Abstract
Variation in serum cholesterol, free-fatty acids, and triglycerides is associated with cardiovascular disease (CVD) risk factors. There is great interest in characterizing the underlying genetic architecture of these risk factors, because they vary greatly within and among human populations and between the sexes. We present results of a genome-wide scan for quantitative trait loci (QTL) affecting serum cholesterol, free-fatty acids, and triglycerides in an F16 advanced intercross line of LG/J and SM/J (Wustl:LG,SM-G16). Half of the population was fed a high-fat diet and half was fed a relatively low-fat diet. Context-dependent genetic (additive and dominance) and epigenetic (imprinting) effects were characterized by partitioning animals into sex, diet, and sex-by-diet cohorts. Here we examine genetic, environmental, and genetic-by-environmental interactions of QTL overlapping previously identified loci associated with CVD risk factors, and we add to the serum lipid QTL landscape by identifying new loci.
Keywords: LG/J by SM/J advanced intercross, cholesterol, free-fatty acid;, triglyceride, imprinting, quantitative trait locus
Cardiovascular disease (CVD) has a complex etiology and is the leading cause of death in the United States (1). Risk factors for CVD include dyslipidemia (e.g., high plasma cholesterol and triglycerides), elevated blood pressure (e.g., hypertension), and obesity [e.g., body mass index (BMI) > 30.0 kg/m2]. These factors have strong environmental contributions, including whether an individual smokes, activity level, and percentage of dietary saturated fat (2). Heritability estimates for the risk factors of CVD indicate there is a strong genetic contribution as well, and heritabilities vary between the sexes as well as within and across ethnic populations (3). Many candidate genes for CVD risk factors have been identified in human linkage and in genome-wide association studies (GWAS) (4). However, these genes account for a very small proportion of the overall heritable variation of risk, approximately 5–10% cumulatively (5). This is due partly to the confounding factors of genetic and environmental heterogeneity in human populations and partly to the lack of statistical power to detect genes with small phenotypic effects, those genes that underlie much of the variation in complex traits, such as circulating lipid levels, blood pressure, and obesity.
Despite not developing CVD per se, mice have nevertheless made major contributions to our knowledge of disease etiology, particularly in our understanding of disease physiology and in our identification of genetic risk factors. This is because phenotypes are ascertained in controlled environments and large numbers of offspring are generated from a set of founder animals of known genomic background. For example, after the mouse leptin and the leptin receptor pathways were characterized, subsequent human familial studies identified over 600 mutations in the homologous LEPR gene (6). Additionally, mutations in other genes in the LEP and LEPR pathways, e.g., apolipoprotein B (7) and the ATP-binding cassette (ABCG5) (8, 9), have been characterized in humans and associated with CVD risk. Recently, a QTL associated with variation in blood pressure in a mouse model was used to identify the candidate gene uredopropionase (Upb1). Studies of the human homolog revealed this locus to be a determinant of variation in both systolic and diastolic blood pressures (10). Currently there are approximately 250 different mouse strains used for CVD risk research, including 27 to model hypertension, 57 to model hypercholesterolemia, and 17 to model hypertriglyceremia (www.jaxmice.org/research/index/html).
Here we present results of a study examining variation in serum cholesterol, free-fatty acids, and triglycerides in an F16 generation of the LG/J×SM/J advanced intercross line (Wustl:LG,SM-G16). The LG/J×SM/J cross has proven to be an excellent system for identifying QTL associated with variation in serum lipid levels and with variation in other metabolic traits, such as obesity and glucose tolerance (11, 12). Genetic responses to high- and low-fat diets between these two strains, as well as trait heritabilities, have been reported elsewhere (13–15). Here we utilize the LG/J×SM/J cross to dissect the complex interactions of genetic effects, environmental factors, and the interplay between them by examining genome-wide genetic and, for the first time, genomic imprinting effects on serum lipids among different sex, diet, and sex-by-diet cohorts. Understanding how genetic variants interact with the environment is critical for understanding the genetics of CVD risk factors. We examine the genetic architecture of previously identified QTL associated with CVD risk factors, and we add to the serum lipid QTL landscape by identifying new loci.
MATERIALS AND METHODS
Mouse population
The mice used in this study are from the F16 generation of the LG/J×SM/J advanced intercross line (Wustl:LG,SM-G16). The line is managed as a pseudo-randomly mated line starting from the F2 generation. One male and one female are chosen from each family as breeders for the next generation. These animals are randomly mated, except that sibling mating is not allowed. For this study, 71 pairs of F15 animals were double mated, producing an experimental F16 population of 1,002 animals in 76 litters, averaging 6.8 animals per sibship. Pups were housed with their mothers until weaning at three weeks of age, and then they were separated into sex-specific cages of no more than five animals per cage [details of the animal husbandry are described in Ehrich et al. (16)]. At this time, one-half of the animals from each litter were fed a high-fat diet (253 males, 248 females), and one-half were fed relatively low-fat diet (247 males, 254 females). The two diets were chosen to be as nutritionally similar as possible (supplementary Table I), with the exception of percentage of calories obtained from fat, which is 15% in the low-fat diet (catalog D12284, Research Diets, New Brunswick, NJ) and 43% in the high-fat diet (catalog TD88137, Harlan Teklad, Madison, WI). All animals were fed ad libitum.
Phenotyping
At 20 weeks of age, animals were fasted for 4 h and anesthetized with sodium pentobarbital. A terminal blood sample was collected via cardiac puncture. Serum was frozen at −20°C until assayed. Concentrations of cholesterol, free-fatty acids, triglycerides, glucose, and insulin were measured by the Nutrition Obesity Research Center, Animal Model Research Core at Washington University. Additionally, fat pads (inguinal, mesenteric, renal, and reproductive) and internal organs (heart, kidneys, liver, and spleen) were removed and weighed. Genetic mapping of the fat pads and of glucose and insulin levels, as well as response to glucose stress, is reported in Cheverud et al. (11) and Lawson, Lee, Fawcett, et al. (unpublished observations).
Genotyping
DNA was extracted from liver tissue using the QIAGEN kit, and 1536 single nucleotide polymorphisms (SNP) were selected from the CTC/Oxford SNP survey (www.well.ox.ac.uk/mouse/INBREDS/) for scoring with the Illumina Golden Gate Bead Array. SNP genotyping was performed at the Washington University Genome Sequencing Center. At total of 1,402 autosomal SNPs were reliably scored and used for this analysis (supplementary Table II). Recombination fractions between the markers were estimated using the package R/qtl (17), and a genetic map was created for the SNPs based on their physical order along the autosomes (mm9; NCBI build 37).
Ordered genotypes were reconstructed at each marker for all F16 animals from familial SNP data (F15 parents and their F16 offspring) using the integer linear programming algorithm as implemented in PedPhase 2.1 (18). Due to the computational intensity of the algorithm, it was necessary to partition the larger chromosomes. Additive (Xa) and dominance (Xd) genotypic scores were assigned at each marker: Xa = 1, 0, −1 and Xd = 0, 1, 0 for the LG/LG, LG/SM and SM/LG, and SM/SM genotypes, respectively. “LG” refers to an allele derived from the LG/J strain, and “SM” refers to an allele derived from the SM/J strain. Further, we assigned imprinting genotypic scores (Xi) to distinguish between the two reciprocal heterozygotes, LG/SM and SM/LG, where the first allele is inherited from the father, and the second from the mother. For the four ordered genotypes, LG/LG, LG/SM, SM/LG, and SM/SM, Xi = 0, +1, −1, 0, respectively (19). Additional genotypes were imputed at 1cM intervals between the most proximal and the most distal SNP on each autosome using the equations of Haley and Knott (20), with the inclusion of newly derived equations for imputing imprinting genotypic scores (supplementary Table III).
QTL analysis
Single locus analyses were performed across each autosome using maximum likelihood in the Mixed Procedure in SAS (version 9.2; SAS Institute, Cary, NC). Our full mapping model included sex, diet, the direct effects of the genomic locations (Xa, Xd, Xi), and their two- and three-way interactions with sex and diet as fixed effects. We included family, sex, diet, and their two- and three-way interactions as random effects in the model. Inclusion of these random effects accounts for the influence of family structure, which could inflate the results. The full model explains variation in trait (Y) using the linear equation:
Yijklm = μ + Sexi + Dietj + aXak + dXdl + iXim + sd(Sexi×Dietj) + as(Xak×Sexi) + ds(Xdl×Sexi) + is(Xim×Sexi) + ad(Xak×Dietj) + dd(Xdl×Dietj) + id(Xim×Dietj) + asd(Xak×Sexi×Dietj) + dsd(Xdl×Sexi×Dietj) + isd(Xim×Sexi×Dietj) + eijklm
where μ is the population mean and e is the residual. The −2 ln (likelihood) of this model was compared with a null model:
Yijklm =μ + Sexi + Dietj + sd(Sexi×Dietj) + eijklm
using a chi-square test with 12 degrees of freedom. Probabilities were transformed into logarithm of odds (LOD) = −log10(Pr). The regression coefficients are the additive [a = (GLG/LG−GSM/SM)/2], dominance {d = [(GLG/SM+GSM/LG)−(GLG/LG−GSM/SM)]/2} and imprinting [i = (GLG/SM−GSM/LG)/2] genotypic values, where G refers to the average phenotypic value of all individuals sharing the subscripted genotype. The coefficients are combined, when appropriate, with the interacting factors of sex (as, ds, is), of diet (ad, dd, id), and of sex-by-diet (asd, dsd, isd). If the full model fit the data better than the null model, we examined the coefficients at the locus post hoc to identify the genetic effects and any significant interactions with sex, diet, and/or sex-by-diet.
The number of independent tests, both genome-wide and chromosome-wise, was calculated using the eigenvalues of the correlation matrix of the marker additive genotypic scores as described in Li and Ji (21). This was then used to calculate Bonferroni adjusted significance thresholds, 1−(1−α)1/M, where M is the number of independent tests, at the genome-wide level (LOD ≥ 3.97) as well as separately for each autosome (supplementary Table IV). The chromosome-wise threshold is less conservative than the genome-wide threshold and has been shown to increase discovery of true positives while avoiding problems using the false discovery rate in linkage mapping (22). A standard one LOD drop from the peak of the QTL was used to determine the 95% confidence intervals.
RESULTS
Mapping results
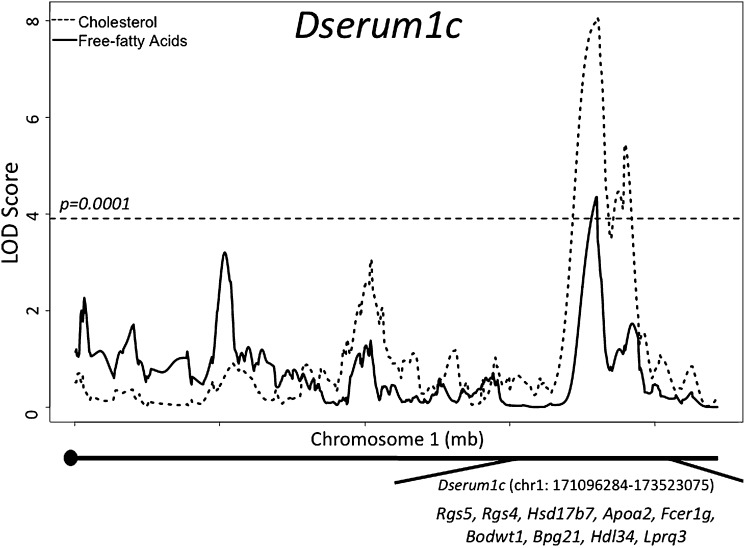
We identified 25 trait-specific loci for serum cholesterol, free-fatty acids, and triglycerides mapping to 23 locations across the genome. Of these 25 QTL, 4 are highly significant by the genome-wide threshold of LOD ≥ 3.97, and 21 pass chromosome-wise significance levels. The most commonly mapped trait is cholesterol with 10 QTL. Triglycerides have 9 QTL, followed by free-fatty acids with 6 QTL. The average QTL spans 4 Mb and contains 51 genes. Many of these genes have been demonstrated to affect serum chemistry and are well-studied positional candidates for susceptibility to dyslipidemia and hypertension. We find that 14 of our QTL correspond to known QTL previously mapped in mouse models of dyslipidemia, hypertension, and atherosclerosis that utilized strains both related and unrelated to LG/J and SM/J (Mouse Genome Database queried October 10, 2009). For example, we find a highly significant QTL on chromosome 1, Dserum1c, which contains the candidate genes Rgs5, Rgs4, Hsd17b7, Apoa2, and Fcer1g, and which overlaps previously identified QTL, Bodwt1, Bpg21, Hdl34, and Lprq3 (Fig. 1). Additionally, we identify 9 novel QTL, each of which contains fruitful candidates for further investigation (Table 1).
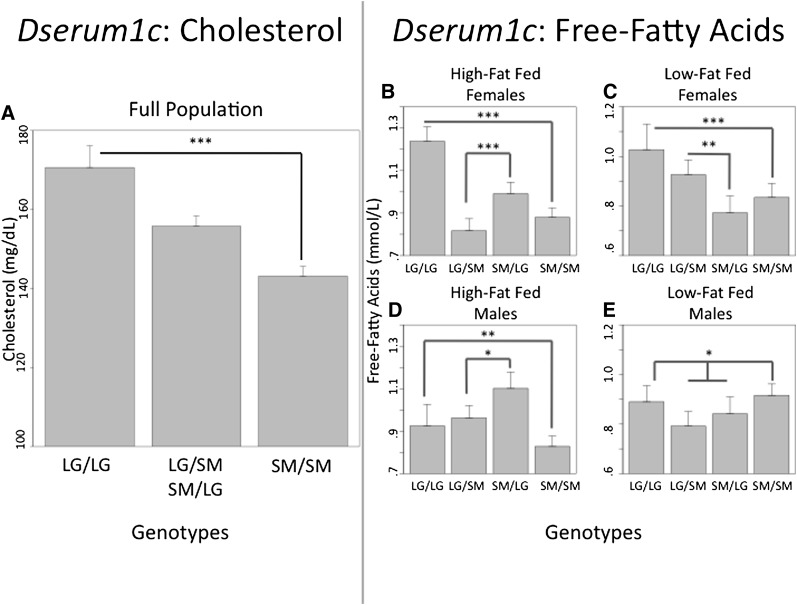
Fig.1.
A highly significant QTL mapped to chr1: 171096284-176523075, Dserum1c. We find this locus is pleiotropic, affecting variation in both cholesterol and free-fatty acids. This QTL contains a number of candidate genes that are well studied in association with CVD risk factors. Additionally, this QTL overlaps previously identified QTL. CVD, cardiovascular disease; QTL, quantitative trait loci.
TABLE 1.
Breakdown of the genome-wide QTL mapped in the study
| Interaction |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | QTL | Trait | LOD | POS (Mb) | Proximal CI | Distal CI | ADD | DOM | IMP | MAT | TotalGenes | Candidate Genes | Known QTL |
| 1 | Dserum1a | FFA | 3.19 | 42.7 | 42.2 | 47.0 | F | Full, HM | 25 | ||||
| Dserum1b | Chol | 3.05 | 85.6 | 82.0 | 90.8 | Full | 77 | Akp3, Dgkd, Trpm8 | |||||
| Dserum1c | FFA | 4.34 | 173.0 | 171.1 | 173.5 | Full, F, HM | HF, HM, LM | H, LF | H,LF | 48 | Rgs5, Rgs4, Hsd17b7, Apoa2, Fcer1g | Bodwt1, Bpq21, Hdl34, Lprq3 | |
| Chol | 8.05 | 173.1 | Full | ||||||||||
| Dserum1d | Chol | 5.45 | 179.1 | 177.8 | 180.6 | Full | 15 | Pcho1, Bw8q1 | |||||
| 3 | Dserum3a | Chol | 3.05 | 93.8 | 89.0 | 96.1 | Full | 199 | Shc1, Kcnn3, Npr1, S100a1, Ctss | Idd10, Hdlq49 | |||
| 4 | Dserum4a | TG | 3.28 | 155.1 | 155.1 | 155.5 | HM | Full, H | 36 | ||||
| 5 | Dserum5a | Chol | 3.26 | 104.1 | 100.5 | 107.1 | Full, H | 49 | Hsd17b13, Hsd17b11, Spp1 | Lprq1, Hdl2 | |||
| 6 | Dserum6a | TG | 2.88 | 3.8 | 3.8 | 8.6 | Full | 30 | Pdk4, Tac1 | Nhdlq5, Lith10 Axtq2 | |||
| 7 | Dserum7a | Chol | 4.40 | 88.9 | 88.1 | 91.2 | Full, HM | Full | 29 | Stard5 | |||
| Dserum7b | TG | 3.00 | 111.4 | 107.2 | 112.5 | LF, LM, HM | LM | 196 | Kcne3, Ucp3, Ucp3 | Chldq4, Nidd1k, Elpt | |||
| 8 | Dserum8a | Chol | 2.82 | 86.3 | 85.4 | 87.1 | HF | HF, LM | 36 | Ucp1, Ptger1 | |||
| Dserum8b | FFA | 3.72 | 104.4 | 100.2 | 108.1 | Full | LF | 45 | Rrad, Nol3 | Tgl1, Hdlq50, Plbcq4 | |||
| Dserum8c | TG | 2.63 | 131.0 | 128.0 | 131.0 | Full | LF | 16 | |||||
| 9 | Dserum9a | FFA | 3.85 | 118.8 | 118.3 | 119.5 | HF | Full | 20 | Scn5a | Obq18, Tgq2 | ||
| 10 | Dserum10a | Chol | 2.78 | 74.6 | 73.1 | 79.1 | Full, F | HF | 101 | Adora2a, Mif, Lss | |||
| Dserum10b | TG | 2.71 | 98.7 | 94.9 | 101.2 | Full | Full | 39 | Dcn | Lith7 | |||
| 11 | Dserum11a | TG | 3.81 | 45.7 | 44.2 | 46.8 | Full, HF | LF, LM | LF, LM | 25 | Adam19 | Hdlq51, Gwth4 | |
| 13 | Dserum13a | FFA | 2.72 | 36.0 | 32.1 | 36.7 | Full | Full, F | 36 | Hypt | |||
| 16 | Dserum16a | TG | 2.67 | 85.1 | 83.8 | 88.2 | Full | Full | 18 | ||||
| 17 | Dserum17b | TG | 3.36 | 9.6 | 6.2 | 11.3 | HF | Full, H | 33 | Idd16a, Idd23, Obq19, Obq4, Adip18 | |||
| Dserum17a | Chol | 2.83 | 31.8 | 31.4 | 35.2 | Full | Full | 135 | Cbs, Angptl4, Ager, Cfb, | ||||
| 18 | Dserum18a | Chol | 2.772 | 72.6 | 71.4 | 74.2 | Full, HF | HM | 7 | Idd1, Idd16, Idd16a | |||
| 19 | Dserum19a | TG 2.719 | FFA 2.495 | 28.5 | 30.1 | Full | Full | Full | 24 | Jak2 | Idd21b | ||
Bold LOD scores indicate QTL significant at the genome-wide threshold, all others are significant at their respective chromosome-wise thresholds (see supplementary Table II). ADD, additive genotypic effect; Chol, cholesterol; Chr, chromosome; CI, confidence interval; DOM, dominance genotypic effect; Full, full F16 population; H, all high-fat fed individuals; HF, high-fat fed females; HM, high-fat fed males; IMP, imprinting genotypic effect; L, all low-fat fed individuals; LF, low-fat fed females; LM, low-fat fed males; LOD, logarithm of odds; MAT, maternal effect; POS, genomic position; QTL, quantitative trait loci; TG, triglyceride.
Genetic effects of QTL
The genetic effects of these QTL are small, which is the general case for genes underlying variation in complex traits such as serum lipids. Significant additive effects average approximately 0.25 SD for cholesterol (range, 0.10–0.43 SD) so that, on average, the two homozygotes are approximately 0.50 SD units or 28.05 mg/dl apart. For free-fatty acids, significant additive effects average approximately 0.27 SD (range, 0.13–0.46 SD) so that, on average, the two homozygotes are approximately 0.54 SD units or 0.262 mmol/l apart. For triglycerides, significant additive effects average approximately 0.22 SD (range, 0.14–0.42 SD) so that, on average, the two homozygotes are approximately 0.44 SD units or 20.03 mg/dl apart. Significant dominance effects also tend to be relatively small, averaging approximately 0.27 absolute SD units for cholesterol, approximately 0.32 absolute SD units for free-fatty acids, and approximately 0.32 absolute SD units for triglycerides. The same holds true for significant imprinting effects, which average approximately 0.18 absolute SD units for cholesterol, approximately 0.24 absolute SD units for free-fatty acids, and approximately 0.21 absolute SD units for triglycerides.
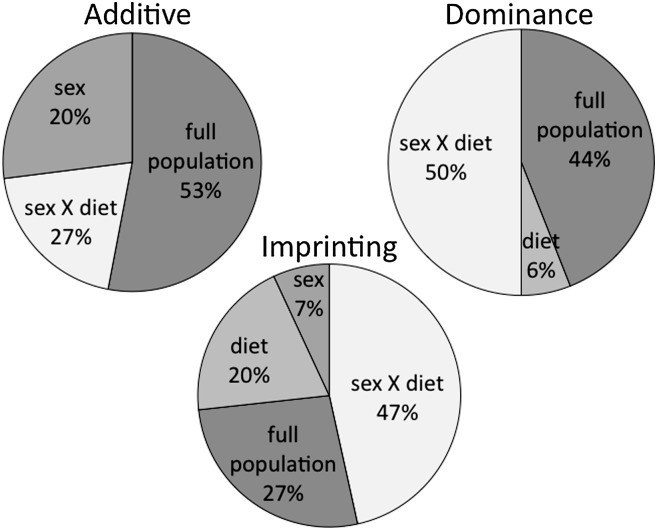
Surprisingly, we find that dominance and imprinting effects occur as frequently as additive effects. Further, many of these QTL have significant interactions with sex, with diet, and/or with sex and diet jointly (Fig. 2). On average, for QTL with additive effects among the nine cohorts (the full F16 population, sex, diet, or sex-by-diet cohorts), animals that are LG homozygotes have higher levels of cholesterol, free-fatty acids, and triglycerides. For QTL with dominance effects among the cohorts, the LG allele is dominant to the SM allele 57% of the time for cholesterol, 20% of the time for free-fatty acids, and 50% of the time for triglycerides. Additionally, we see 7 examples of loci showing under-dominance effects, where heterozygote animals have significantly lower serum lipids than either of the two homozygotes, and 13 examples of loci showing over-dominance effects, where heterozygote animals have significantly higher serum lipids than either of the two homozygotes. For QTL with imprinting effects among the cohorts, 72% of imprinting values are positive for cholesterol, 53% are positive for free-fatty acids, and 50% are positive for triglycerides, indicating that most often, heterozygote animals that inherit their LG allele from their fathers and their SM allele from their mothers have higher serum lipids.
Fig.2.
Relative occurrences of genetic effects in the full population, and of genetic effects interacting with sex, diet, and/or sex-by-diet.
Maternal effects (i.e., the effect of the maternal genotype and, hence, maternal environment, on the expression of traits in her offspring) have been shown to produce genetic patterns similar to imprinting (23). To determine whether maternal effects contribute to the imprinting patterns we identify, we reran the full model, including maternal additive and dominance scores, and their 2- and 3-way interactions with diet and sex at loci showing significant imprinting effects. Of the 13 QTL showing imprinting effects, 2 loci, Dserum1c and Dserum11a, show maternal effects in addition to the imprinted effects. One locus, Dserum8c, shows significant maternal effects with no imprinting (Table 1).
The imprinting patterns among the cohorts are complex, with 4 examples of paternal expression imprinting, 6 examples of maternal expression imprinting, 14 examples of polar dominance imprinting (no additive effects), and 17 examples of bipolar dominance imprinting (no additive or dominance effects) at the locus. Detailed descriptions of the varied patterns of imprinting effects can be found in Wolf et al. (24) and in Cheverud et al. (25). Supplementary Table V lists genotypic values for all 25 trait-specific QTL for all cohorts.
Context dependency of QTL
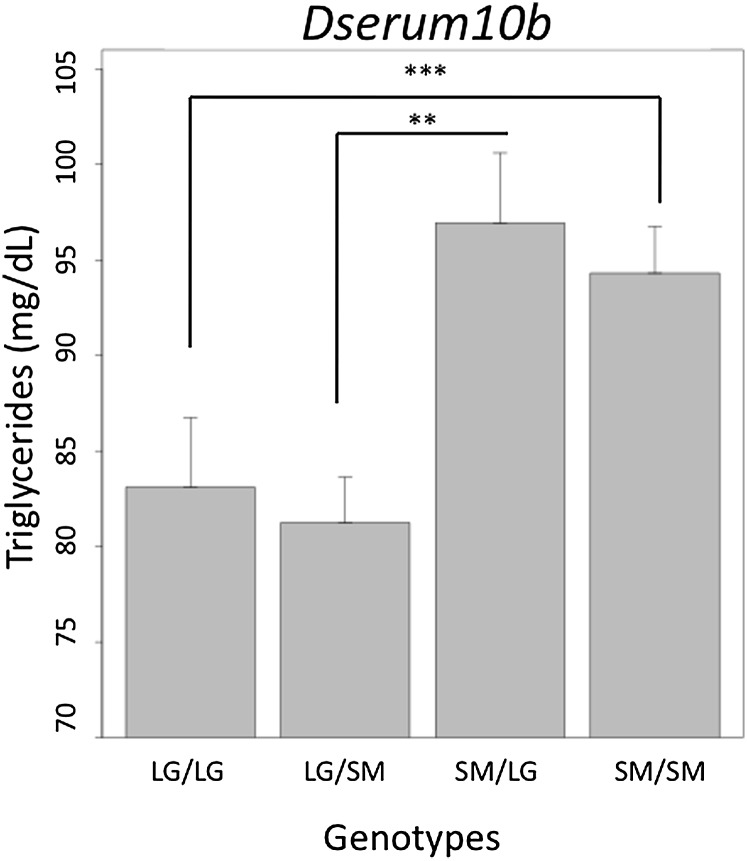
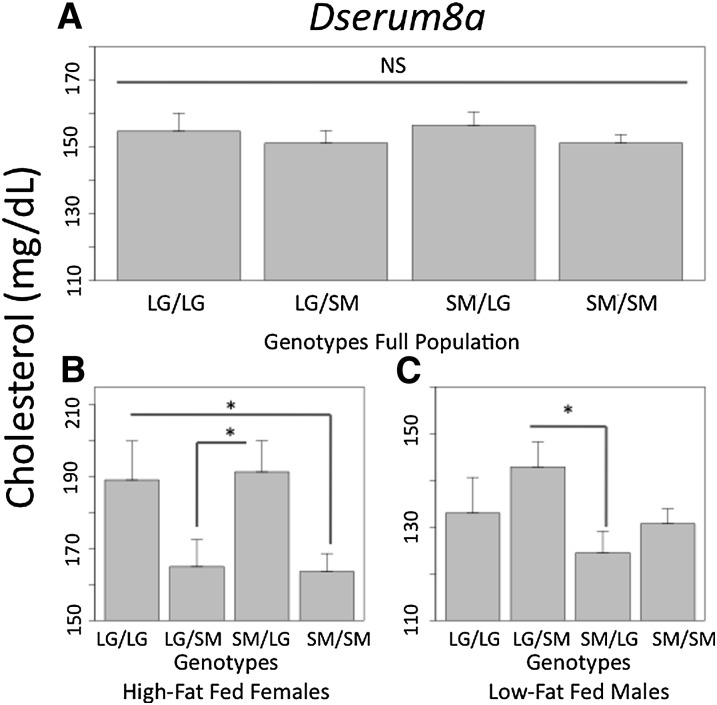
An intriguing result of this study is the importance of context to the underlying genetic architecture of serum levels. While it is well known that sex and diet are important factors contributing to heritable variation in CVD risk factors, we show that the underlying genetic effects themselves are highly context-dependent. For example, Fig. 3 illustrates a QTL, Dserum10b, which is significant in the full F16 population. The locus has an additive effect, where animals homozygous for the SM allele have higher triglycerides than animals homozygous for the LG allele. Additionally, this locus has paternal expression imprinting, where heterozygote animals that inherit their SM allele from their fathers have higher triglycerides than heterozygote animals inheriting their SM allele from their mothers. We find that 15 of the 25 trait-specific QTL show genotypic effects in multiple cohorts. Often, when genotypic effects are found in multiple cohorts, they affect the cohorts in different ways, and the effects are not always seen in the full population (Table 1). For example, at a novel QTL identified on chromosome 8, Dserum8a, which is associated with variation in cholesterol, we find a significant gene-by-sex-by-diet interaction (Fig. 4). In females fed a high-fat diet, there is a significant additive effect where animals homozygous with the LG allele have higher cholesterol. An additive effect is not seen in any cohort besides the high-fat fed females and does not register as significant in the full population. Additionally, there is significant maternal expression imprinting in the high-fat fed females, where heterozygote animals inheriting their LG allele from their mothers have higher cholesterol than heterozygote animals inheriting their LG allele from their fathers. In low-fat fed males, there is significant bipolar dominance imprinting (no additive or dominance effects), where heterozygote animals inheriting their LG allele from their father have higher cholesterol than heterozygote animals inheriting their LG allele from their mothers. The imprinting effects seen in the high-fat fed females and in the low-fat fed males are of the opposite signs. These effects do not register as significant in the full population because they cancel each other.
Fig.3.
A QTL significant in the full population, Dserum10b, associated with variation in triglycerides. This locus has additive effects, where SM homozygotes have higher triglycerides than LG homozygotes. Additionally, this locus has paternal expression imprinting, where heterozygote animals inheriting their SM allele from their fathers have higher triglycerides. ***P < 0.001; **P < 0.01. LG, allele derived from the LG/J strain; QTL, quantitative trait loci; SM, allele derived from the SM/J strain.
Fig.4.
A novel QTL, Dserum8a, associated with variation in cholesterol. In high-fat fed females, there is a significant additive effect, where LG homozygotes have higher cholesterol than SM homozygotes (B). This effect does not register as significant in the full population because the signal is washed out by absence of genetic effects in the other sex-by-diet cohorts (A). Further, high-fat fed females have significant maternal expression imprinting, where animals inheriting their LG allele from their mothers have higher cholesterol (B). In low-fat fed males, there is significant bipolar dominance imprinting (C). Imprinting effects do not register in the full population because the negative value in the females and the positive value in the males cancel each other out (A). *P < 0.05. LG, allele derived from the LG/J strain; QTL, quantitative trait loci; SM, allele derived from the SM/J strain.
The complexity of this context dependency is further illustrated at Dserum1c discussed above and shown in Fig. 1. This highly significant pleiotropic locus is associated with variation in both cholesterol and free-fatty acids. However, the genetic architecture of the locus is different for each trait (Fig. 5). For cholesterol, there is a highly significant additive effect in the full F16 population, where animals homozygous for the LG allele have higher cholesterol. Heterozygote genotypic values fall between the two homozygotes, and there is no significant difference between the two reciprocal heterozygotes. At this same locus, for free-fatty acids, the genotypic effect is dependent on an animal's sex and diet. All sex-by-diet cohorts, except the low-fat fed males, have significant additive effects, where animals homozygous for the LG allele have higher free-fatty acids. High-fat fed females have significant dominance effects, with the SM allele dominant to the LG. Additionally, high-fat fed females have significant maternal expression imprinting, where heterozygote animals have higher free-fatty acids when they inherit their LG allele from their mothers than when they inherit their LG allele from their fathers. Low-fat fed females have significant paternal expression imprinting, where heterozygote animals inheriting their LG allele from their father have higher free-fatty acids than when they inherit their LG allele from their mothers. High-fat fed males have significant over-dominance effects, with the heterozyotes having higher free-fatty acid levels than either of the two homozygotes at this locus. Additionally, high-fat fed males have significant maternal expression imprinting. In low-fat fed males, there are no significant imprinting effects. Rather, this cohort has significant under-dominance, where heterozygote animals have lower levels of free-fatty acids than either of the two homozygotes at this locus, and there is no significant difference between the two reciprocal heterozygotes, LG/SM and SM/LG.
Fig.5.
A highly significant pleiotropic QTL, Dserum1c, associated with variation in both cholesterol and free-fatty acids. For cholesterol, the genetic effects fit an additive model, and animals that are LG homozygotes have higher cholesterol than SM homozygotes. The two reciprocal heterozygotes fall at the midpoint between the homozygotes and are pooled in the graph for display purposes (A). For free-fatty acids, all sex-by-diet cohorts, except the low-fat fed males, have significant additive effects, where LG homozygotes have higher free-fatty acids than SM homozygotes (B-E). High-fat fed females have significant dominance effects and maternal expression imprinting (B). Low-fat fed females have significant paternal expression imprinting (C). High-fat fed males have significant over-dominance effects and maternal expression imprinting (D). Low-fat fed males have significant under-dominance effects and no imprinting (E). ***P < 0.001; **P < 0.01; *P < 0.05. LG, allele derived from the LG/J strain; QTL, quantitative trait loci; SM, allele derived from the SM/J strain.
DISCUSSION
Mouse models of cardiovascular disease are an integral part of the genetic mapping toolbox, and the LG/J×SM/J cross has been well characterized with respect to CVD-related risk factors (26). We have taken advantage of the genotypic and phenotypic differences between these two strains to identify both genetic variation and gene-by-environmental variation in serum lipids. The QTL described here have been mapped with a higher resolution than in previous studies, because an F16 advanced intercross population has approximately eight times the recombination of an F2 intercross, which is the experiment by which most mouse QTL have been found. Further, by dividing the litters into high- and low-fat dietary treatments, we are able to tease apart the context dependency of gene-by-environmental interactions. A number of QTL have been identified in crosses between inbred mouse strains fed a high-fat diet, either throughout or at some point in their lives (27–30), and some of these QTL show sex-specificity (31). These studies have proven invaluable for characterizing individual response in serum lipids to a high-fat environment. However most studies do not examine these genetic responses relative to a low-fat diet fed in the same manner. In this study, we have improved mapping resolution and knowledge of the genetic architecture of previously detected QTL. Additionally, we add to the QTL landscape by identifying nine novel loci on chromosomes 1, 4, 7, 8, 10, 16, and 17.
One striking result of this study is the percentage of loci that deviate from a strictly additive model and the overall prevalence of epigenetic genomic imprinting effects. Our knowledge of the influence of epigenetic factors, cell-specific heritable changes in gene expression that occur in the absence of DNA mutation, on complex traits is limited. However, the various risk factors for CVD have some non-Mendelian features, such as some disassociation among twins, male and female differences in prevalence, as well as individual variation in both healthy and disease state. Each of these features is consistent with epigenetic mechanisms (32). Imprinting occurs when the effect of an allele depends on whether it is maternally or paternally inherited. More than 80 imprinted genes have been identified in both mice and humans, and it has been estimated that the imprinting effects of approximately 30% of these genes overlap (33, 34). Computational tools have been developed to predict imprinted genes based on genomic imprinting signatures, such as methylation and histone modification, and bioinformatic scans suggest that hundreds of genes are likely to be imprinted across the genome (www.har.mrc.ac.uk/research/genomic_imprinting/citation.html). It is becoming apparent that imprinting is an important aspect of the architecture of many quantitative traits, including CVD risk factors, such as obesity (35–37), dyslipidemia (38), and blood pressure (39). Our results indicate that, in addition to maternal and paternal expression imprinting patterns, more complicated patterns of polar dominance imprinting and bipolar dominance imprinting commonly affect variation in serum lipids. Further, we show that an individual's maternal environment can affect variation in these traits later in life. These patterns are highly context-dependent, a result that is consistent with previous analyses showing epigenetic patterns are not fixed across all genotypes and all environments (19, 33, 40).
Another striking result of this study is the nearly ubiquitous context dependency of the genetic effects underlying these traits. Fig. 2 illustrates that 47% of additive effects depend on sex and/or diet, that 56% of dominance effects depend on sex and/or diet, and that 73% of imprinting effects depend on sex and/or dietary environment. Context was found to be an important factor underlying variation in both obesity and diabetes-related traits mapped in this same population and described by Cheverud et al. (11) and Lawson, Lee, Fawcett, et al. (unpublished observations). For variation in serum lipids, a majority of loci show genetic effects in multiple cohorts, and most effects are seen in high-fat fed females. The same trend is found for variation in obesity, and for variation in diabetes-related traits, most effects are seen in high-fat fed males. Taken together, these studies highlight the complex genetic architecture underlying the suite of metabolic disorders (obesity, type-2 diabetes, dyslipidemia) composing metabolic syndrome (41). Individuals diagnosed with metabolic syndrome have a 2–3 times higher rate of CVD than the general population (42).
This context dependency is illustrated by the highly significant QTL (Dserum1c, discussed above and in Figs. 1 and 5) associated with variation in both serum cholesterol and free-fatty acids and overlapping a frequently mapped cholesterol QTL on distal chromosome 1 (28, 43). For cholesterol, the genetic effects fit an additive model in the full population: there is a significant difference in levels between animals with the two homozygote genotypes, and heterozygote animals’ cholesterol levels fall at the midpoint. For free-fatty acid, the genetic effects are a complex combination of additive, dominance, and imprinting effects. The dominance effect seen and the imprinting pattern detected depend on an animal's sex and diet (see Fig. 5).
This same QTL region has been mapped in approximately 15 different crosses of mouse strains (28), and the candidate genes in the region are well studied in mouse models of CVD risk (44–48). The gene Hsd17b7 located in this region has been shown to play a role in cholesterol biosynthesis in both mice and humans (46). Additionally, variations in the homologous human APOA2 sequence (also located in this QTL region) have been well studied for their association with CVD risk factors in and among human populations (49–52). Our results extend these previous studies by showing that not only is this genomic region pleiotropic, contributing to multiple phenotypes, but also this same region affects these multiple phenotypes differently depending on an animal's sex and diet. This result is consistent with the varying penetrance and complexity of CVD, and with the varying heritabilities of CVD risk factors seen among human populations and between the sexes. Context-dependent effects have been proposed to be a mechanism by which genetic variation in quantitative traits is maintained in natural populations (53). We find this same level of complexity at other known QTL, as well as at novel loci detected in this study. Our results indicate that if context such as sex and/or diet are not accounted for, not only can genetic signals in specific cohorts be masked or even cancelled in the full study population, but also they can be erroneously assigned to specific cohorts if only the full population is considered. Mouse models are especially appropriate for this type of study because the confounding factor of genetic heterogeneity that plagues human studies is overcome through crosses between animals of known genomic background and with measurable phenotypic differences. This not only increases the power to detect QTL, and eventually quantitative trait genes (QTG) or quantitative trait nucleotides (QTN) having small effects (54), but it also allows for detailed analysis of the architecture of gene-by-environmental interactions, which for practical reasons is not possible in large-scale human population studies. We propose that a candidate gene approach, where candidates are identified independently in mouse models, can be used to protect genomic regions from strict thresholds and increase the power of GWAS, allowing for dissection of the context dependency of the genetic architecture of CVD risk factors. Results such as those presented here, which tease apart gene-by-environmental interactions, can be used to inform study design in human population studies, where little is known about the context dependency of genes that contribute to inter- and intrapopulation variability in CVD risk factors.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Seth Crosby and the Washington University Genome Sequencing Center for their help in SNP genotyping.
Footnotes
Abbreviations:
- CVD
- cardiovascular disease
- GWAS
- genome-wide association study
- QTL
- quantitative trait loci
- SNP
- single nucleotide polymorphism
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant R01 DK-055736 (J.M.C. and C.F.S.); National Institutes of Health, Nutrition Obesity Research Center (NORC) Grant P30 DK-056341; National Institutes of Health, National Heart, Lung and Blood Institute (NHLBI) Grant T32-HL-091823 (H.A.L.); and Biotechnology and Biological Sciences Research Council Grant BBSRC-BB/C/516936 (J.B.W.) Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and three tables.
REFERENCES
- 1.Lloyd-Jones D., Adams R., Carnethon M., De Simone G., Ferguson T. B., Flegal K., Ford E., Furie K., Go A., Greenlund K., et al. 2009. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 119: 480–486. [DOI] [PubMed] [Google Scholar]
- 2.Kromhout D., Menotti A., Kesteloot H., Sans S. 2002. Prevention of coronary heart disease by diet and lifestyle: evidence from prospective cross-cultural, cohort, and intervention studies. Circulation. 105: 893–898. [DOI] [PubMed] [Google Scholar]
- 3.Elder S. J., Lichtenstein A. H., Pittas A. G., Roberts S. B., Fuss P. J., Greenberg A. S., McCrory M. A., Bouchard T. J., Jr, Saltzman E., Neale M. C. 2009. Genetic and environmental influences on factors associated with cardiovascular disease and the metabolic syndrome. J. Lipid Res. 50: 1917–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chico T. J., Milo M., Crossman D. C. 2010. The genetics of cardiovascular disease: new insights from emerging approaches. J. Pathol. 220: 186–197. [DOI] [PubMed] [Google Scholar]
- 5.Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S., Manolio T. A. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein J. L., Hobbs H. H., Brown M. S. 2001. Familial hypercholesterolemia. In The Metabolic and Molecular Basis of Inherited Disease. Scriver C. R., Beaudet A. L., Sly W. S., Vall D., McGraw-Hill, New York: 2863–2913. [Google Scholar]
- 7.Goldstein J. L., Brown M. S. 2001. Molecular medicine. The cholesterol quartet. Science. 292: 1310–1312. [DOI] [PubMed] [Google Scholar]
- 8.Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. 2000. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 290: 1771–1775. [DOI] [PubMed] [Google Scholar]
- 9.Nabel E. G. 2003. Cardiovascular disease. N. Engl. J. Med. 349: 60–72. [DOI] [PubMed] [Google Scholar]
- 10.Koutnikova H., Laakso M., Lu L., Combe R., Paananen J., Kuulasmaa T., Kuusisto J., Haring H. U., Hansen T., Pedersen O., et al. 2009. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet. 5: e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheverud J. M., Lawson H. A., Fawcett G., Wang B., Pletscher L. S., Fox A., Maxwell T. J., Ehrich T. H., Kenney-Hunt J., Wolf J., et al. Diet-dependent genetic and genomic imprinting effects on obesity in mice. Obesity (Silver Spring). Epub ahead of print. June 10, 2010; doi: 10.1038/oby.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheverud J. M., Fawcett G. L., Jarvis J. P., Norgard E. A., Pavlicev M., Pletscher L. S., Polonsky K. S., Ye H., Bell G. I., Semenkovich C. F. 2010. Calpain-10 is a component of the obesity-related quantitative trait locus, Adip1. J. Lipid Res. 51: 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrich T. H., Hrbek T., Kenney-Hunt J. P., Pletscher L. S., Wang B., Semenkovich C. F., Cheverud J. M. 2005. Fine-mapping gene-by-diet interactions on chromosome 13 in a LG/J x SM/J murine model of obesity. Diabetes. 54: 1863–1872. [DOI] [PubMed] [Google Scholar]
- 14.Fawcett G. L., Roseman C. C., Jarvis J. P., Wang B., Wolf J. B., Cheverud J. M. 2008. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring). 16: 1861–1868. [DOI] [PubMed] [Google Scholar]
- 15.Cheverud J. M., Ehrich T. H., Hrbek T., Kenney J. P., Pletscher L. S., Semenkovich C. F. 2004. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 53: 3328–3336. [DOI] [PubMed] [Google Scholar]
- 16.Ehrich T. H., Kenney-Hunt J. P., Pletscher L. S., Cheverud J. M. 2005. Genetic variation and correlation of dietary response in an advanced intercross mouse line produced from two divergent growth lines. Genet. Res. 85: 211–222. [DOI] [PubMed] [Google Scholar]
- 17.Broman K. W., Saunak S. 2009. A Guide to QTL Mapping with R/qtl. Springer, New York. [Google Scholar]
- 18.Li J., Jiang T. 2005. Computing the minimum recombinant haplotype configuration from incomplete genotype data on a pedigree by integer linear programming. J. Comput. Biol. 12: 719–739. [DOI] [PubMed] [Google Scholar]
- 19.Wolf J. B., Hager R., Cheverud J. M. 2008. Genomic imprinting effects on complex traits: a phenotype-based perspective. Epigenetics. 3: 295–299. [DOI] [PubMed] [Google Scholar]
- 20.Haley C. S., Knott S. A. 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 69: 315–324. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Ji L. 2005. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 95: 221–227. [DOI] [PubMed] [Google Scholar]
- 22.Chen L., Storey J. D. 2006. Relaxed significance criteria for linkage analysis. Genetics. 173: 2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hager R., Cheverud J. M., Wolf J. B. 2008. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics. 178: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf J. B., Cheverud J. M., Roseman C., Hager R. 2008. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 4: e1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheverud J. M., Hager R., Roseman C., Fawcett G., Wang B., Wolf J. B. 2008. Genomic imprinting effects on adult body composition in mice. Proc. Natl. Acad. Sci. USA. 105: 4253–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrich T. H., Kenney J. P., Vaughn T. T., Pletscher L. S., Cheverud J. M. 2003. Diet, obesity, and hyperglycemia in LG/J and SM/J mice. Obes. Res. 11: 1400–1410. [DOI] [PubMed] [Google Scholar]
- 27.Su Z., Wang X., Tsaih S. W., Zhang A., Cox A., Sheehan S., Paigen B. 2009. Genetic basis of HDL variation in 129/SvImJ and C57BL/6J mice: importance of testing candidate genes in targeted mutant mice. J. Lipid Res. 50: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Z., Cox A., Shen Y., Stylianou I. M., Paigen B. 2009. Farp2 and Stk25 are candidate genes for the HDL cholesterol locus on mouse chromosome 1. Arterioscler. Thromb. Vasc. Biol. 29: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Le Roy I., Nicodeme E., Li R., Wagner R., Petros C., Churchill G. A., Harris S., Darvasi A., Kirilovsky J., et al. 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13: 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svenson K. L., Von Smith R., Magnani P. A., Suetin H. R., Paigen B., Naggert J. K., Li R., Churchill G. A., Peters L. L. 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 102: 2369–2378. [DOI] [PubMed] [Google Scholar]
- 31.Korstanje R., Li R., Howard T., Kelmenson P., Marshall J., Paigen B., Churchill G. 2004. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J. Lipid Res. 45: 881–888. [DOI] [PubMed] [Google Scholar]
- 32.Junien C., Nathanielsz P. 2007. Report on the IASO Stock Conference 2006: early and lifelong environmental epigenomic programming of metabolic syndrome, obesity and type II diabetes. Obes. Rev. 8: 487–502. [DOI] [PubMed] [Google Scholar]
- 33.Luedi P. P., Hartemink A. J., Jirtle R. L. 2005. Genome-wide prediction of imprinted murine genes. Genome Res. 15: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantey C., Brockmann G. A., Kalm E., Reinsch N. 2005. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J. Hered. 96: 329–338. [DOI] [PubMed] [Google Scholar]
- 35.Rampersaud E., Mitchell B. D., Naj A. C., Pollin T. I. 2008. Investigating parent of origin effects in studies of type 2 diabetes and obesity. Curr. Diabetes Rev. 4: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie T., Chen M., Gavrilova O., Lai E. W., Liu J., Weinstein L. S. 2008. Severe obesity and insulin resistance due to deletion of the maternal Gsalpha allele is reversed by paternal deletion of the Gsalpha imprint control region. Endocrinology. 149: 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstein L. S., Xie T., Qasem A., Wang J., Chen M. 2009. The role of GNAS and other imprinted genes in the development of obesity. Int. J. Obes. 34: 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snell-Bergeon J. K., Dabelea D. 2009. The infant of the diabetic mother: metabolic imprinting. In Diabetes in Women: Pathophysiology and Therapy. Tsatsoulis A, editor Springer, New York: 359–376. [Google Scholar]
- 39.Yang J., Lin S. 2009. Detection of imprinting and heterogeneous maternal effects on high blood pressure using Framingham Heart Study data. BMC Proc. 3 (Suppl 7): S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hager R., Cheverud J. M., Wolf J. B. 2009. Relative contribution of additive, dominance, and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution. 63: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawson H. A., Cheverud J. M. 2010. Metabolic syndrome components in murine models. Endocr. Metab. Immune Disord. Drug Targets. 10: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeppesen J., Hansen T. W., Rasmussen S., Ibsen H., Torp-Pedersen C. 2006. Metabolic syndrome, low-density lipoprotein cholesterol, and risk of cardiovascular disease: a population-based study. Atherosclerosis. 189: 369–374. [DOI] [PubMed] [Google Scholar]
- 43.Rollins J., Chen Y., Paigen B., Wang X. 2006. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc. Med. 16: 220–234. [DOI] [PubMed] [Google Scholar]
- 44.Cho H., Park C., Hwang I. Y., Han S. B., Schimel D., Despres D., Kehrl J. H. 2008. Rgs5 targeting leads to chronic low blood pressure and a lean body habitus. Mol. Cell. Biol. 28: 2590–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iankova I., Chavey C., Clape C., Colomer C., Guerineau N. C., Grillet N., Brunet J. F., Annicotte J. S., Fajas L. 2008. Regulator of G protein signaling-4 controls fatty acid and glucose homeostasis. Endocrinology. 149: 5706–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohnesorg T., Adamski J. 2006. Analysis of the 5′ flanking regions of human and murine HSD17B7: identification of a cholesterol dependent enhancer region. Mol. Cell. Endocrinol. 248: 164–167. [DOI] [PubMed] [Google Scholar]
- 47.Broedl U. C., Jin W., Fuki I. V., Millar J. S., Rader D. J. 2006. Endothelial lipase is less effective at influencing HDL metabolism in vivo in mice expressing apoA-II. J. Lipid Res. 47: 2191–2197. [DOI] [PubMed] [Google Scholar]
- 48.Sumiyoshi K., Mokuno H., Iesaki T., Shimada K., Miyazaki T., Kume A., Kiyanagi T., Kuremoto K., Watanabe Y., Tada N., et al. 2008. Deletion of the Fc receptors gamma chain preserves endothelial function affected by hypercholesterolaemia in mice fed on a high-fat diet. Cardiovasc. Res. 80: 463–470. [DOI] [PubMed] [Google Scholar]
- 49.Chen S. N., Cilingiroglu M., Todd J., Lombardi R., Willerson J. T., Gotto A. M., Jr, Ballantyne C. M., Marian A. J. 2009. Candidate genetic analysis of plasma high-density lipoprotein-cholesterol and severity of coronary atherosclerosis. BMC Med. Genet. 10: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao J., Zhang F., Wiltshire S., Hung J., Jennens M., Beilby J. P., Thompson P. L., McQuillan B. M., McCaskie P. A., Carter K. W., et al. 2008. The apolipoprotein AII rs5082 variant is associated with reduced risk of coronary artery disease in an Australian male population. Atherosclerosis. 199: 333–339. [DOI] [PubMed] [Google Scholar]
- 51.Lara-Castro C., Hunter G. R., Lovejoy J. C., Gower B. A., Fernandez J. R. 2005. Apolipoprotein A-II polymorphism and visceral adiposity in African-American and white women. Obes. Res. 13: 507–512. [DOI] [PubMed] [Google Scholar]
- 52.Han Z., Heath S. C., Shmulewitz D., Li W., Auerbach S. B., Blundell M. L., Lehner T., Ott J., Stoffel M., Friedman J. M., et al. 2002. Candidate genes involved in cardiovascular risk factors by a family-based association study on the island of Kosrae, Federated States of Micronesia. Am. J. Med. Genet. 110: 234–242. [DOI] [PubMed] [Google Scholar]
- 53.Gillespie J. H., Turelli M. 1989. Genotype-environment interactions and the maintenance of polygenic variation. Genetics. 121: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackay T. F., Stone E. A., Ayroles J. F. 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.