Abstract
Drosophila melanogaster has been considered a model organism for investigating human diseases and genetic pathways. Whether Drosophila is an ideal model for nutrigenomics, especially for FA metabolism, however, remains to be illustrated. The aim of this study was to examine the metabolism of C20 and C22 PUFAs in Drosophila. Analysis of FA composition revealed a complete lack of C20 and C22 PUFAs in the body tissue of larvae, pupae, and adult flies fed either a base or supplemented diet abundant in the PUFA precursors linoleic acid and α-linolenic acid. PUFA with >C20 could only be found in flies supplemented with specific FAs. Interestingly, the supplemented C22 PUFAs docosahexaenoic acid (22:6n-3) and docosatetraenoic acid (22:4n-6) were largely converted to the shorter chain C20 PUFAs eicosapentaenoic acid (20:5n-3) and arachidonic acid (20:4n-6), respectively. Furthermore, a genome sequence scan indicated that no gene encoding Δ-6/ Δ-5 desaturases, the key enzymes for the synthesis of C20/C22 PUFA, was present in Drosophila. These findings demonstrate that Drosophila lacks the capability to synthesize the biologically important C20 and C22 PUFAs, and thereby argue that Drosophila is not a valid model for the study of lipid metabolism and related diseases.
Keywords: D. melanogaster, fatty acid metabolism, desaturases, β-oxidation, model organism
The fruit fly Drosophila melanogaster is one of the most intensively studied organisms in biology and serves as a model system for the investigation of many developmental and cellular processes common to more-complex eukaryotes, including humans (1). Drosophila distinguishes itself from yeast (Saccharomyces cerevisiae) and nematodes (Caenorhabditis elegans) with its adipose-like tissues and a lipid transport system, making it a closer model to humans. Drosophila has been considered an ideal model since around 1915, when it was first used to study aging (2, 3). Since the complete genomic sequence of the fly was released in early 2000, 177 out of 289 (∼61%) of the human genes responsible for disease have been found to have strong homologs or orthologs in the fly (4). Two-thirds of genes implicated in human cancers also have a counterpart in the fly genome. Furthermore, the use of Drosophila as a model has many advantages, including its sophisticated genetics, small genome size, high fecundity, low cost, and short generation time (4). Candidate pathways influenced by nutrients can be identified first in Drosophila, and later verified in mouse models and humans. Drosophila is thus currently used as a model organism to study human diseases, including obesity, diabetes, cancer, cardiovascular disease, age-related diseases, and longevity.
Lipids come in a wide variety and have many functions in cells besides simply being used for energy. Not only do they make up cell membranes, but they and their products serve as intra- and extracellular messengers that control and regulate many vital bodily functions. Imbalances of lipids cause or play a role in diseases that affect millions worldwide, such as heart disease, cancer, diabetes, Alzheimer's, etc. Certain PUFAs are especially crucial components of human nutrition, including ω-6 (n-6) and ω-3 (n-3) FAs, which are essential nutrients and important determinants of the structure and function of mammalian cells (5). In particular, the dietary essential FAs linoleic acid (LA,18:2n-6) and α-linolenic acid (ALA, 18:3n-3) are precursors for the important long-chain PUFAs arachidonic acid (AA, 20:4n-6), eicosapentaenoic acid (EPA, 20:5n-3), and docosahexaenoic acid (DHA, 22:6n-3), which are synthesized through a series of desaturation, catalyzed sequentially by Δ-6 desaturase and Δ-5 desaturase and chain-elongation enzyme systems. The C20 and C22 PUFAs (AA, EPA, and DHA) are critical as structural components of membrane phospholipids and as precursors of metabolites, including prostaglandins (PGs), prostacyclins (PCs), thromboxanes (TXs), leukotrienes (LTs), lipoxins (LXs), resolvins (Rvs), and protectins (PDs). These PUFA-derived metabolites play important roles in modulating a wide range of physiological and pathophysiological processes, particularly inflammatory responses (6). In addition, the tissue content of DHA is highly related to the retinal and brain development of humans, especially children (7). Thus, the tissue status and metabolism of PUFAs have a significant impact on both physiology and pathology, and the identification of valid models for understanding PUFA metabolism is warranted.
In the current era of genomics and proteomics, the emerging field of lipidomics presents powerful techniques and technology for understanding and manipulating the vital role of lipids in cells, which may further the elucidation of the mechanisms of relevant diseases (8). The use of lipidomics to understand lipid metabolism in Drosophila will identify how qualified Drosophila is to be an experimental system for nutrigenomics, particularly lipid metabolism, and related human disease. To further examine the metabolic characteristics of n-3 and n-6 PUFAs in Drosophila, we determined their FA composition and metabolite profile in different life stages and genders after supplementation with different types of long-chain PUFAs.
MATERIALS AND METHODS
Reagents
PUFA, 20:4n-6, 22:6n-3, 20:5n-3, docosatetraenoic acid (DTA, 22:4n-6), 18:2n-6, and 18:3n-3 were purchased from Nu-Chek-Prep, Inc. All PUFAs were aliquoted into GC sample vials with tight caps and were stored with nitrogen below −20°C before use.
Drosophila diets
Based on the traditional corn-yeast fly medium (9.6 g agar, 72.3 g glucose, 36.3 g sucrose, 22.5 g wheat germ, 36.8 g yeast, 62.5 g corn meal, and 15 ml acid A were mixed with 817.5 ml hot water and processed into 1,000-ml diet), the base diet was made once a week. After the base diet cooled to 60°C, individual FAs were added to the base media at 50 mg/100 g, respectively; the media was then mixed using a Hamilton Beach® hand mixer and aliquoted into vials or bottles. The experimental diets in the vials and bottles were packed with dark plastic bags and stored in a cold room (4°C) before use. In addition, experimental diets were sampled and immediately frozen and stored at −80°C for future GC analysis.
Drosophila stocks
Four Drosophila laboratory stocks, Canton-S, Oregon, PR, and W1118, were used for analysis of general FA composition. A sample of flies from each stock fed on base diets was collected randomly from fly breeding bottles. Canton-S cohorts were used to investigate the metabolism of C20 and C22 PUFAs in Drosophila. All experimental flies were maintained on a 12/12-h light:dark cycle at 25°C and 68% humidity, according to Troen et al. (9). Canton-S cohorts were established by allowing flies of the same age to mate at a density of 20 males and 40 females per bottle. Flies were provided with the base diet and six diets supplemented with six types of PUFA, then removed after 5 days, leaving their eggs to develop into adult flies. Newly emerged flies of the same ages were transferred into new bottles with the tested diets above and maintained for 5 days, and larvae and pupae in every dietary group were sampled at the same age. Five-day-old female and male flies were collected over light CO2. Samples of larvae, pupae, and adult flies were frozen in liquid nitrogen and stored at −80°C for future analysis. For each parameter measured, three replicates were used from each treatment.
GC Analysis of FA compositions
Analysis of the FA compositions of Drosophila and diets was carried out by GC, according to the method of Kang and Wang (10). Twenty male flies (10–15 mg), 15 female flies (15–20 mg), or 100-mg diet samples were mixed with 5 ml hexane and 5 ml BF3/MeOH reagent (14% boron trifluoride methanol solution) in 16-ml glass tubes. The mixture was heated at 100°C for 1 h and then cooled to room temperature. Methyl esters were extracted in the hexane phase following addition of 1 ml H2O. The samples were centrifuged for 5 min at 3,000 rpm. The upper hexane layer was then removed and concentrated under nitrogen. FA methyl esters were analyzed by the Agilent 6890N GC system with the 7683 auto-injector. An Omegawax 250 capillary column (30 m × 0.25 mm inner diameter) was used with helium as a carrier. Peaks were identified by comparing their retention times to those of mixture standards (Nu-Chek-Prep, Elysian, MN5608). The relative percentage for each composition was calculated with the area of each resolved peak against the total area of all peaks. Areas of identified peaks were quantified using 20:0 or 23:0 as internal standard, corrected for molecular weight, and mol% distributions and quantitative yields were calculated.
Liquid chromatography-mass spectrometry determination of 20:5n-3 in Drosophila fed a 22:6n-3 diet
A Waters 625 liquid chromatography (LC) system connected with Finnigan LCQ classic electrospray ionization tandem mass spectrometry (ESI-MS/MS) was applied for determination of 20:5n-3 in the dietary 22:6n-3 group. Four to five adults (3–6 mg) were homogenized in liquid nitrogen. The powder was then transferred into a clean glass tube with a screw cap. The powder was mixed with 5 ml chloroform-methanol (2:1) for extraction overnight. The mixture was centrifuged at 2,000 rpm for 10 min. The chloroform layer was transferred into a 16-ml screw cap glass bottle and dried under nitrogen. One milliliter of 0.5 M NaOH solution was added to the glass bottle, which was then filled with nitrogen, heated at 100°C for 1 h, and cooled to room temperature; the pH was adjusted to 3.52 with 2 N HCl, 2 ml hexane was added, and the mixture was vortexed vigorously for about 1 min. The mixture was centrifuged at 2,000 rpm for 10 min and frozen at −80°C. The top hexane layer was transferred into a clean tube and dried under nitrogen, and 100 μl methanol:H2O (50/50) was added. An aliquot of a 20-μl sample was injected onto the LC column for analysis. LC was performed using a binary gradient elution system consisting of solution A: 65% methanol with 0.1% glacial acid, and solution B: 100% methanol with 0.1% glacial acid. Separation was achieved using the following gradient program: 0–8 min 100% A, 8–35 min 100% A to 100% B, 35–36 min 100% B to 100% A, 36–40 min 100% A. The flow rate was set at 0.2 ml/min. According to the total ion chromatogram (TIC) of 20:5n-3 on selected reaction monitoring (SRM) mode and the fragment information of 20:5n-3, the presence of 20:5n-3 was identified.
Bioinformatics search of Drosophila FA desaturase and elongase enzymes
To assess the occurrence of FA desaturases and elongases in the D. melanogaster genome, a collection of human enzymes served as queries in protein sequence similarity searches. The queries were Δ-5 desaturases FADS1 and SC5DL, Δ-6 desaturase FADS2, stearoyl-CoA desaturases (SCDs) and SCD5, and elongase ELOVL6 in BLAST (11) searches against D. melanogaster genome build 5.2 at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with default parameters. Using the top Drosophila hit as a query against the human genome allowed assignment of the gene pairs as homolog or ortholog. Mutual best hits were considered orthologs.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 (SPSS, Inc., Chicago, IL). In biochemical analyses, the relative amount represents at least three independent experiments, and results are reported as mean ± SD. For statistical analyses, we conducted one-way ANOVA with Dunnett's test with p < 0.05 being statistically significant. Means denoted by the same letter do not significantly differ from each other, p > 0.05.
RESULTS
FA composition of diet and different Drosophila stocks
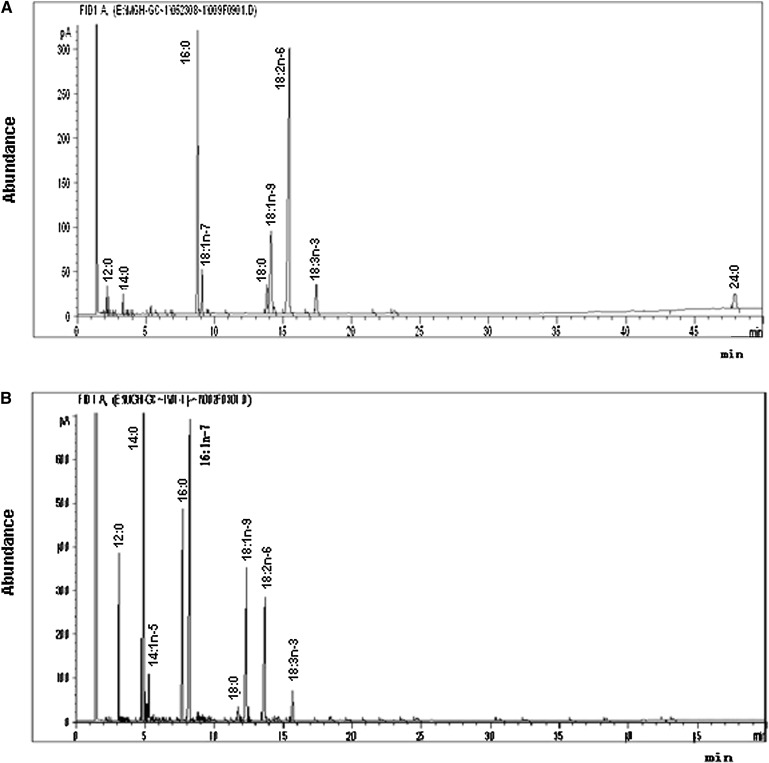
GC analysis showed that the base diet contained nine FAs: C12:0, C14:0, C16:0, 16:1n-7, C18:0, 18:1 n-9, 18:2 n-6, 18:3 n-3, and lignoceric acid (C24:0) (Fig. 1A). The relative amounts (RAs) in total FAs (RA% ± SD) of these components were: 0.81 ± 0.24, 0.77 ± 0.30, 23.78 ± 0.43, 3.73 ± 0.13, 4.87 ± 1.58, 15.07 ± 0.93, 44.28 ± 2.96, 2.23 ± 1.64, and 4.42 ± 0.21, respectively, with 18:2n-6, C16:0, and 18:1n-9 being predominant. Thus, the base diet contained five saturated FAs (SFAs), two monounsaturated fatty acids (MUFAs), and two C18 PUFAs; and no C20 or C22 PUFAs were detected.
Fig. 1.
Representative chromatograms of the FA composition in (A) base diet and (B) Canton-S female fly of Drosophila. Total lipids from fly samples were extracted and methylated with 14% boron trifluoride methanol solution and hexane. FA methyl esters were analyzed by an Agilent 6890N GC system with an Omegawax 250 capillary column. Peaks were identified by comparison with FA standards.
GC analysis of the extracts from four Drosophila stock adults (Canton-S, W1118, Oregon, and PR) showed that Drosophila contains nine FAs: C12:0, C14:0, 14:1n-5, C16:0, 16:1n-7, 18:0, 18:1n-9, 18:2n-6, and 18:3n-3 (Fig. 1B), with C14:0 (RA, 16.67 ± 3.49), C16:0 (16.11 ± 0.97), 16:1n-7 (21.81 ± 1.62), 18:1n-9 (20.03 ± 2.57), and 18:2n-6 (14.01 ± 1.42) being the five major FAs (Table 1). These nine FAs included four SFAs, three MUFAs, and two C18 PUFAs. However, no C20 or C22 PUFAs were found in Drosophila.
TABLE 1.
The profiles of FAs in different Drosophila stocks
| Relative Amount (% ± SD) |
||||
|---|---|---|---|---|
| FA | Canton-S | W1118 | Oregon | PR |
| 12:0 | 4.74 ± 1.05d | 3.84 ± 0.93f | 3.67 ± 0.44d,e | 5.07 ± 0.51e |
| 14:0 | 20.16 ± 4.00a,b | 13.61 ± 0.87e | 13.73 ± 2.80b,c | 19.18 ± 2.13a,b |
| 16:0 | 15.40 ± 0.44b,c | 17.51 ± 0.52c | 16.00 ± 1.52b | 15.53 ± 3.44b,c |
| 18:0 | 1.51 ± 0.34d | 1.96 ± 0.43g | 2.13 ± 0.20e | 1.70 ± 0.28f |
| 14:1n-5 | 1.41 ± 0.20d | 1.23 ± 0.44g | 1.28 ± 0.29e | 1.37 ± 0.10f |
| 16:1n-7 | 22.24 ± 0.91a | 19.87 ± 1.22b | 23.75 ± 2.19a | 21.38 ± 1.99a |
| 18:1n-9 | 18.03 ± 3.07a,b,c | 22.63 ± 0.49a | 21.85 ± 2.22a | 17.63 ± 1.70b,c |
| 18:2n-6 | 13.51 ± 1.66c | 15.94 ± 1.42d | 12.58 ± 2.18c | 14.03 ± 1.07d |
| 18:3n-3 | 3.02 ± 0.96d | 3.42 ± 0.73f | 5.01 ± 1.19d | 2.90 ± 0.76e,f |
| SFA | 41.80 ± 4.33 | 36.91 ± 2.07 | 35.53 ± 3.28 | 41.47 ± 0.90 |
| USFA | 58.20 ± 4.33 | 63.09 ± 2.08 | 64.46 ± 3.83 | 57.36 ± 2.32 |
The values (% of total FAs) are expressed as the means ± SD (n = 4). SFA, saturated FA; USFA, unsaturated FA. Means denoted with the same letter (a-g) do not significantly differ from each other.
GC analysis of Canton-S at different life stages and genders showed no significant difference in PUFA levels between male and female, but some marked differences among different stages (Table 2). For example, the levels of C12:0, 18:2n-6, and 18:3n-3 in all stages were nearly constant, C16:0 in larvae was less than that in pupae and adults, 18:1n-9 in pupae was lower than that in larvae and adults, and C14:0 and C18:0 in larvae and pupae were significantly higher than those in adults. Overall, the levels of unsaturated FAs (USFAs) in adults were significantly higher than those in larvae and pupae.
TABLE 2.
The profiles of FAs in different developmental stages of Canton-S Drosophila
| FA | Larvae | Pupae | Female | Male |
|---|---|---|---|---|
| 12:0 | 2.57 ± 0.54c,d | 2.14 ± 0.24d | 3.67 ± 0.91e | 3.64 ± 1.16c |
| 14:0 | 21.17 ± 4.50a | 17.77 ± 1.66b | 14.95 ± 1.61d | 14.83 ± 3.74b |
| 16:0 | 1.73 ± 0.98c | 25.29 ± 0.76a | 18.02 ± 1.85b | 17.08 ± 2.81b |
| 18:0 | 13.86 ± 3.53b | 14.22 ± 2.79b | 1.90 ± 0.26f,g | 1.97 ± 0.44c |
| 14:1n-5 | 12.32 ± 3.94d | 2.28 ± 0.47c | 1.16 ± 0.31g | 1.17 ± 0.22c |
| 16:1n-7 | 6.15 ± 2.22b | 10.73 ± 2.71c | 19.87 ± 2.14b | 20.11 ± 2.41a |
| 18:1n-9 | 24.42 ± 1.50a | 5.35 ± 2.18d | 21.64 ± 1.47a | 20.54 ± 2.60a |
| 18:2 n-6 | 16.07 ± 4.10b | 16.09 ± 4.74b | 14.85 ± 1.16d | 15.66 ± 2.87b |
| 18:3 n-3 | 2.73 ± 0.74c,d | 4.34 ± 1.86d | 3.38 ± 0.86e,f | 3.58 ± 1.26c |
| SFA | 54.10 ± 5.54 | 50.53 ± 0.11 | 38.54 ± 2.78 | 43.59 ± 7.03 |
| USFA | 46.70 ± 4.02 | 47.64 ± 0.29 | 60.90 ± 3.33 | 61.07 ± 5.45 |
The values (% of total FAs) are expressed as mean ± SD (n = 3).Means denoted with the same letter (a-g) do not significantly differ from each other.
Metabolism of supplemented PUFA in Drosophila
GC analyses were performed on Drosophila fed diets supplemented with six different PUFAs: 18:2n-6, 18:3n-3, 20:5n-3, 22:6n-3, 22:4n-6, and 20:4n-6. The levels (%) of PUFA in supplemented diets (22:6n-3, 20:5n-3, 22:4n-6, 20:4n-6, 18:2n-6, and 18:3n-3), were 12.59, 23.88, 14.04, 16.62, 56.21 (44.28 in base diet), and 37.10 (2.23 in base diet), respectively. Results showed that flies fed with 22:6n-3, 20:5n-3, 22:4n-6, and 20:4n-6 contained C20 and C22 PUFAs, whereas flies fed with the base diet did not. To determine whether the C20 and C22 PUFAs could be synthesized from 18:2n-6 and 18:3n-3 in Drosophila, the FA profile of flies fed with the diets supplemented with 18:2n-6 and 18:3n-3 was analyzed. The levels of the PUFAs (18:2n-6 and 18:3n-3) were significantly enhanced in the supplemented flies 27.99 ± 0.36 (14.42 ± 1.10 in control) and 12.26 ± 0.06 (2.47 ± 0.24 in control), respectively, when compared with control groups fed with the base diet. But no C20 or C22 PUFAs were detected from larvae, pupae, or adults fed with the diet supplemented with 18:2n-6 and 18:3n-3. These results suggest that C20 and C22 PUFAs cannot be endogenously synthesized from 18:2n-6 and 18:3n-3 in Drosophila.
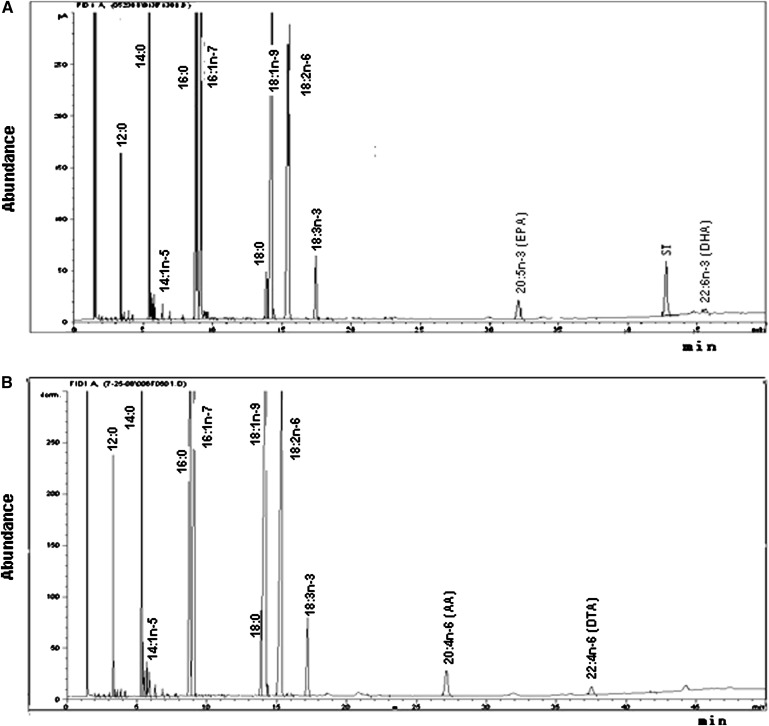
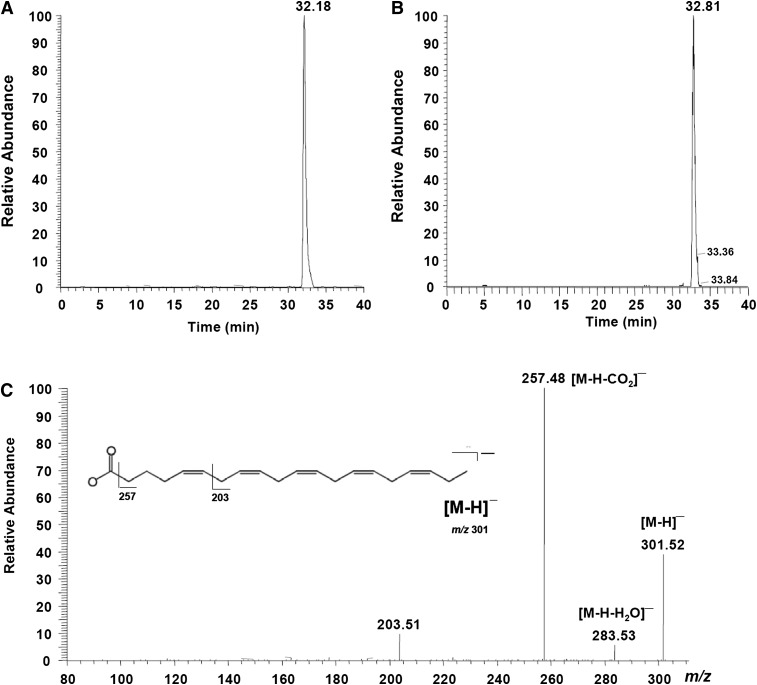
Interestingly, a distinct 20:5n-3 peak and a lower 22:6n-3 peak were observed in all three developmental stages of Drosophila that were fed with 22:6n-3 (Fig. 2A, Table 3). The peaks in Drosophila were identical to the 20:5n-3 standard (Fig. 3B). The relative levels of 20:5n-3 and 22:6n-3 in the 22:6n-3-supplemented flies were 1.76 and 0.31, suggesting that about 85% of 22:6n-3 in Drosophila was converted into 20:5n-3. The identity of 20:5n-3 was confirmed by an LC-MS analysis (Fig. 3A, C). As shown in Table 4, female, male, pupae, and larvae have a differential ability of converting C22 PUFA into C20 PUFA. Adults exhibited a significantly higher 20:5n-3/22:6n-3 ratio, compared with larvae and pupae, suggesting greater conversion ability. In adults, the ability to convert 22:6n-3 to 20:5n-3 was significantly higher in males than in females. A similar conversion was observed in flies fed 22:4n-6. The distinct peak of 20:4n-6 was found in all samples of adult (Fig. 2B, Table 3), larvae, and pupae supplemented with 22:4n-6 (Table 4). The levels of 20:4n-6 and 22:4n-6 in adults fed with 22:4n-6 were 1.37 and 0.22, respectively, indicating about an 86% conversion of 22:4n-6 to 20:4n-6 in Drosophila. Again, adults showed a significantly higher ability to convert C22 PUFA to C20 PUFA than did larvae and pupae (Table 4). However, no reduced (converted) products were found in Drosophila supplemented with C20 PUFA (20:4n-6 and 20:5n-3).
Fig. 2.
Representative chromatograms of FA composition of Drosophila adults supplemented with 22:6n-3 docosahexaenoic acid (DHA) (A) and Drosophila adults supplemented with 22:4n-6 docosatetraenoic acid (B). Total lipids from fly samples were extracted and methylated with 14% boron trifluoride methanol solution and hexane. FA methyl esters were analyzed by the Agilent 6890N GC system. Areas of identified peaks were quantified using 23:0 as internal standard, corrected for molecular weight, and mol% distributions and quantitative yields were calculated.
TABLE 3.
Fatty acid composition of Drosophila adults fed with 22:6n-3 (DHA) and 22:4n-6 (DTA) (n = 4)
| Relative Amount (% ± SD) |
||
|---|---|---|
| FA | Fed on DHA | Fed on DTA |
| 12:0 | 2.32 ± 0.32 | 3.14 ± 0.82 |
| 14:0 | 13.31 ± 1.33 | 13.10 ± 3.27 |
| 16:0 | 20.98 ± 0.93 | 17.61 ± 3.59 |
| 18:0 | 3.65 ± 3.18 | 2.70 ± 0.49 |
| 14:1n-5 | 0.63 ± 0.17 | 0.85 ± 0.15 |
| 16:1n-7 | 14.41 ± 1.70 | 17.20 ± 2.40 |
| 18:1n-9 | 21.64 ± 4.01 | 21.04 ± 2.69 |
| 18:2n-6 | 16.02 ± 1.67 | 19.02 ± 2.58 |
| 20:4n-6 | ND | 1.37 ± 0.62 |
| 22:4n-6 | ND | 0.22 ± 0.24 |
| 18:3n-3 | 4.21 ± 1.35 | 4.03 ± 1.39 |
| 20:5n-3 | 1.76 ± 0.23 | ND |
| 22:6n-3 | 0.31 ± 0.26 | ND |
DHA, docosahexaenoic acid; DTA, docosatetraenoic acid; ND, not detected.
Fig. 3.
Eicosapentaenoic acid (EPA) identification by liquid chromatography-mass spectrometry. (A) SRM mode of 20:5n-3 (EPA) in Drosophila adults fed with 22:6n-3 (DHA) and (B) EPA standard; (C) the MS fragment information of EPA in Drosophila adults fed with DHA.
TABLE 4.
Composition of C20 and C22 PUFAs in Drosophila of different stages fed with 22:6n-3 (DHA) and 22:4n-6 (DTA)
| Relative Amount % |
|||||
|---|---|---|---|---|---|
| Diets | C20 and C22 PUFA in Flies | Female | Male | Larvae | Pupae |
| Added DHA | EPA | 1.90 | 1.46 | 0.79 | 0.85 |
| DHA | 0.11 | 0.06 | 0.22 | 0.7 | |
| EPA/DHA | 17.27 | 24.33 | 3.59 | 1.21 | |
| Added DTA | AA | 1.98 | 2.35 | 1.24 | 0.41 |
| DTA | 0.16 | 0.23 | 0.41 | 0.52 | |
| AA/DTA | 12.38 | 10.22 | 3.02 | 0.79 | |
AA, arachidonic acid.
Bioinformatics of Drosophila enzyme
To examine whether Drosophila carries genes encoding Δ-6 and Δ-5 desaturases, the key enzymes for the synthesis of C20 and C22 PUFAs, we performed genetic searches for sequence similarity to known Δ-5 and Δ-6 desaturases. The results of sequence similarity searching strongly suggest that both Δ-5 and Δ-6 desaturases are not encoded by the D. melanogaster genome (Table 5). Human FADS1, FADS2, and SC5DL queries returned fly sequences that possess greater similarity over longer sequence length to sterol-C4-methyl oxidases and cytochrome b5 (Cyt-b5)-type proteins than to the desaturases. Specifically, this comparison showed that human Δ-5 desaturases FADS1 and SC5DL matched most closely to Drosophila proteins CG11162 and CG1998, encoding FA hydroxylase, and CG6870 and CG2140, encoding Cyt-b5-like heme/steroid binding domain, while Δ-6 desaturase most closely matched Drosophila Cyt-5b genes CG3566 and CG2140. These searches showed low to moderate sequence conservation (amino acid identity ranging from 33–44% for the top hit) over a short length of the query sequence (18–54% of query sequence aligned), indicating probable sharing of a short functional domain and, importantly, a lack of the Δ-5 and Δ-6 desaturases in the fly genome. In contrast, searches against the fly genome with human SCD and SCD5 and elongase ELOVL6 resulted in longer alignments (82–100% of query sequence aligned) of higher quality (46–58% amino acid identity) (Table 5). These results provide genetic evidence to support the notion that Drosophila does not have the ability to synthesize PUFAs, but is able to produce MUFAs.
TABLE 5.
Gene ontology hit of D -5 and D -6 desaturases
| Human Gene Term | Gene Symbol | D. melanogaster Hit # 1 (Gene Function) | D. melanogaster Hit # 2 (Gene Function) | Human Aligned |
|---|---|---|---|---|
| % | ||||
| Δ-5 Desaturase | FADS1 | CG11162:72.4:3e-13 (FA hydroxylase) | CG1998:71.6:5e-13(FA hydroxylase) | 56 |
| FADS1 | CG6870:64.7:1e-10 (Cyt-b5) | CG2140:55.1:9e-08 (Cyt-b5) | 20 | |
| Δ-6 Desaturase | FADS2 | CG3566:62.4:5e-10 (Cyt-b5) | CG2140:57.4:2e-08 (Cyt-b5) | 16 |
| Stearoyl-CoA desaturase ( D -9-desaturase) | SCD | CG5887 (FA desaturase) | CG5925 (FA desaturase) | 82 |
| Stearoyl-CoA desaturase-5 | SCD5 | CG5887 (FA desaturase) | CG5925 (FA desaturase) | 92 |
| Enlongase | ELOVL6 | CG3971 (very long chain FA elongase) | CG33110 (long-chain FA elongase) | 96 |
Enlongase, ELOVL family member 6, elongation of long-chain FAs (FEN1/Elo2, SUR4/Elo3-like, yeast); FADS, FA desaturases; SCD, stearoyl-CoA desaturase.
DISCUSSION
Given the fact that mammalian cells are able to convert the essential FAs 18:2n-6 and 18:3n-3 to corresponding C20 and C22 PUFAs (20:4n-6, 20:5n-3, 22:6n-3, etc.), which are the major components of cell membrane phospholipids and the precursors of important signaling molecules, determining whether or not Drosophila has the same capability would indicate its validity as an experimental model for the study of lipid metabolism and lipid-related diseases. Thus far, there has been no direct evidence of PUFA biosynthesis in Drosophila. In the present study, our results demonstrate that Drosophila cannot synthesize C20 and C22 PUFAs from 18:2n-6 and 18:3n-3, respectively, as evidenced by both the lipid profile of flies raised on 18:2n-6/18:3n-3-supplemented diets, which showed no C20 and C22 PUFAs in their body tissues, and by bioinformatic analysis of the Drosophila genome, which indicated a lack of Δ-6 and Δ-5 desaturases (the key enzymes for C20 and C22 PUFA biosynthesis). Our findings add new evidence to the notion that Drosophila flies do not require or biosynthesize C20 PUFA, as proposed by Stanley-Samuelson et al. (12), based on their observation that 10 consecutive generations of wild-type flies could be maintained on a synthetic diet lacking added FAs (13). However, we cannot exclude 18:2n-6 and 18:3n-3 as essential for other functions besides conversion to C20 and C22.
The C20 and C22 PUFA biosynthesis machinery, such as Δ-6/ Δ-5 desaturation, exists in various organisms, including mammals, nematodes, fungi, yeast, marine protists, microalgae, and moss (12). Many insects also require C20 and C22 PUFAs and have the elongation/desaturation pathways to produce these PUFAs from 18:2n-6 and 18:3n-3 (12). The lack of C20 PUFA in Drosophila makes it a special species that does not require C20 PUFA for formation of eicosanoids and other oxygenated metabolites, which are considered to be of broad physiological significance in animals. However, our data from the present study show that when C20 and C22 PUFAs are added to the diet, flies appear to absorb and incorporate the dietary PUFAs into body tissues. Interestingly, the majority of dietary C22 PUFAs, 22:6n-3 DHA and 22:4n-6 DTA, are quickly shortened into 20:5n-3 EPA and 20:4n-6 AA, respectively, in Drosophila. The conversion rate of C22 to C20 PUFAs seems to be exceptionally higher in Drosophila than that in mammals, such as mice, in which 22:6n-3 DHA content is normally much more than that of 20:5n-3 EPA. The biochemical mechanism(s) and physiological significance of this phenomenon in Drosophilia are unclear. It should be noted that the degradation of C20 and C22 PUFAs in vertebrates primarily occurs through the β-oxidation pathway. Whether Drosophila possesses a greater β-oxidation system or alternative mechanisms to metabolize PUFAs requires further investigation.
According to the report of Gutierrez et al. (14), Drosophila has a fat body filled with an adipose-like tissue and a lipid transport system similar to humans, which is considered one of the major advantages of it serving as a unique model for nutrigenomics research (15). However, Gutierrez et al. also found that the lipid metabolism in Drosophila requires directional coupling between fat and oenocytes, which synthesize, modify, and oxidize FAs. It is possible that FAs in Drosophila could be shortened, at least partially, by the peroxisomal β-oxidation enzyme system. Several genes, including CG11151 (similar to Sterol carrier protein 2), CG12428 (Carnitin O-octanoyl transferase), and CG9527 (Pristanoyl-CoA oxidase), etc., have been found to encode enzymes involved in peroxisomal β-oxidation. Larval oenocytes in insects were first described over 140 years ago, but their functions remain unclear (14). It is likely that the β-oxidation enzymatic system in Drosophila might exist in their fat and oenocytes, which is different from mammals. The greater shortening of 22:6n-3 and 22:4n-6 in Drosophila suggests that Drosophila has a unique lipid metabolism system.
On the basis of the FA composition, genetic information, and metabolism of supplemented PUFAs in Drosophila observed in our study, we conclude that Drosophila lacks C20 and C22 PUFAs and appears to have a special lipid metabolism system that is quite different from that in mammals. Thus, Drosophila has certain limitations and might not be a valid experimental model for the study of lipid metabolism and related diseases.
Acknowledgments
The authors thank Mary Roberts, Xi-Ying Qu, Lan Ma, and Bai Zhen-Zhong for their excellent technical assistance, and Chengwei He, Jian Shen, and Yu-Chi Lee for helpful discussions.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- ALA
- α-linolenic acid
- Cyt-b5
- cytochrome b5
- DHA
- docosahexaenoic acid
- DTA
- docosatetraenoic acid
- ELOV
- elongation of very-long-chain FAs
- EPA
- eicosapentaenoic acid
- FADS
- FA desaturases
- LA
- linoleic acid
- LT
- leukotriene
- LX
- lipoxin
- PC
- prostacyclin
- PD
- protectin
- PG
- prostaglandin
- RA
- relative amount
- Rv
- resolvin
- SFA
- saturated fatty acid
- SC5DL
- sterol-C5-desaturase (ERG3 D -5-desaturase homolog, Saccharomyces cerevisiae)-like)
- SCD
- stearoyl-CoA desaturase
- TIC
- total ion chromatogram
- TX
- thromboxane
- USFA
- unsaturated FA
This study was supported by National Institutes of Health Grant CA-113605 and grant 2007AA10Z324 from The Ministry of Science and Technology of China. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., Scherer S. E., Li P. W., Hoskins R. A., Galle R. F. 2000. The genome sequence of Drosophila melanogaster. Science. 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- 2.Helfand S. L., Rogina B. 2003. Molecular genetics of aging in the fly: is this the end of the beginning? Bioessays. 25: 134–141. [DOI] [PubMed] [Google Scholar]
- 3.Piper M. D., Skorupa D., Partridge L. 2005. Diet, metabolism and lifespan in Drosophila. Exp. Gerontol. 40: 857–862. [DOI] [PubMed] [Google Scholar]
- 4.Ruden D. M., Lu X. 2006. Evolutionary conservation of metabolism explains how Drosophila nutrigenomics can help us understand human nutrigenomics. Genes Nutr. 1: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salem N. J., Simopoulos A. P., Galli C., Lagarde M., Knapp H. R., eds 1996. Fatty acids and lipids from cell biology to human disease. Lipids. 31 (Suppl.): 1–326.8649226 [Google Scholar]
- 6.Weylandt K. H., Kang J. X. 2005. Rethinking lipid mediators. Lancet. 366: 618–620. [DOI] [PubMed] [Google Scholar]
- 7.Schuchardt J. P., Huss M., Stauss-Grabo M., Hahn A. 2008. Significance of long-chain polyunsaturated fatty acids (PUFA) for the development and behaviour of children. Eur. J. Pediatr. 169: 149–164. [DOI] [PubMed] [Google Scholar]
- 8.Oresic M., Hanninen V. A., Vidal-Puig A. 2008. Lipidomics: a new window to biomedical frontiers. Trends Biotechnol. 26: 647–652. [DOI] [PubMed] [Google Scholar]
- 9.Troen A. M., French E. E., Roberts J. F., Selhub J., Ordovas J. M., Parnell L. D., Lai C. Q. 2007. Lifespan modification by glucose and methionine in Drosophila melanogaster fed a chemically defined diet. Age (Dordr.). 29: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang J. X., Wang J. 2005. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley-Samuelson D. W., Jurenka R. A., Cripps C. 1988. Fatty acids in insects: composition, metabolism, and biological significance. Arch. Insect Biochem. Physiol. 9: 1–33. [Google Scholar]
- 13.Rapport E. W., Stanley-Samuelson D., Dadd R. H. 1984. Ten generations of Drosophila melanogaster reared axenically on a fatty acid-free holidic diet. Arch. Insect Biochem. Physiol. 1: 243–250. [Google Scholar]
- 14.Gutierrez E., Wiggins D., Fielding B., Gould A. P. 2007. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 445: 275–280. [DOI] [PubMed] [Google Scholar]
- 15.Ruden D. M., De Luca M., Garfinkel M. D., Bynum K. L., Lu X. 2005. Drosophila nutrigenomics can provide clues to human gene-nutrient interactions. Annu. Rev. Nutr. 25: 499–522. [DOI] [PubMed] [Google Scholar]