The three currently known peroxisomal members, ABCD1 (ALDP), ABCD2 (ALDRP), and ABCD3 (PMP70), belong to the same subfamily and are half transporters. A fourth half transporter, ABCD4 (P70R), initially reported as peroxisomal was shown recently to be ER localized (196). Perhaps this discrepancy is due to cell or species differences, but ABCD4 was also not identified during proteomic investigations (2, 4, 197). To be functional, the peroxisomal halftransporters have to dimerize, either as homo- or heteromers. Heterodimerization is certainly possible in vitro or upon overexpression (198). The most physiological approaches suggest that ABCD1 and ABCD3 would form homodimers (198, 199). Given the topology of the transporters and their ATP binding site, located at the cytosolic site, the direction of transport is toward the matrix (Fig. 6). The ligands are considered to be acyl-CoA esters, VLC-acyl-CoA for ABCD1 and (putatively) pristanoyl-CoA/phytanoyl-CoA for ABCD3, although direct proof is missing so far. Knockout mouse models for all three transporters have been created and might be helpful to answer some intriguing questions.
The main biochemical abnormality in X-ALD, due to mutations in ABCD1, is the accumulation of C24:0 and C26:0 in tissues and fluids. In X-ALD fibroblasts, oxidation of VLCFA is reduced (to 30%) as well as their activation. Expression of wild-type ABCD1 restores VLCFA oxidation in X-ALD fibroblasts. Similarly, overexpression of ABCD3 or ABCD2 corrects partially, respectively completely, the defect in X-ALD cells, indicative that the functions of the transporters are partially redundant. Why this redundancy is not operative in X-ALD is not known. One possibility might be spatial separation. Expression of the transporters is tissue specific in humans and mice, and a marked cell specificity is seen in some tissues (brain and adrenals; so-called mirror expression). Based on mouse models, ABCD2 would play a role in the uptake of VLCFA in dorsal root ganglia but not in fibroblasts. Another peculiarity is the fact that all other lipophilic acids, known to be degraded by peroxisomes, appear normal in X-ALD (phytanic acid, pristanic acid, dicarboxylic acids, leukotrienes, PUFAs). Finally, it seems that VLCFA levels are not per se linked to β-oxidation; the latter can be reduced without causing VLCFA accumulation in mice (141).
Diseases linked to β-oxidation
In PBD, a plethora of abnormalities due to deficient β-oxidation have been documented: accumulations of abnormal bile acids, VLCFA, pristanic acid, dicarboxylic acids, and metabolites of prostanoids and leukotrienes and decreased PUFA (10). In fact, these apply only to the ZS-spectrum. In RCDP type 1, no signs for a β-oxidation defect are seen. This can be explained by the broad spectrum of SCPx taking over the function of the nonimported ACAA1. Given the complexity of the β-oxidation, enzymes with overlapping substrate specificities and opposite stereospecificities, correct diagnosis sometimes took several years and likely some defects might not show up (e.g., ACAA1). Today, although more than 25 proteins are related to peroxisomal β-oxidation (Table 2), only six disorders with a single protein deficiency have been clearly diagnosed. The best characterized one is X-ALD, also the most common peroxisomal β-oxidation disease. The other ones, in sequence of their discovery, are ACOX1, MFP2, AMACR, and SCPX deficiencies. One should notice that the previously described MFP1 (200) and ACAA1 deficiencies (201) were wrongly diagnosed (202, 203). The number of reported patients is low, the largest group being linked to MFP2. In Table 1, a compilation is given with common clinical and biochemical abnormalities. For more information on these disorders, see (204).
X-ALD.
Adrenoleukodystrophy can present with different manifestations depending on the presence and type of neurologic findings (194). Based on the age of onset and affected organs, patients are classified in six groups. The most affected group, also with the earliest manifestations, is childhood cerebral adrenoleukodystrophy. Around the first decade, symptoms appear in boys, starting with altered behavior, visual and auditory problems, and motor incoordination progressing to a vegetative state.
The first abnormalities reported in X-ALD were cholesteryl esters with abnormal long FAs (C24:0 to C30:0) in brain, adrenals, and testis. Due to a defective ABCD1 transporter, VLCFA in fact accumulate in all tissues, but the disease affects primarily brain myelin, adrenal cortex, and testis. As seen in PBD and other β-oxidation disorders, these VLCFA can be further elongated (to C30:0) (205). Currently, 1058 mutations of ABCD1 have been reported, 519 of which are unique (see www.X-ald.nl). The real trigger for perturbation of the adrenal steroidogenesis, axonal degeneration in spinal cord, and the progressive inflammatory demyelination in brain is not known. One hypothesis is that VLCFA cause a change in lipid antigens [gangliosides; (lyso)phospholipids] that are presented via CD1 molcules to the immune system (206).
ACOX1 deficiency.
Since the initial description of two siblings by Poll-The et al. (207) in 1988 as pseudo-neonatal ALD, about 25 patients have now been described (103). Accumulation of VLCFA and impaired cellular degradation of VLCFA is the main biochemical abnormality. In one patient, however, plasma VLCFAs were normal (208). With more elaborated analysis, abnormal metabolites of epoxyeicosatetraenoic are revealed (186). All patients show neurological abnormalities such as hypotonia, seizures, hearing loss, retardation, and retinopathy. Sometimes facial dysmorphism is reported. Developmental regression starts around 2 to 3 years, with progressive brain white matter demyelination, psychomotor problems, and peripheral neuropathy. The mean age at death is 5 years. When investigated, peroxisomes are less in number but are enlarged.
About 20 mutations have been mapped, including a large deletion. One interesting patient is affected in the alternatively spliced exon 3, such that only isoform a is produced (103). Also, VLCFA accumulate in this patient. The missing isoform is active on VLC-acyl-CoA, while the produced isoform prefers medium-chain acyl-CoA (in vitro). In a mouse model, not only saturated VLCFA were documented, but also presence of VLC-PUFA (C24-30 with five and six double bonds) (160).
MFP2 deficiency.
Although the first patient was investigated in 1989, in fact before the discovery of MFP2, the correct diagnosis was only published in 1997 (209). MFP2 or D-bifunctional protein deficiency is a relatively frequent disorder and currently the largest group of patients, second to X-ALD (210). About 130 patients have been diagnosed. Most children suffer from hypotonia, infantile seizures, and delayed development and die within the first 2 years of life, pneumonia being the main cause of death. In about two-thirds of the cases, facial dysmorphia is present. Depending on which domain of MFP2 is affected, one can subdivide these patients into three groups with a total absence of enzyme based on immunotechniques (type 1), deficiency of the 2-enoyl-CoA hydratase activity (type 2), or a problem with the dehydrogenase reaction (type 3). Type 1 patients have a more severe phenotype, and their expected life span is shorter (<14 months). Longer living patients, mostly type 3 patients (eldest 8–10 year; mean age at death, 17.6 months) can achieve psychomotor development. C26:0 oxidation activity in skin fibroblasts seems to be predictive for life expectancy.
Biochemically, VLCFA and abnormal C27-bile acids accumulate in plasma, as does pristanic acid (and secondarily, phytanic acid); however, not all patients show all these abnormalities. Depending on the defect, the composition of bile acids differs. In addition to THCA and 24E-THCA, 24S-hydoxy,25S-THCA, likely formed via MFP1 (Fig. 5), is present when the hydratase domain is affected. In type III, the main accumulating bile acid is 24R-hydroxy,25R-THCA. In urine, taurine conjugates of hydroxylated THCA and 24E-THCA are characteristic. VLCFA and pristanic acid degradation is impaired in fibroblasts.
2-Methylacyl-CoA racemase deficiency.
In 1998, Sequeira et al. (211) described a case with malabsorption and steatorrhea and elevated levels of THCA and pristanic acid, which responded to bile acid therapy. It was classified as “Niemann-Pick disease type C with a defective peroxisomal β-oxidation.” Subsequent genetic analysis (86) showed a mutation in AMACR. Presently, seven patients have been described in the literature (86, 212–216). It cannot be excluded that older descriptions of cases with bile acid abnormalities represent AMACR deficiency. As predicted, the accumulating isomer is 25R-THCA. Moreover, a peculiar C29-dicarboxy bile acid, originally identified in ZS, is also found (139, 213).
The clinical picture is diverse and, despite the small number of cases, might be divided in two age groups. The younger patients had a shortage of soluble vitamins, coagulopathy, and cholestatic liver disease, with or without hypotonia and motor delay. The malabsorption can be linked to the accumulation of mainly unconjugated 25R-THCA and the lack of conjugated primary bile acids in the intestines. At a later age, in addition to bile acid abnormalities, elevations in pristanic acid become evident. Due to the pristanic acid elevation, these patients can resemble Refsum disease (pigmentary retinopathy). Adult patients can become quite old, with mild symptoms. One female patient was even asymptomatic till the age of 48 (86). Common signs are adult-onset sensorimotor neuropathy, seizures, and visual problems, but tremor and cognitive decline also have been reported. In addition to bile acid therapy, restriction of phytanic acid/pristanic acid is recommended. A child who was not treated died at 6 months from intracranial bleed (202, 213).
SCPX deficiency.
The single report so far (217) concerns a 44-year-old male presenting with torticollis, dystonic head tremor, hyposmia, and nystagmus. MRI revealed leukencephalopathy and impaired nerve conduction, predominantly motor and slight sensory neuropathy. Hypergonadotrophic hypogonadism and azoospermia were noticed at a younger age. Pristanic acid was elevated in plasma and its oxidation in fibroblasts impaired. In urine, large amounts of bile alcohols were noticed and glucuronides of penta- and hexahydroxy-27-nor-5β-cholestane-24-one. Similar ketones were seen in the SCP2/SCPx knockout mouse (218) and are formed by decarboxylation of 3-oxo-THCA (or its CoA-ester). The bile alcohols indicate the upregulation of an alternative microsomal 25-hydroxylase pathway. The mutation affected only SCPX; SCP2 was still present. A brother with similar neurological and fertility problems was likely affected as well.
BAAT deficiency.
Familial hypercholanemia, a syndrome characterized by fat malabsorption, elevated serum bile acid concentrations, and itching, can be caused by mutations in BAAT (219) as well as in other genes, including TJP2 (219) and EPHX1 (220), encoding tight junction protein-2 and microsomal epoxide hydrolase 1 (EPHX1), respectively. In serum of children with a mutated BAAT (M76V), taurine- and glycine-conjugated bile acids were absent. Malabsorption, due to lack of conjugated bile acids in the gastrointestinal tract, causes failure to thrive and vitamin K shortage with coagulopathy. Because BAAT is clearly associated with peroxisomes in liver (144), this entity can be considered a novel peroxisomal disorder. The symptoms can be treated with ursodeoxycholic acid supplementation.
Perspectives
Clearly, the key enzymes of peroxisomal β-oxidation are important to human health, but various deficiencies remain to be discovered and diagnozed. Insight in the phenotype of these disorders can be provided by mouse models. Presently, these include ABCD2 (221), ABCD3 (222), MFP1 (223), and ACAA1b (224), and SLC25A17 (E. Van Ael, M. Baes, and P.P. Van Veldhoven, unpublished observations) knockout models, all present, however, with mild or no pathology under normal conditions.
An obvious hiatus in the list of human disorders is ACOX2 deficiency. Based on the phenotype of AMACR deficiency, such patients will suffer from neonatal cholestasis, fat malabsorption, and coagulation problems. Furthermore, it is clear that different (peroxisomal) acyl-CoA hydrolase and acyl-CoA synthetase deficiencies might influence peroxisomal metabolism. ACSVL5 knockout mice, with a decreased VLCFA oxidation in ACSVL5-expressing tissues, present with a tight rigid skin, causing breathing problems and neonatal death (153). This phenotype resembles ichthyosis prematurity syndrome (OMIM #608649). Similarly, in skin fibroblasts of ACSVL5 (FATP4)-deficient patients, VLCFA-CoA synthetase activity is reduced (225). Whether the skin abnormalities in these patients are caused only by a reduced synthesis of epidermal ceramides, rich in 2OH-VLCFA and VLCFA, remains to be proven.
To conclude this section, evidence is emerging that an impaired β-oxidation is linked to cancer and that β-oxidation is upregulated in certain cancers. In rodents, links among hypolipidemic fibrates, peroxisome proliferation, peroxisomal β-oxidation, and hepatocarcinoma are well established (226). More striking is the spontaneous development of liver tumors in Acox1−/− mice, which is due to sustained activation of PPARα by unmetabolized substrates of ACOX1 (227). Also in MFP2 knockout mice, PPARα activation was noticed (112). On the other hand, in many cancers, enzymes acting on branched substrates are upregulated. A remarkable example is the overexpression of AMACR in prostate cancer and different other neoplasms (96, 228). Also, MFP2 and ACOX3 are upregulated in transformed tissues such as prostate and breast cancer (107). Although the role and exact transcriptional regulation of these enzymes in cancer is not known, AMACR is currently already employed as a marker (P504S) for prostate cancer (228) and expression of MFP2 as a predictor of poor patient outcome (229). Given the correlation between consumption of high amounts of red meat and some cancers, it is thought that phytanic acid and pristanic acid are the responsible dietary factors, acting via nuclear receptors (96). This might be an overly simplistic view, because these branched FAs are also present in fish, a potentially cancer protective food, beneficial to general health, and the apparent trouble-free life of ruminants.
Supplementary Material
Acknowledgments
The author is grateful to Dr. J. Jaeken (UZ-Leuven, Leuven) and Dr. L. Van Maldergem (Centre de Génétique Humaine, Liège) for access to patient-related material and thanks his colleagues Dr. M. Fransen, Dr. M. Casteels, and Dr. M. Baes, and the referees for their valuable comments. He would like to dedicate this paper to one of his mentors, Dr. G. Mannaerts, on the occasion of his retirement.
Footnotes
Abbreviations:
- 2OH-FA
- 2-hydroxy FA
- ABC
- ATP binding cassette
- ACSL
- long-chain acyl-CoA synthetase
- ACSVL
- very long-chain acyl-CoA synthetase
- CMC
- critical micellar concentration
- PTS
- peroxisome targeting signal
- PMP
- peroxisomal integral membrane protein
- RCDP
- rhizomelic chondrodysplasia punctata
- SCP
- sterol carrier protein
- SLC
- solute carrier
- SLS
- Sjögren-Larsson syndrome
- THCA
- 3α,7α,12α-trihydroxycholestanoic acid
- TPP
- thiamine-pyrophosphate
- VLC
- very long-chain
- ZS
- Zellweger syndrome
- X-ALD
- X-linked adrenoleukodystrophy
Financial support was received from the Flemish government (Geconcerteerde Onderzoeksacties 2004/08), Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (G.0721.10N), and K.U.Leuven (OT/09/045).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
Latency refers to the fact that an enzyme activity cannot be measured unless the membrane is broken. Catalase is an exception but this is due to its kinetics whereby diffusion becomes a limiting factor.
The nomenclature of acyl-CoA synthetases has been revised considerably during the last years; symbols according to current guidelines (31, 32). ACSL stands for acyl-CoA synthetase long-chain.
The approved gene symbol for this enzyme is SLC27A2. In this paper, the acyl-CoA synthetase designation proposed by Watkins (34) is followed for the members of the SLC27A family, ACSVL standing for acyl-CoA synthetase VLC. This provides more consistency with the ACSL symbols used. Moreover, recent data indicate that these proteins do not act as transporters as suggested by their current gene symbol, but enhance transport by transforming FAs into CoA-esters (35, 36).
At physiological pH, formyl-CoA has a limited stability and might chemically decompose (48).
15-p-[123I]iodophenyl-3(R,S)-methylpentadecanoic acid (3MIPP) is an approved radiotracer for clinical use.
Based on similar experiments an alternative route for phytanic acid breakdown involving PHYH, hydrolysis and oxidation by HAO2 is considered to be minor or not existing.
Initially, these enzymes were named according their (assumed specific) substrate(s), however this created substantial confusion since these appear to differ between species. In concert with HUGO, a more systematic nomenclature was introduced, based on ACOX as acronym followed by a number.
Defined as having no methyl group in the first 3 carbon atoms.
The gene acronym is EHHADH (enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase).
The official gene acronym HSD17B4 (17-hydroxysteroid dehydrogenase type IV) refers to a postulated role in steroid metabolism. Although a low activity is seen in vitro with 17β-estradiol, the MFP2-deficiency phenotype points to a major role of the gene product in β-oxidation, and not in sterol dehydrogenation.
Contrary to rodents, which express two peroxisomal isoforms encoded by separate homologous genes, only one gene is present in man. Likely different proteins can be generated by splicing.
Based on the full-length protein.
The short-branched chain acyl-CoA dehydrogenase can act on 2R-methylacyl-CoA but will form 2-alkylacrylyl-CoA.
For recent changes in nomenclature of ACOTs, see (128).
The original description (130) of a peroxisomal specific mRNA in man is not supported by EST analysis. Instead an exon skipping mechanism as operational in mice, is responsible.
Position of the hydroxyl-group was not analyzed but presumably a mixture of different acids.
Human ACOT4 combines the activities of three separate peroxisomal murine esterase encoded by Acot3, Acot4 and Acot5 (174).
REFERENCES
- 1.Goldfischer S., Moore C. L., Johnson A. B., Spiro A. J., Valsamis M. P., Wisniewski H. K., Ritch R. H., Norton W. T., Rapin I., Gartner L. M. 1973. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 182: 62–64. [DOI] [PubMed] [Google Scholar]
- 2.Kikuchi M., Hatano N., Yokota S., Shimozawa N., Imanaka T., Taniguchi H. 2004. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 279: 421–428. [DOI] [PubMed] [Google Scholar]
- 3.Mi J., Kirchner E., Cristobal S. 2007. Quantitative proteomic comparison of mouse peroxisomes from liver and kidney. Proteomics. 7: 1916–1928. [DOI] [PubMed] [Google Scholar]
- 4.Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., et al. 2007. Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell. Proteomics. 6: 2045–2057. [DOI] [PubMed] [Google Scholar]
- 5.Ma C., Subramani S. 2009. Peroxisome matrix and membrane protein biogenesis. IUBMB Life. 61: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platta H. W., Erdmann R. 2007. The peroxisomal protein import machinery. FEBS Lett. 581: 2811–2819. [DOI] [PubMed] [Google Scholar]
- 7.Brocard C., Hartig A. 2006. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta. 1763: 1565–1573. [DOI] [PubMed] [Google Scholar]
- 8.Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., Fujiki Y. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275: 21703–21714. [DOI] [PubMed] [Google Scholar]
- 9.Van Ael E., Fransen M. 2006. Targeting signals in peroxisomal membrane proteins. Biochim. Biophys. Acta. 1763: 1629–1638. [DOI] [PubMed] [Google Scholar]
- 10.Wanders R. J., Waterham H. R. 2005. Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67: 107–133. [DOI] [PubMed] [Google Scholar]
- 11.de Duve C., Baudhuin P. 1966. Peroxisomes (microbodies and related particles). Physiol. Rev. 46: 323–357. [DOI] [PubMed] [Google Scholar]
- 12.Van Veldhoven P. P., Just W. W., Mannaerts G. P. 1987. Permeability of the peroxisomal membrane to cofactors of beta-oxidation. Evidence for the presence of a pore-forming protein. J. Biol. Chem. 262: 4310–4318. [PubMed] [Google Scholar]
- 13.Antonenkov V. D., Rokka A., Sormunen R. T., Benz R., Hiltunen J. K. 2005. Solute traffic across mammalian peroxisomal membrane–single channel conductance monitoring reveals pore-forming activities in peroxisomes. Cell. Mol. Life Sci. 62: 2886–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rokka A., Antonenkov V. D., Soininen R., Immonen H. L., Pirila P. L., Bergmann U., Sormunen R. T., Weckstrom M., Benz R., Hiltunen J. K. 2009. Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS ONE. 4: e5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonenkov V. D., Hiltunen J. K. 2006. Peroxisomal membrane permeability and solute transfer. Biochim. Biophys. Acta. 1763: 1697–1706. [DOI] [PubMed] [Google Scholar]
- 16.Visser W. F., van Roermund C. W., Ijlst L., Waterham H. R., Wanders R. J. 2007. Metabolite transport across the peroxisomal membrane. Biochem. J. 401: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber G., Islinger M., Weber P., Eckerskorn C., Volkl A. 2004. Efficient separation and analysis of peroxisomal membrane proteins using free-flow isoelectric focusing. Electrophoresis. 25: 1735–1747. [DOI] [PubMed] [Google Scholar]
- 18.McLennan A. G. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63: 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foulon V., Sniekers M., Huysmans E., Asselberghs S., Mahieu V., Mannaerts G. P., Van Veldhoven P. P., Casteels M. 2005. Breakdown of 2-hydroxylated straight chain fatty acids via peroxisomal 2-hydroxyphytanoyl-CoA lyase: a revised pathway for the alpha-oxidation of straight chain fatty acids. J. Biol. Chem. 280: 9802–9812. [DOI] [PubMed] [Google Scholar]
- 20.Casteels M., Sniekers M., Fraccascia P., Mannaerts G. P., Van Veldhoven P. P. 2007. The role of 2-hydroxyacyl-CoA lyase, a thiamin pyrophosphate-dependent enzyme, in the peroxisomal metabolism of 3-methyl-branched fatty acids and 2-hydroxy straight-chain fatty acids. Biochem. Soc. Trans. 35: 876–880. [DOI] [PubMed] [Google Scholar]
- 21.Lough A. K. 1977. The phytanic acid content of the lipids of bovine tissues and milk. Lipids. 12: 115–119. [DOI] [PubMed] [Google Scholar]
- 22.van den Brink D. M., Wanders R. J. 2006. Phytanic acid: production from phytol, its breakdown and role in human disease. Cell. Mol. Life Sci. 63: 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mize C. E., Avigan J., Steinberg D., Pittman R. C., Fales H. M., Milne G. W. 1969. A major pathway for the mammalian oxidative degradation of phytanic acid. Biochim. Biophys. Acta. 176: 720–739. [DOI] [PubMed] [Google Scholar]
- 24.Poulos A., Sharp P., Singh H., Johnson D. W., Carey W. F., Easton C. 1993. Formic acid is a product of the alpha-oxidation of fatty acids by human skin fibroblasts: deficiency of formic acid production in peroxisome-deficient fibroblasts. Biochem. J. 292: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihalik S. J., Rainville A. M., Watkins P. A. 1995. Phytanic acid alpha-oxidation in rat liver peroxisomes. Production of alpha-hydroxyphytanoyl-CoA and formate is enhanced by dioxygenase cofactors. Eur. J. Biochem. 232: 545–551. [DOI] [PubMed] [Google Scholar]
- 26.Foulon V., Antonenkov V. D., Croes K., Waelkens E., Mannaerts G. P., Van Veldhoven P. P., Casteels M. 1999. Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc. Natl. Acad. Sci. USA. 96: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Veldhoven P. P., Casteels M., Mannaerts G. P., Baes M. 2001. Further insights into peroxisomal lipid breakdown via alpha- and beta-oxidation. Biochem. Soc. Trans. 29: 292–298. [DOI] [PubMed] [Google Scholar]
- 28.Wanders R. J., Komen J. C. 2007. Peroxisomes, Refsum's disease and the alpha- and omega-oxidation of phytanic acid. Biochem. Soc. Trans. 35: 865–869. [DOI] [PubMed] [Google Scholar]
- 29.Wierzbicki A. S. 2007. Peroxisomal disorders affecting phytanic acid alpha-oxidation: a review. Biochem. Soc. Trans. 35: 881–886. [DOI] [PubMed] [Google Scholar]
- 30.Van Veldhoven P. P., Huang S., Eyssen H. J., Mannaerts G. P. 1993. The deficient degradation of synthetic 2- and 3-methyl-branched fatty acids in fibroblasts from patients with peroxisomal disorders. J. Inherit. Metab. Dis. 16: 381–391. [DOI] [PubMed] [Google Scholar]
- 31.Mashek D. G., Bornfeldt K. E., Coleman R. A., Berger J., Bernlohr D. A., Black P., DiRusso C. C., Farber S. A., Guo W., Hashimoto N., et al. 2004. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J. Lipid Res. 45: 1958–1961. [DOI] [PubMed] [Google Scholar]
- 32.Watkins P. A., Maiguel D., Jia Z., Pevsner J. 2007. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48: 2736–2750. [DOI] [PubMed] [Google Scholar]
- 33.Watkins P. A., Howard A. E., Gould S. J., Avigan J., Mihalik S. J. 1996. Phytanic acid activation in rat liver peroxisomes is catalyzed by long-chain acyl-CoA synthetase. J. Lipid Res. 37: 2288–2295. [PubMed] [Google Scholar]
- 34.Watkins P. A. 2008. Very-long-chain acyl-CoA synthetases. J. Biol. Chem. 283: 1773–1777. [DOI] [PubMed] [Google Scholar]
- 35.Milger K., Herrmann T., Becker C., Gotthardt D., Zickwolf J., Ehehalt R., Watkins P. A., Stremmel W., Fullekrug J. 2006. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J. Cell Sci. 119: 4678–4688. [DOI] [PubMed] [Google Scholar]
- 36.Jia Z., Pei Z., Maiguel D., Toomer C. J., Watkins P. A. 2007. The fatty acid transport protein (FATP) family: very long chain acyl-CoA synthetases or solute carriers? J. Mol. Neurosci. 33: 25–31. [DOI] [PubMed] [Google Scholar]
- 37.Steinberg S. J., Wang S. J., Kim D. G., Mihalik S. J., Watkins P. A. 1999. Human very-long-chain acyl-CoA synthetase: cloning, topography, and relevance to branched-chain fatty acid metabolism. Biochem. Biophys. Res. Commun. 257: 615–621. [DOI] [PubMed] [Google Scholar]
- 38.Foulon V., Asselberghs S., Geens W., Mannaerts G. P., Casteels M., Van Veldhoven P. P. 2003. Further studies on the substrate spectrum of phytanoyl-CoA hydroxylase: implications for Refsum disease? J. Lipid Res. 44: 2349–2355. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd M. D., Mukherji M., Kershaw N. J., Chien W., Wierzbicki A. S., Schofield C. J. 2003. Role of phytanoyl-CoA 2-hydroxylase in phytanic acid metabolism. Adv. Exp. Med. Biol. 544: 303–304. [DOI] [PubMed] [Google Scholar]
- 40.Seedorf U., Raabe M., Ellinghaus P., Kannenberg F., Fobker M., Engel T., Denis S., Wouters F., Wirtz K. W., Wanders R. J., et al. 1998. Defective peroxisomal catabolism of branched fatty acyl co-enzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes Dev. 12: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Searls T., Butler D., Chien W., Mukherji M., Lloyd M. D., Schofield C. J. 2005. Studies on the specificity of unprocessed and mature forms of phytanoyl-CoA 2-hydroxylase and mutation of the iron binding ligands. J. Lipid Res. 46: 1660–1667. [DOI] [PubMed] [Google Scholar]
- 42.Croes K., Foulon V., Casteels M., Van Veldhoven P. P., Mannaerts G. P. 2000. Phytanoyl-CoA hydroxylase: recognition of 3-methyl-branched acyl-coAs and requirement for GTP or ATP and Mg(2+) in addition to its known hydroxylation cofactors. J. Lipid Res. 41: 629–636. [PubMed] [Google Scholar]
- 43.McDonough M. A., Kavanagh K. L., Butler D., Searls T., Oppermann U., Schofield C. J. 2005. Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease. J. Biol. Chem. 280: 41101–41110. [DOI] [PubMed] [Google Scholar]
- 44.Rizzo W. B. 2007. Sjogren-Larsson syndrome: molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol. Genet. Metab. 90: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verhoeven N. M., Jakobs C., Carney G., Somers M. P., Wanders R. J., Rizzo W. B. 1998. Involvement of microsomal fatty aldehyde dehydrogenase in the alpha-oxidation of phytanic acid. FEBS Lett. 429: 225–228. [DOI] [PubMed] [Google Scholar]
- 46.Ashibe B., Hirai T., Higashi K., Sekimizu K., Motojima K. 2007. Dual subcellular localization in the endoplasmic reticulum and peroxisomes and a vital role in protecting against oxidative stress of fatty aldehyde dehydrogenase are achieved by alternative splicing. J. Biol. Chem. 282: 20763–20773. [DOI] [PubMed] [Google Scholar]
- 47.Jansen G. A., van den Brink D. M., Ofman R., Draghici O., Dacremont G., Wanders R. J. 2001. Identification of pristanal dehydrogenase activity in peroxisomes: conclusive evidence that the complete phytanic acid alpha-oxidation pathway is localized in peroxisomes. Biochem. Biophys. Res. Commun. 283: 674–679. [DOI] [PubMed] [Google Scholar]
- 48.Croes K., Van Veldhoven P. P., Mannaerts G. P., Casteels M. 1997. Production of formyl-CoA during peroxisomal alpha-oxidation of 3-methyl-branched fatty acids. FEBS Lett. 407: 197–200. [DOI] [PubMed] [Google Scholar]
- 49.Try K. 1968. The in vitro omega-oxidation of phytanic acid and other branched chain fatty acids by mammalian liver. Scand. J. Clin. Lab. Invest. 22: 224–230. [DOI] [PubMed] [Google Scholar]
- 50.Stokke O. 1969. The degradation of a branched chain fatty acid by alternations between alpha- and beta-oxidations. Biochim. Biophys. Acta. 176: 54. [DOI] [PubMed] [Google Scholar]
- 51.Tamaki N., Morita K., Kuge Y., Tsukamoto E. 2000. The role of fatty acids in cardiac imaging. J. Nucl. Med. 41: 1525–1534. [PubMed] [Google Scholar]
- 52.Fliesler S. J., Keller R. K. 1995. Metabolism of [3H]farnesol to cholesterol and cholesterogenic intermediates in the living rat eye. Biochem. Biophys. Res. Commun. 210: 695–702. [DOI] [PubMed] [Google Scholar]
- 53.Kodaira Y., Usui K., Kon I., Sagami H. 2002. Formation of (R)-2,3-dihydrogeranylgeranoic acid from geranylgeraniol in rat thymocytes. J. Biochem. 132: 327–334. [DOI] [PubMed] [Google Scholar]
- 54.Steen L., Van Dessel G., de Wolf M., Lagrou A., Hilderson H. J., De Keukeleire D., Pinkse F. A., Fokkens R. H., Dierick W. S. 1984. Identification and characterization of dolichyl dolichoate, a novel isoprenoic derivative in bovine thyroid. Biochim. Biophys. Acta. 796: 294–303. [DOI] [PubMed] [Google Scholar]
- 55.Ward W. C., Guan Z., Zucca F. A., Fariello R. G., Kordestani R., Zecca L., Raetz C. R., Simon J. D. 2007. Identification and quantification of dolichol and dolichoic acid in neuromelanin from substantia nigra of the human brain. J. Lipid Res. 48: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 56.Van Houte H. A., Van Veldhoven P. P., Mannaerts G. P., Baes M. I., Declercq P. E. 1997. Metabolism of dolichol, dolichoic acid and nordolichoic acid in cultured cells. Biochim. Biophys. Acta. 1347: 93–100. [DOI] [PubMed] [Google Scholar]
- 57.Zoller I., Meixner M., Hartmann D., Bussow H., Meyer R., Gieselmann V., Eckhardt M. 2008. Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration. J. Neurosci. 28: 9741–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edvardson S., Hama H., Shaag A., Gomori J. M., Berger I., Soffer D., Korman S. H., Taustein I., Saada A., Elpeleg O. 2008. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 83: 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sniekers M., Foulon V., Mannaerts G. P., Van M. L., Mandel H., Gelb B. D., Casteels M., Van Veldhoven P. P. 2006. Thiamine pyrophosphate: an essential cofactor for the alpha-oxidation in mammals–implications for thiamine deficiencies? Cell. Mol. Life Sci. 63: 1553–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrero A. B., Astudillo A. M., Balboa M. A., Cuevas C., Balsinde J., Moreno S. 2008. Levels of SCS7/FA2H-mediated fatty acid 2-hydroxylation determine the sensitivity of cells to antitumor PM02734. Cancer Res. 68: 9779–9787. [DOI] [PubMed] [Google Scholar]
- 61.Alderson N. L., Rembiesa B. M., Walla M. D., Bielawska A., Bielawski J., Hama H. 2004. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 279: 48562–48568. [DOI] [PubMed] [Google Scholar]
- 62.Kishimoto Y., Akanuma H., Singh I. 1979. Fatty acid alpha-hydroxylation and its relation to myelination. Mol. Cell. Biochem. 28: 93–105. [DOI] [PubMed] [Google Scholar]
- 63.van den Brink D. M., van Miert J. N., Dacremont G., Rontani J. F., Jansen G. A., Wanders R. J. 2004. Identification of fatty aldehyde dehydrogenase in the breakdown of phytol to phytanic acid. Mol. Genet. Metab. 82: 33–37. [DOI] [PubMed] [Google Scholar]
- 64.Kowalik D., Haller F., Adamski J., Moeller G. 2009. In search for function of two human orphan SDR enzymes: hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O). J. Steroid Biochem. Mol. Biol. 117: 117–124. [DOI] [PubMed] [Google Scholar]
- 65.Lei Z., Chen W., Zhang M., Napoli J. L. 2003. Reduction of all-trans-retinal in the mouse liver peroxisome fraction by the short-chain dehydrogenase/reductase RRD: induction by the PPAR alpha ligand clofibrate. Biochemistry. 42: 4190–4196. [DOI] [PubMed] [Google Scholar]
- 66.Seedorf U., Ellinghaus P., Roch N. J. 2000. Sterol carrier protein-2. Biochim. Biophys. Acta. 1486: 45–54. [DOI] [PubMed] [Google Scholar]
- 67.Monnig G., Wiekowski J., Kirchhof P., Stypmann J., Plenz G., Fabritz L., Bruns H. J., Eckardt L., Assmann G., Haverkamp W., et al. 2004. Phytanic acid accumulation is associated with conduction delay and sudden cardiac death in sterol carrier protein-2/sterol carrier protein-x deficient mice. J. Cardiovasc. Electrophysiol. 15: 1310–1316. [DOI] [PubMed] [Google Scholar]
- 68.Westin M. A., Hunt M. C., Alexson S. E. 2007. Peroxisomes contain a specific phytanoyl-CoA/pristanoyl-CoA thioesterase acting as a novel auxiliary enzyme in alpha- and beta-oxidation of methyl-branched fatty acids in mouse. J. Biol. Chem. 282: 26707–26716. [DOI] [PubMed] [Google Scholar]
- 69.Fraccascia P., Sniekers M., Casteels M., Van Veldhoven P. P. 2007. Presence of thiamine pyrophosphate in mammalian peroxisomes. BMC Biochem. 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewin T. M., Van Horn C. G., Krisans S. K., Coleman R. A. 2002. Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch. Biochem. Biophys. 404: 263–270. [DOI] [PubMed] [Google Scholar]
- 71.Jia Z., Moulson C. L., Pei Z., Miner J. H., Watkins P. A. 2007. Fatty acid transport protein 4 is the principal very long chain fatty acyl-CoA synthetase in skin fibroblasts. J. Biol. Chem. 282: 20573–20583. [DOI] [PubMed] [Google Scholar]
- 72.Van Veldhoven P. P., Mannaerts G. P. 1986. Coenzyme A in purified peroxisomes is not freely soluble in the matrix but firmly bound to a matrix protein. Biochem. Biophys. Res. Commun. 139: 1195–1201. [DOI] [PubMed] [Google Scholar]
- 73.Wylin T., Baes M., Brees C., Mannaerts G. P., Fransen M., Van Veldhoven P. P. 1998. Identification and characterization of human PMP34, a protein closely related to the peroxisomal integral membrane protein PMP47 of Candida boidinii. Eur. J. Biochem. 258: 332–338. [DOI] [PubMed] [Google Scholar]
- 74.Visser W. F., van Roermund C. W., Waterham H. R., Wanders R. J. 2002. Identification of human PMP34 as a peroxisomal ATP transporter. Biochem. Biophys. Res. Commun. 299: 494–497. [DOI] [PubMed] [Google Scholar]
- 75.Komen J. C., Wanders R. J. 2006. Identification of the cytochrome P450 enzymes responsible for the omega-hydroxylation of phytanic acid. FEBS Lett. 580: 3794–3798. [DOI] [PubMed] [Google Scholar]
- 76.Xu F., Ng V. Y., Kroetz D. L., de Montellano P. R. 2006. CYP4 isoform specificity in the omega-hydroxylation of phytanic acid, a potential route to elimination of the causative agent of Refsum's disease. J. Pharmacol. Exp. Ther. 318: 835–839. [DOI] [PubMed] [Google Scholar]
- 77.Jansen G. A., Denis S., Verhoeven N. M., Jakobs C., Wanders R. J. 2000. Phytanic acid alpha-oxidation in man: identification of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal enzyme with normal activity in Zellweger syndrome. J. Inherit. Metab. Dis. 23: 421–424. [DOI] [PubMed] [Google Scholar]
- 78.Dulaney J. T., Williams M., Evans J. E., Costello C. E., Kolodny E. H. 1978. Occurrence of novel branched-chain fatty acids in Refsum's disease. Biochim. Biophys. Acta. 529: 1–12. [DOI] [PubMed] [Google Scholar]
- 79.Evans J. E., Dulaney J. T. 1983. Location of double bonds in two unsaturated forms of phytanic acid from Refsum disease as determined by mass spectrometry. Biochim. Biophys. Acta. 752: 346–352. [DOI] [PubMed] [Google Scholar]
- 80.Jansen G. A., Hogenhout E. M., Ferdinandusse S., Waterham H. R., Ofman R., Jakobs C., Skjeldal O. H., Wanders R. J. 2000. Human phytanoyl-CoA hydroxylase: resolution of the gene structure and the molecular basis of Refsum's disease. Hum. Mol. Genet. 9: 1195–1200. [DOI] [PubMed] [Google Scholar]
- 81.Schofield C. J., McDonough M. A. 2007. Structural and mechanistic studies on the peroxisomal oxygenase phytanoyl-CoA 2-hydroxylase (PhyH). Biochem. Soc. Trans. 35: 870–875. [DOI] [PubMed] [Google Scholar]
- 82.Mukherji M., Kershaw N. J., MacKinnon C. H., Clifton I. J., Wierzbicki A. S., Schofield C. J., Lloyd M. D. 2001. ‘Chemical co-substrate rescue’ of phytanoyl-CoA 2-hydroxylase mutants causing Refsum's Disease. Chem. Commun. (Camb.). 972–973. [Google Scholar]
- 83.van den Brink D. M., Brites P., Haasjes J., Wierzbicki A. S., Mitchell J., Lambert-Hamill M., de Belleroche J., Jansen G. A., Waterham H. R., Wanders R. J. 2003. Identification of PEX7 as the second gene involved in Refsum disease. Am. J. Hum. Genet. 72: 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wierzbicki A. S., Mitchell J., Lambert-Hammill M., Hancock M., Greenwood J., Sidey M. C., de Belleroche J., Gibberd F. B. 2000. Identification of genetic heterogeneity in Refsum's disease. Eur. J. Hum. Genet. 8: 649–651. [DOI] [PubMed] [Google Scholar]
- 85.Horn M. A., van den Brink D. M., Wanders R. J., Duran M., Poll-The B. T., Tallaksen C. M., Stokke O. H., Moser H., Skjeldal O. H. 2007. Phenotype of adult Refsum disease due to a defect in peroxin 7. Neurology. 68: 698–700. [DOI] [PubMed] [Google Scholar]
- 86.Ferdinandusse S., Denis S., Clayton P. T., Graham A., Rees J. E., Allen J. T., McLean B. N., Brown A. Y., Vreken P., Waterham H. R., et al. 2000. Mutations in the gene encoding peroxisomal alpha-methylacyl-CoA racemase cause adult-onset sensory motor neuropathy. Nat. Genet. 24: 188–191. [DOI] [PubMed] [Google Scholar]
- 87.Fiskerstrand T., Knappskog P., Majewski J., Wanders R. J., Boman H., Bindoff L. A. 2009. A novel Refsum-like disorder that maps to chromosome 20. Neurology. 72: 20–27. [DOI] [PubMed] [Google Scholar]
- 88.MacBrinn M. C., O'Brien J. S. 1968. Lipid composition of the nervous system in Refsum's disease. J. Lipid Res. 9: 552–561. [PubMed] [Google Scholar]
- 89.Kahlert S., Schonfeld P., Reiser G. 2005. The Refsum disease marker phytanic acid, a branched chain fatty acid, affects Ca2+ homeostasis and mitochondria, and reduces cell viability in rat hippocampal astrocytes. Neurobiol. Dis. 18: 110–118. [DOI] [PubMed] [Google Scholar]
- 90.Ferdinandusse S., Zomer A. W., Komen J. C., van den Brink C. E., Thanos M., Hamers F. P., Wanders R. J., van Der Saag P. T., Poll-The B. T., Brites P. 2008. Ataxia with loss of Purkinje cells in a mouse model for Refsum disease. Proc. Natl. Acad. Sci. USA. 105: 17712–17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schonfeld P., Kahlert S., Reiser G. 2006. A study of the cytotoxicity of branched-chain phytanic acid with mitochondria and rat brain astrocytes. Exp. Gerontol. 41: 688–696. [DOI] [PubMed] [Google Scholar]
- 92.Ronicke S., Kruska N., Kahlert S., Reiser G. 2009. The influence of the branched-chain fatty acids pristanic acid and Refsum disease-associated phytanic acid on mitochondrial functions and calcium regulation of hippocampal neurons, astrocytes, and oligodendrocytes. Neurobiol. Dis. 36: 401–410. [DOI] [PubMed] [Google Scholar]
- 93.Atshaves B. P., Storey S. M., Petrescu A., Greenberg C. C., Lyuksyutova O. I., Smith R., III, Schroeder F. 2002. Expression of fatty acid binding proteins inhibits lipid accumulation and alters toxicity in L cell fibroblasts. Am. J. Physiol. Cell Physiol. 283: C688–C703. [DOI] [PubMed] [Google Scholar]
- 94.Bunik V. I., Raddatz G., Wanders R. J., Reiser G. 2006. Brain pyruvate and 2-oxoglutarate dehydrogenase complexes are mitochondrial targets of the CoA ester of the Refsum disease marker phytanic acid. FEBS Lett. 580: 3551–3557. [DOI] [PubMed] [Google Scholar]
- 95.Singh H., Poulos A. 1995. Substrate specificity of rat liver mitochondrial carnitine palmitoyl transferase I: evidence against alpha-oxidation of phytanic acid in rat liver mitochondria. FEBS Lett. 359: 179–183. [DOI] [PubMed] [Google Scholar]
- 96.Lloyd M. D., Darley D. J., Wierzbicki A. S., Threadgill M. D. 2008. Alpha-methylacyl-CoA racemase–an ‘obscure’ metabolic enzyme takes centre stage. FEBS J. 275: 1089–1102. [DOI] [PubMed] [Google Scholar]
- 97.Lazarow P. B., de Duve C. 1976. A fatty acyl-CoA oxidizing system in rat liver peroxisomes; enhancement by clofibrate, a hypolipidemic drug. Proc. Natl. Acad. Sci. USA. 73: 2043–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bronfman M., Inestrosa N. C., Leighton F. 1979. Fatty acid oxidation by human liver peroxisomes. Biochem. Biophys. Res. Commun. 88: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 99.Singh H., Beckman K., Poulos A. 1994. Peroxisomal beta-oxidation of branched chain fatty acids in rat liver. Evidence that carnitine palmitoyltransferase I prevents transport of branched chain fatty acids into mitochondria. J. Biol. Chem. 269: 9514–9520. [PubMed] [Google Scholar]
- 100.Vamecq J., Draye J. P., Brison J. 1989. Rat liver metabolism of dicarboxylic acids. Am. J. Physiol. 256: G680–G688. [DOI] [PubMed] [Google Scholar]
- 101.Wanders R. J. 2004. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol. Genet. Metab. 83: 16–27. [DOI] [PubMed] [Google Scholar]
- 102.Morais S., Knoll-Gellida A., Andre M., Barthe C., Babin P. J. 2007. Conserved expression of alternative splicing variants of peroxisomal acyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol. Genomics. 28: 239–252. [DOI] [PubMed] [Google Scholar]
- 103.Ferdinandusse S., Denis S., Hogenhout E. M., Koster J., van Roermund C. W., Ijlst L., Moser A. B., Wanders R. J., Waterham H. R. 2007. Clinical, biochemical, and mutational spectrum of peroxisomal acyl-coenzyme A oxidase deficiency. Hum. Mutat. 28: 904–912. [DOI] [PubMed] [Google Scholar]
- 104.Nakajima Y., Miyahara I., Hirotsu K., Nishina Y., Shiga K., Setoyama C., Tamaoki H., Miura R. 2002. Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J. Biochem. 131: 365–374. [DOI] [PubMed] [Google Scholar]
- 105.Van Veldhoven P. P., Vanhove G., Assselberghs S., Eyssen H. J., Mannaerts G. P. 1992. Substrate specificities of rat liver peroxisomal acyl-CoA oxidases: palmitoyl-CoA oxidase (inducible acyl-CoA oxidase), pristanoyl-CoA oxidase (non-inducible acyl-CoA oxidase), and trihydroxycoprostanoyl-CoA oxidase. J. Biol. Chem. 267: 20065–20074. [PubMed] [Google Scholar]
- 106.Van Veldhoven P. P., Croes K., Asselberghs S., Herdewijn P., Mannaerts G. P. 1996. Peroxisomal beta-oxidation of 2-methyl-branched acyl-CoA esters: stereospecific recognition of the 2S-methyl compounds by trihydroxycoprostanoyl-CoA oxidase and pristanoyl-CoA oxidase. FEBS Lett. 388: 80–84. [DOI] [PubMed] [Google Scholar]
- 107.Zha S., Ferdinandusse S., Hicks J. L., Denis S., Dunn T. A., Wanders R. J., Luo J., De Marzo A. M., Isaacs W. B. 2005. Peroxisomal branched chain fatty acid beta-oxidation pathway is upregulated in prostate cancer. Prostate. 63: 316–323. [DOI] [PubMed] [Google Scholar]
- 108.Kim J. J., Miura R. 2004. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences KIM2004. Eur. J. Biochem. 271: 483–493. [DOI] [PubMed] [Google Scholar]
- 109.Kurochkin I. V., Mizuno Y., Konagaya A., Sakaki Y., Schonbach C., Okazaki Y. 2007. Novel peroxisomal protease Tysnd1 processes. EMBO J. 26: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van Veldhoven P. P. 1995. Activity measurements of acyl-CoA oxidases in human liver. J. Inherit. Metab. Dis. 18(Suppl. 1): 125–134. [DOI] [PubMed] [Google Scholar]
- 111.Kiema T. R., Taskinen J. P., Pirila P. L., Koivuranta K. T., Wierenga R. K., Hiltunen J. K. 2002. Organization of the multifunctional enzyme type 1: interaction between N- and C-terminal domains is required for the hydratase-1/isomerase activity. Biochem. J. 367: 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huyghe S., Mannaerts G. P., Baes M., Van Veldhoven P. P. 2006. Peroxisomal multifunctional protein-2: the enzyme, the patients and the knockout mouse model. Biochim. Biophys. Acta. 1761: 973–994. [DOI] [PubMed] [Google Scholar]
- 113.Gallegos A. M., Atshaves B. P., Storey S. M., Starodub O., Petrescu A. D., Huang H., McIntosh A. L., Martin G. G., Chao H., Kier A. B., et al. 2001. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog. Lipid Res. 40: 498–563. [DOI] [PubMed] [Google Scholar]
- 114.Antonenkov V. D., Van Veldhoven P. P., Waelkens E., Mannaerts G. P. 1997. Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase purified from normal rat liver peroxisomes. Sterol carrier protein 2/3-oxoacyl-CoA thiolase is involved in the metabolism of 2-methyl-branched fatty acids and bile acid intermediates. J. Biol. Chem. 272: 26023–26031. [DOI] [PubMed] [Google Scholar]
- 115.Roff C. F., Pastuszyn A., Strauss J. F., III, Billheimer J. T., Vanier M. T., Brady R. O., Scallen T. J., Pentchev P. G. 1992. Deficiencies in sex-regulated expression and levels of two hepatic sterol carrier proteins in a murine model of Niemann-Pick type C disease. J. Biol. Chem. 267: 15902–15908. [PubMed] [Google Scholar]
- 116.Atshaves B. P., Payne H. R., McIntosh A. L., Tichy S. E., Russell D., Kier A. B., Schroeder F. 2004. Sexually dimorphic metabolism of branched-chain lipids in C57BL/6J mice. J. Lipid Res. 45: 812–830. [DOI] [PubMed] [Google Scholar]
- 117.Schmitz W., Albers C., Fingerhut R., Conzelmann E. 1995. Purification and characterization of an alpha-methylacyl-CoA racemase from human liver. Eur. J. Biochem. 231: 815–822. [DOI] [PubMed] [Google Scholar]
- 118.Bhaumik P., Schmitz W., Hassinen A., Hiltunen J. K., Conzelmann E., Wierenga R. K. 2007. The catalysis of the 1,1-proton transfer by alpha-methyl-acyl-CoA racemase is coupled to a movement of the fatty acyl moiety over a hydrophobic, methionine-rich surface. J. Mol. Biol. 367: 1145–1161. [DOI] [PubMed] [Google Scholar]
- 119.Cuebas D. A., Phillips C., Schmitz W., Conzelmann E., Novikov D. K. 2002. The role of alpha-methylacyl-CoA racemase in bile acid synthesis. Biochem. J. 363: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reichel C., Brugger R., Bang H., Geisslinger G., Brune K. 1997. Molecular cloning and expression of a 2-arylpropionyl-coenzyme A epimerase: a key enzyme in the inversion metabolism of ibuprofen. Mol. Pharmacol. 51: 576–582. [DOI] [PubMed] [Google Scholar]
- 121.Elkrief L., Chryssostalis A., Moachon L., Franck N., Terris B., Chaussade S., Sogni P. 2007. [Severe cholestatic hepatitis associated with Stevens-Johnson syndrome after taking ibuprofen] [article in French]. Gastroenterol. Clin. Biol. 31: 1043–1045. [DOI] [PubMed] [Google Scholar]
- 122.Van Veldhoven P. P., Mannaerts G. P. 1999. Role and organization of peroxisomal beta-oxidation. Adv. Exp. Med. Biol. 466: 261–272. [DOI] [PubMed] [Google Scholar]
- 123.Baes M., Van Veldhoven P. P. 2003. Lessons from knockout mice. I: phenotypes of mice with peroxisome biogenesis disorders. Adv. Exp. Med. Biol. 544: 113–122. [DOI] [PubMed] [Google Scholar]
- 124.Berger J., Kunze M., Forss-Petter S. 2003. Lessons from knockout mice II: mouse models for peroxisomal disorders with single protein deficiency. Adv. Exp. Med. Biol. 544: 123–134. [DOI] [PubMed] [Google Scholar]
- 125.Kurosawa T., Sato M., Watanabe T., Suga T., Tohma M. 1997. Effect of the side-chain structure on the specificity of beta-oxidation in bile acid biosynthesis in rat liver homogenates. J. Lipid Res. 38: 2589–2602. [PubMed] [Google Scholar]
- 126.Wouters F. S., Bastiaens P. I., Wirtz K. W., Jovin T. M. 1998. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. EMBO J. 17: 7179–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Makkar R. S., Contreras M. A., Paintlia A. S., Smith B. T., Haq E., Singh I. 2006. Molecular organization of peroxisomal enzymes: protein-protein interactions in the membrane and in the matrix. Arch. Biochem. Biophys. 451: 128–140. [DOI] [PubMed] [Google Scholar]
- 128.Hunt M. C., Yamada J., Maltais L. J., Wright M. W., Podesta E. J., Alexson S. E. 2005. A revised nomenclature for mammalian acyl-CoA thioesterases/hydrolases. J. Lipid Res. 46: 2029–2032. [DOI] [PubMed] [Google Scholar]
- 129.Hunt M. C., Alexson S. E. 2008. Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog. Lipid Res. 47: 405–421. [DOI] [PubMed] [Google Scholar]
- 130.Corti O., DiDonato S., Finocchiaro G. 1994. Divergent sequences in the 5′ region of cDNA suggest alternative splicing as a mechanism for the generation of carnitine acetyltransferases with different subcellular localizations. Biochem. J. 303: 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Westin M. A., Hunt M. C., Alexson S. E. 2008. Short- and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of beta-oxidation products out of peroxisomes. Cell. Mol. Life Sci. 65: 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leighton F., Bergseth S., Rortveit T., Christiansen E. N., Bremer J. 1989. Free acetate production by rat hepatocytes during peroxisomal fatty acid and dicarboxylic acid oxidation. J. Biol. Chem. 264: 10347–10350. [PubMed] [Google Scholar]
- 133.Tran T. N., Christophersen B. O. 2001. Studies on the transport of acetyl groups from peroxisomes to mitochondria in isolated liver cells oxidizing the polyunsaturated fatty acid 22:4n-6. Biochim. Biophys. Acta. 1533: 255–265. [DOI] [PubMed] [Google Scholar]
- 134.Reszko A. E., Kasumov T., David F., Jobbins K. A., Thomas K. R., Hoppel C. L., Brunengraber H., Des R. C. 2004. Peroxisomal fatty acid oxidation is a substantial source of the acetyl moiety of malonyl-CoA in rat heart. J. Biol. Chem. 279: 19574–19579. [DOI] [PubMed] [Google Scholar]
- 135.Hayashi H., Hara M. 1997. 1-Alkenyl group of ethanolamine plasmalogen derives mainly from de novo-synthesized fatty alcohol within peroxisomes, but not extraperoxisomal fatty alcohol or fatty acid. J. Biochem. 121: 978–983. [DOI] [PubMed] [Google Scholar]
- 136.Das A. K., Uhler M. D., Hajra A. K. 2000. Molecular cloning and expression of mammalian peroxisomal trans-2-enoyl-coenzyme A reductase cDNAs. J. Biol. Chem. 275: 24333–24340. [DOI] [PubMed] [Google Scholar]
- 137.Hayashi H., Sato A. 1997. Fatty alcohol synthesis accompanied with chain elongation in liver peroxisomes. Biochim. Biophys. Acta. 1346: 38–44. [DOI] [PubMed] [Google Scholar]
- 138.Youssef J. A., Song W. O., Badr M. Z. 1997. Mitochondrial, but not peroxisomal, beta-oxidation of fatty acids is conserved in coenzyme A-deficient rat liver. Mol. Cell. Biochem. 175: 37–42. [DOI] [PubMed] [Google Scholar]
- 139.Ferdinandusse S., Denis S., Faust P. L., Wanders R. J. 2009. Bile acids: role of peroxisomes. J. Lipid Res. 50: 2139–2147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mihalik S. J., Steinberg S. J., Pei Z., Park J., Kim D. G., Heinzer A. K., Dacremont G., Wanders R. J., Cuebas D. A., Smith K. D., et al. 2002. Participation of two members of the very long-chain acyl-CoA synthetase family in bile acid synthesis and recycling. J. Biol. Chem. 277: 24771–24779. [DOI] [PubMed] [Google Scholar]
- 141.Heinzer A. K., Watkins P. A., Lu J. F., Kemp S., Moser A. B., Li Y. Y., Mihalik S., Powers J. M., Smith K. D. 2003. A very long-chain acyl-CoA synthetase-deficient mouse and its relevance to X-linked adrenoleukodystrophy. Hum. Mol. Genet. 12: 1145–1154. [DOI] [PubMed] [Google Scholar]
- 142.Hubbard B., Doege H., Punreddy S., Wu H., Huang X., Kaushik V. K., Mozell R. L., Byrnes J. J., Stricker-Krongrad A., Chou C. J., et al. 2006. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology. 130: 1259–1269. [DOI] [PubMed] [Google Scholar]
- 143.Hunt M. C., Solaas K., Kase B. F., Alexson S. E. 2002. Characterization of an acyl-coA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. J. Biol. Chem. 277: 1128–1138. [DOI] [PubMed] [Google Scholar]
- 144.Pellicoro A., van den Heuvel F. A., Geuken M., Moshage H., Jansen P. L., Faber K. N. 2007. Human and rat bile acid-CoA:amino acid N-acyltransferase are liver-specific peroxisomal enzymes: implications for intracellular bile salt transport. Hepatology. 45: 340–348. [DOI] [PubMed] [Google Scholar]
- 145.Steinberg S. J., Mihalik S. J., Kim D. G., Cuebas D. A., Watkins P. A. 2000. The human liver-specific homolog of very long-chain acyl-CoA synthetase is cholate:CoA ligase. J. Biol. Chem. 275: 15605–15608. [DOI] [PubMed] [Google Scholar]
- 146.Bjorkhem I., Andersson U., Ellis E., Alvelius G., Ellegard L., Diczfalusy U., Sjovall J., Einarsson C. 2001. From brain to bile. Evidence that conjugation and omega-hydroxylation are important for elimination of 24S-hydroxycholesterol (cerebrosterol) in humans. J. Biol. Chem. 276: 37004–37010. [DOI] [PubMed] [Google Scholar]
- 147.Ferdinandusse S., Denis S., Overmars H., Van E. L., Van Veldhoven P. P., Duran M., Wanders R. J., Baes M. 2005. Developmental changes of bile acid composition and conjugation in L- and D-bifunctional protein single and double knockout mice. J. Biol. Chem. 280: 18658–18666. [DOI] [PubMed] [Google Scholar]
- 148.Ferdinandusse S., Mulders J., Ijlst L., Denis S., Dacremont G., Waterham H. R., Wanders R. J. 1999. Molecular cloning and expression of human carnitine octanoyltransferase: evidence for its role in the peroxisomal beta-oxidation of branched-chain fatty acids. Biochem. Biophys. Res. Commun. 263: 213–218. [DOI] [PubMed] [Google Scholar]
- 149.Ofman R., el Mrabet L., Dacremont G., Spijer D., Wanders R. J. 2002. Demonstration of dimethylnonanoyl-CoA thioesterase activity in rat liver peroxisomes followed by purification and molecular cloning of the thioesterase involved. Biochem. Biophys. Res. Commun. 290: 629–634. [DOI] [PubMed] [Google Scholar]
- 150.Uchiyama A., Aoyama T., Kamijo K., Uchida Y., Kondo N., Orii T., Hashimoto T. 1996. Molecular cloning of cDNA encoding rat very long-chain acyl-CoA synthetase. J. Biol. Chem. 271: 30360–30365. [DOI] [PubMed] [Google Scholar]
- 151.Steinberg S. J., Morgenthaler J., Heinzer A. K., Smith K. D., Watkins P. A. 2000. Very long-chain acyl-CoA synthetases. Human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids. J. Biol. Chem. 275: 35162–35169. [DOI] [PubMed] [Google Scholar]
- 152.Pei Z., Jia Z., Watkins P. A. 2006. The second member of the human and murine bubblegum family is a testis- and brainstem-specific acyl-CoA synthetase. J. Biol. Chem. 281: 6632–6641. [DOI] [PubMed] [Google Scholar]
- 153.Hall A. M., Wiczer B. M., Herrmann T., Stremmel W., Bernlohr D. A. 2005. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J. Biol. Chem. 280: 11948–11954. [DOI] [PubMed] [Google Scholar]
- 154.Cheng J. B., Russell D. W. 2004. Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 279: 37789–37797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reilly S. J., O'Shea E. M., Andersson U., O'Byrne J., Alexson S. E., Hunt M. C. 2007. A peroxisomal acyltransferase in mouse identifies a novel pathway for taurine conjugation of fatty acids. FASEB J. 21: 99–107. [DOI] [PubMed] [Google Scholar]
- 156.Sprecher H., Chen Q. 1999. Polyunsaturated fatty acid biosynthesis: a microsomal-peroxisomal process. Prostaglandins Leukot. Essent. Fatty Acids. 60: 317–321. [DOI] [PubMed] [Google Scholar]
- 157.Martinez M. 1992. Abnormal profiles of polyunsaturated fatty acids in the brain, liver, kidney and retina of patients with peroxisomal disorders. Brain Res. 583: 171–182. [DOI] [PubMed] [Google Scholar]
- 158.Baes M., Gressens P., Baumgart E., Carmeliet P., Casteels M., Fransen M., Evrard P., Fahimi D., Declercq P. E., Collen D., et al. 1997. A mouse model for Zellweger syndrome. Nat. Genet. 17: 49–57. [DOI] [PubMed] [Google Scholar]
- 159.Ferdinandusse S., Denis S., Mooijer P. A., Zhang Z., Reddy J. K., Spector A. A., Wanders R. J. 2001. Identification of the peroxisomal beta-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 42: 1987–1995. [PubMed] [Google Scholar]
- 160.Infante J. P., Tschanz C. L., Shaw N., Michaud A. L., Lawrence P., Brenna J. T. 2002. Straight-chain acyl-CoA oxidase knockout mouse accumulates extremely long chain fatty acids from alpha-linolenic acid: evidence for runaway carousel-type enzyme kinetics in peroxisomal beta-oxidation diseases. Mol. Genet. Metab. 75: 108–119. [DOI] [PubMed] [Google Scholar]
- 161.Huyghe S., Schmalbruch H., De G. K., Verhoeven G., Guillou F., Van Veldhoven P. P., Baes M. 2006. Peroxisomal multifunctional protein 2 is essential for lipid homeostasis in Sertoli cells and male fertility in mice. Endocrinology. 147: 2228–2236. [DOI] [PubMed] [Google Scholar]
- 162.De Nys K. 2002. Peroxisomal beta-oxidation of unsaturated fatty acids. Acta Biomedica Lovaniensia. 265 PhD Thesis. Leuven University Press, Leuven. [Google Scholar]
- 163.De Nys K., Meyhi E., Mannaerts G. P., Fransen M., Van Veldhoven P. P. 2001. Characterisation of human peroxisomal 2,4-dienoyl-CoA reductase. Biochim. Biophys. Acta. 1533: 66–72. [DOI] [PubMed] [Google Scholar]
- 164.Poulos A., Sharp P., Johnson D., Easton C. 1988. The occurrence of polyenoic very long chain fatty acids with greater than 32 carbon atoms in molecular species of phosphatidylcholine in normal and peroxisome-deficient (Zellweger's syndrome) brain. Biochem. J. 253: 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hardwick J. P. 2008. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 75: 2263–2275. [DOI] [PubMed] [Google Scholar]
- 166.Dhar M., Sepkovic D. W., Hirani V., Magnusson R. P., Lasker J. M. 2008. Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. J. Lipid Res. 49: 612–624. [DOI] [PubMed] [Google Scholar]
- 167.Sanders R. J., Ofman R., Dacremont G., Wanders R. J., Kemp S. 2008. Characterization of the human omega-oxidation pathway for omega-hydroxy-very-long-chain fatty acids. FASEB J. 22: 2064–2071. [DOI] [PubMed] [Google Scholar]
- 168.Rocchiccioli F., Aubourg P., Bougneres P. F. 1986. Medium and long-chain dicarboxylic aciduria in patients with Zellweger syndrome and neonatal adrenoleukodystrophy. Pediatr. Res. 20: 62–66. [DOI] [PubMed] [Google Scholar]
- 169.Rizzo C., Boenzi S., Wanders R. J., Duran M., Caruso U., Dionisi-Vici C. 2003. Characteristic acylcarnitine profiles in inherited defects of peroxisome biogenesis: a novel tool for screening diagnosis using tandem mass spectrometry. Pediatr. Res. 53: 1013–1018. [DOI] [PubMed] [Google Scholar]
- 170.Ferdinandusse S., Denis S., van Roermund C. W., Wanders R. J., Dacremont G. 2004. Identification of the peroxisomal beta-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J. Lipid Res. 45: 1104–1111. [DOI] [PubMed] [Google Scholar]
- 171.Dirkx R., Meyhi E., Asselberghs S., Reddy J., Baes M., Van Veldhoven P. P. 2007. Beta-oxidation in hepatocyte cultures from mice with peroxisomal gene knockouts. Biochem. Biophys. Res. Commun. 357: 718–723. [DOI] [PubMed] [Google Scholar]
- 172.Westin M. A., Hunt M. C., Alexson S. E. 2005. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. J. Biol. Chem. 280: 38125–38132. [DOI] [PubMed] [Google Scholar]
- 173.Pourfarzam M., Bartlett K. 1992. Intermediates of peroxisomal beta-oxidation of [U-14C]hexadecanedionoate. A study of the acyl-CoA esters which accumulate during peroxisomal beta-oxidation of [U-14C]hexadecanedionate and [U-14C]hexadecanedionoyl-mono-CoA. Eur. J. Biochem. 208: 301–307. [DOI] [PubMed] [Google Scholar]
- 174.Hunt M. C., Rautanen A., Westin M. A., Svensson L. T., Alexson S. E. 2006. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 20: 1855–1864. [DOI] [PubMed] [Google Scholar]
- 175.Fang X., Dillon J. S., Hu S., Harmon S. D., Yao J., Anjaiah S., Falck J. R., Spector A. A. 2007. 20-carboxy-arachidonic acid is a dual activator of peroxisome proliferator-activated receptors alpha and gamma. Prostaglandins Other Lipid Mediat. 82: 175–184. [DOI] [PubMed] [Google Scholar]
- 176.Nguyen S. D., Baes M., Van Veldhoven P. P. 2008. Degradation of very long chain dicarboxylic polyunsaturated fatty acids in mouse hepatocytes, a peroxisomal process. Biochim. Biophys. Acta. 1781: 400–405. [DOI] [PubMed] [Google Scholar]
- 177.Buczynski M. W., Dumlao D. S., Dennis E. A. 2009. Thematic review series: proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 50: 1015–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Diczfalusy U., Kase B. F., Alexson S. E., Bjorkhem I. 1991. Metabolism of prostaglandin F2 alpha in Zellweger syndrome. Peroxisomal beta-oxidation is a major importance for in vivo degradation of prostaglandins in humans. J. Clin. Invest. 88: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tsikas D., Schwedhelm E., Fauler J., Gutzki F. M., Mayatepek E., Frolich J. C. 1998. Specific and rapid quantification of 8-iso-prostaglandin F2alpha in urine of healthy humans and patients with Zellweger syndrome by gas chromatography-tandem mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 716: 7–17. [DOI] [PubMed] [Google Scholar]
- 180.Diczfalusy U., Vesterqvist O., Kase B. F., Lund E., Alexson S. E. 1993. Peroxisomal chain-shortening of thromboxane B2: evidence for impaired degradation of thromboxane B2 in Zellweger syndrome. J. Lipid Res. 34: 1107–1113. [PubMed] [Google Scholar]
- 181.de Waart D. R., Koomen G. C., Wanders R. J. 1994. Studies on the urinary excretion of thromboxane B2 in Zellweger patients and control subjects: evidence for a major role for peroxisomes in the beta-oxidative chain-shortening of thromboxane B2. Biochim. Biophys. Acta. 1226: 44–48. [DOI] [PubMed] [Google Scholar]
- 182.Spector A. A., Fang X., Snyder G. D., Weintraub N. L. 2004. Epoxyeicosatrienoic acids (EETs): metabolism and biochemical function. Prog. Lipid Res. 43: 55–90. [DOI] [PubMed] [Google Scholar]
- 183.Jedlitschky G., Mayatepek E., Keppler D. 1993. Peroxisomal leukotriene degradation: biochemical and clinical implications. Adv. Enzyme Regul. 33: 181–194. [DOI] [PubMed] [Google Scholar]
- 184.Mayatepek E., Flock B. 1999. Increased urinary excretion of LTB4 and omega-carboxy-LTB4 in patients with Zellweger syndrome. Clin. Chim. Acta. 282: 151–155. [DOI] [PubMed] [Google Scholar]
- 185.Mayatepek E., Ferdinandusse S., Meissner T., Wanders R. J. 2004. Analysis of cysteinyl leukotrienes and their metabolites in bile of patients with peroxisomal or mitochondrial beta-oxidation defects. Clin. Chim. Acta. 345: 89–92. [DOI] [PubMed] [Google Scholar]
- 186.Fang X., Kaduce T. L., VanRollins M., Weintraub N. L., Spector A. A. 2000. Conversion of epoxyeicosatrienoic acids (EETs) to chain-shortened epoxy fatty acids by human skin fibroblasts. J. Lipid Res. 41: 66–74. [PubMed] [Google Scholar]
- 187.Ferdinandusse S., Meissner T., Wanders R. J., Mayatepek E. 2002. Identification of the peroxisomal beta-oxidation enzymes involved in the degradation of leukotrienes. Biochem. Biophys. Res. Commun. 293: 269–273. [DOI] [PubMed] [Google Scholar]
- 188.Serhan C. N., Levy B. 2003. Novel pathways and endogenous mediators in anti-inflammation and resolution. Chem. Immunol. Allergy. 83: 115–145. [DOI] [PubMed] [Google Scholar]
- 189.Swigonova Z., Mohsen A. W., Vockley J. 2009. Acyl-CoA dehydrogenases: dynamic history of protein family evolution. J. Mol. Evol. 69: 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dansen T. B., Westerman J., Wouters F. S., Wanders R. J., van Hoek A., Gadella T. W., Jr., Wirtz K. W. 1999. High-affinity binding of very-long-chain fatty acyl-CoA esters to the peroxisomal non-specific lipid-transfer protein (sterol carrier protein-2). Biochem. J. 339: 193–199. [PMC free article] [PubMed] [Google Scholar]
- 191.Ofman R., Speijer D., Leen R., Wanders R. J. 2006. Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem. J. 393: 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reilly S. J., Tillander V., Ofman R., Alexson S. E., Hunt M. C. 2008. The nudix hydrolase 7 is an Acyl-CoA diphosphatase involved in regulating peroxisomal coenzyme A homeostasis. J. Biochem. 144: 655–663. [DOI] [PubMed] [Google Scholar]
- 193.Lamhonwah A. M., Ackerley C. A., Tilups A., Edwards V. D., Wanders R. J., Tein I. 2005. OCTN3 is a mammalian peroxisomal membrane carnitine transporter. Biochem. Biophys. Res. Commun. 338: 1966–1972. [DOI] [PubMed] [Google Scholar]
- 194.Berger J., Gartner J. 2006. X-linked adrenoleukodystrophy: clinical, biochemical and pathogenetic aspects. Biochim. Biophys. Acta. 1763: 1721–1732. [DOI] [PubMed] [Google Scholar]
- 195.Wanders R. J., Visser W. F., van Roermund C. W., Kemp S., Waterham H. R. 2007. The peroxisomal ABC transporter family. Pflugers Arch. 453: 719–734. [DOI] [PubMed] [Google Scholar]
- 196.Kashiwayama Y., Seki M., Yasui A., Murasaki Y., Morita M., Yamashita Y., Sakaguchi M., Tanaka Y., Imanaka T. 2009. 70-kDa peroxisomal membrane protein related protein (P70R/ABCD4) localizes to endoplasmic reticulum not peroxisomes, and NH2-terminal hydrophobic property determines the subcellular localization of ABC subfamily D proteins. Exp. Cell Res. 315: 190–205. [DOI] [PubMed] [Google Scholar]
- 197.Islinger M., Luers G. H., Li K. W., Loos M., Volkl A. 2007. Rat liver peroxisomes after fibrate treatment. A survey using quantitative mass spectrometry. J. Biol. Chem. 282: 23055–23069. [DOI] [PubMed] [Google Scholar]
- 198.Hillebrand M., Verrier S. E., Ohlenbusch A., Schafer A., Soling H. D., Wouters F. S., Gartner J. 2007. Live cell FRET microscopy: homo- and heterodimerization of two human peroxisomal ABC transporters, the adrenoleukodystrophy protein (ALDP, ABCD1) and PMP70 (ABCD3). J. Biol. Chem. 282: 26997–27005. [DOI] [PubMed] [Google Scholar]
- 199.Guimaraes C. P., Domingues P., Aubourg P., Fouquet F., Pujol A., Jimenez-Sanchez G., Sa-Miranda C., Azevedo J. E. 2004. Mouse liver PMP70 and ALDP: homomeric interactions prevail in vivo. Biochim. Biophys. Acta. 1689: 235–243. [DOI] [PubMed] [Google Scholar]
- 200.Watkins P. A., Chen W. W., Harris C. J., Hoefler G., Hoefler S., Blake D. C., Jr., Balfe A., Kelley R. I., Moser A. B., Beard M. E. 1989. Peroxisomal bifunctional enzyme deficiency. J. Clin. Invest. 83: 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schram A. W., Goldfischer S., van Roermund C. W., Brouwer-Kelder E. M., Collins J., Hashimoto T., Heymans H. S., van den Bosch H., Schutgens R. B., Tager J. M., et al. 1987. Human peroxisomal 3-oxoacyl-coenzyme A thiolase deficiency. Proc. Natl. Acad. Sci. USA. 84: 2494–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.van Grunsven E. G., van Berkel E., Mooijer P. A., Watkins P. A., Moser H. W., Suzuki Y., Jiang L. L., Hashimoto T., Hoefler G., Adamski J., et al. 1999. Peroxisomal bifunctional protein deficiency revisited: resolution of its true enzymatic and molecular basis. Am. J. Hum. Genet. 64: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ferdinandusse S., van Grunsven E. G., Oostheim W., Denis S., Hogenhout E. M., Ijlst L., van Roermund C. W., Waterham H. R., Goldfischer S., Wanders R. J. 2002. Reinvestigation of peroxisomal 3-ketoacyl-CoA thiolase deficiency: identification of the true defect at the level of d-bifunctional protein. Am. J. Hum. Genet. 70: 1589–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wanders R. J., Waterham H. R. 2006. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta. 1763: 1707–1720. [DOI] [PubMed] [Google Scholar]
- 205.Kemp S., Valianpour F., Denis S., Ofman R., Sanders R. J., Mooyer P., Barth P. G., Wanders R. J. 2005. Elongation of very long-chain fatty acids is enhanced in X-linked adrenoleukodystrophy. Mol. Genet. Metab. 84: 144–151. [DOI] [PubMed] [Google Scholar]
- 206.Ito M., Blumberg B. M., Mock D. J., Goodman A. D., Moser A. B., Moser H. W., Smith K. D., Powers J. M. 2001. Potential environmental and host participants in the early white matter lesion of adreno-leukodystrophy: morphologic evidence for CD8 cytotoxic T cells, cytolysis of oligodendrocytes, and CD1-mediated lipid antigen presentation. J. Neuropathol. Exp. Neurol. 60: 1004–1019. [DOI] [PubMed] [Google Scholar]
- 207.Poll-The B. T., Roels F., Ogier H., Scotto J., Vamecq J., Schutgens R. B., Wanders R. J., van Roermund C. W., van Wijland M. J., Schram A. W. 1988. A new peroxisomal disorder with enlarged peroxisomes and a specific deficiency of acyl-CoA oxidase (pseudo-neonatal adrenoleukodystrophy). Am. J. Hum. Genet. 42: 422–434. [PMC free article] [PubMed] [Google Scholar]
- 208.Rosewich H., Waterham H. R., Wanders R. J., Ferdinandusse S., Henneke M., Hunneman D., Gartner J. 2006. Pitfall in metabolic screening in a patient with fatal peroxisomal beta-oxidation defect. Neuropediatrics. 37: 95–98. [DOI] [PubMed] [Google Scholar]
- 209.Suzuki Y., Jiang L. L., Souri M., Miyazawa S., Fukuda S., Zhang Z., Une M., Shimozawa N., Kondo N., Orii T., et al. 1997. D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein deficiency: a newly identified peroxisomal disorder. Am. J. Hum. Genet. 61: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ferdinandusse S., Denis S., Mooyer P. A., Dekker C., Duran M., Soorani-Lunsing R. J., Boltshauser E., Macaya A., Gartner J., Majoie C. B., et al. 2006. Clinical and biochemical spectrum of D-bifunctional protein deficiency. Ann. Neurol. 59: 92–104. [DOI] [PubMed] [Google Scholar]
- 211.Sequeira J. S., Vellodi A., Vanier M. T., Clayton P. T. 1998. Niemann-Pick disease type C and defective peroxisomal beta-oxidation of branched-chain substrates. J. Inherit. Metab. Dis. 21: 149–154. [DOI] [PubMed] [Google Scholar]
- 212.Van Veldhoven P. P., Meyhi E., Squires R. H., Fransen M., Fournier B., Brys V., Bennett M. J., Mannaerts G. P. 2001. Fibroblast studies documenting a case of peroxisomal 2-methylacyl-CoA racemase deficiency: possible link between racemase deficiency and malabsorption and vitamin K deficiency. Eur. J. Clin. Invest. 31: 714–722. [DOI] [PubMed] [Google Scholar]
- 213.Setchell K. D., Heubi J. E., Bove K. E., O'Connell N. C., Brewsaugh T., Steinberg S. J., Moser A., Squires R. H., Jr 2003. Liver disease caused by failure to racemize trihydroxycholestanoic acid: gene mutation and effect of bile acid therapy. Gastroenterology. 124: 217–232. [DOI] [PubMed] [Google Scholar]
- 214.McLean B. N., Allen J., Ferdinandusse S., Wanders R. J. 2002. A new defect of peroxisomal function involving pristanic acid: a case report. J. Neurol. Neurosurg. Psychiatry. 72: 396–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Clarke C. E., Alger S., Preece M. A., Burdon M. A., Chavda S., Denis S., Ferdinandusse S., Wanders R. J. 2004. Tremor and deep white matter changes in alpha-methylacyl-CoA racemase deficiency. Neurology. 63: 188–189. [DOI] [PubMed] [Google Scholar]
- 216.Thompson S. A., Calvin J., Hogg S., Ferdinandusse S., Wanders R. J., Barker R. A. 2008. Relapsing encephalopathy in a patient with alpha-methylacyl-CoA racemase deficiency. J. Neurol. Neurosurg. Psychiatry. 79: 448–450. [DOI] [PubMed] [Google Scholar]
- 217.Ferdinandusse S., Kostopoulos P., Denis S., Rusch H., Overmars H., Dillmann U., Reith W., Haas D., Wanders R. J., Duran M., et al. 2006. Mutations in the gene encoding peroxisomal sterol carrier protein X (SCPx) cause leukencephalopathy with dystonia and motor neuropathy. Am. J. Hum. Genet. 78: 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kannenberg F., Ellinghaus P., Assmann G., Seedorf U. 1999. Aberrant oxidation of the cholesterol side chain in bile acid synthesis of sterol carrier protein-2/sterol carrier protein-x knockout mice. J. Biol. Chem. 274: 35455–35460. [DOI] [PubMed] [Google Scholar]
- 219.Carlton V. E., Harris B. Z., Puffenberger E. G., Batta A. K., Knisely A. S., Robinson D. L., Strauss K. A., Shneider B. L., Lim W. A., Salen G., et al. 2003. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat. Genet. 34: 91–96. [DOI] [PubMed] [Google Scholar]
- 220.Zhu Q. S., Xing W., Qian B., von Dippe P., Shneider B. L., Fox V. L., Levy D. 2003. Inhibition of human m-epoxide hydrolase gene expression in a case of hypercholanemia ZHU2003. Biochim. Biophys. Acta. 1638: 208–216. [DOI] [PubMed] [Google Scholar]
- 221.Ferrer I., Kapfhammer J. P., Hindelang C., Kemp S., Troffer-Charlier N., Broccoli V., Callyzot N., Mooyer P., Selhorst J., Vreken P., et al. 2005. Inactivation of the peroxisomal ABCD2 transporter in the mouse leads to late-onset ataxia involving mitochondria, Golgi and endoplasmic reticulum damage. Hum. Mol. Genet. 14: 3565–3577. [DOI] [PubMed] [Google Scholar]
- 222.Jimenez-Sanchez G., Hebron K. J., Silva-Zolezzi I., Mihalik S., Watkins P., Espeel M., Moser A., Thomas G., Roels F., Valle D. 2000. Fasting fuel homeostasis triggered by defective phytanicand pristanic acids metabolism in the 70 kDa peroxisomalmembrane protein (PMP70) deficient mice. Am. J. Hum. Genet. 67: 65. [Google Scholar]
- 223.Qi C., Zhu Y., Pan J., Usuda N., Maeda N., Yeldandi A. V., Rao M. S., Hashimoto T., Reddy J. K. 1999. Absence of spontaneous peroxisome proliferation in enoyl-CoA Hydratase/L-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver. Further support for the role of fatty acyl CoA oxidase in PPARalpha ligand metabolism. J. Biol. Chem. 274: 15775–15780. [DOI] [PubMed] [Google Scholar]
- 224.Chevillard G., Clemencet M. C., Latruffe N., Nicolas-Frances V. 2004. Targeted disruption of the peroxisomal thiolase B gene in mouse: a new model to study disorders related to peroxisomal lipid metabolism. Biochimie. 86: 849–856. [DOI] [PubMed] [Google Scholar]
- 225.Klar J., Schweiger M., Zimmerman R., Zechner R., Li H., Torma H., Vahlquist A., Bouadjar B., Dahl N., Fischer J. 2009. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am. J. Hum. Genet. 85: 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gonzalez F. J., Shah Y. M. 2008. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 246: 2–8. [DOI] [PubMed] [Google Scholar]
- 227.Meyer K., Lee J. S., Dyck P. A., Cao W. Q., Rao M. S., Thorgeirsson S. S., Reddy J. K. 2003. Molecular profiling of hepatocellular carcinomas developing spontaneously in acyl-CoA oxidase deficient mice: comparison with liver tumors induced in wild-type mice by a peroxisome proliferator and a genotoxic carcinogen. Carcinogenesis. 24: 975–984. [DOI] [PubMed] [Google Scholar]
- 228.Thornburg T., Turner A. R., Chen Y. Q., Vitolins M., Chang B., Xu J. 2006. Phytanic acid, AMACR and prostate cancer risk. Future Oncol. 2: 213–223. [DOI] [PubMed] [Google Scholar]
- 229.Rasiah K. K., Gardiner-Garden M., Padilla E. J., Moller G., Kench J. G., Alles M. C., Eggleton S. A., Stricker P. D., Adamski J., Sutherland R. L., et al. 2009. HSD17B4 overexpression, an independent biomarker of poor patient outcome in prostate cancer. Mol. Cell. Endocrinol. 301: 89–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.