Summary
PTEN (Phosphatase and tensin homolog on chromosome ten) is a tumor suppressor whose cellular regulation remains incompletely understood. We identified Phosphatidylinositol-3,4,5-trisphosphate RAC Exchanger 2a (P-REX2a) as a PTEN-interacting protein. P-REX2a mRNA was more abundant in cancer, and significantly increased in tumors with wild type PTEN that expressed an activated mutant of PIK3CA encoding the p110 subunit of phosphoinositide 3-kinase-α (PI3Kα). P-REX2a inhibited PTEN lipid phosphatase activity and stimulated the PI3K pathway only in the presence of PTEN. P-REX2a stimulated cell growth and cooperated with a PIK3CA mutant to promote growth factor-independent proliferation and transformation. Depletion of P-REX2a reduced amounts of phosphorylated AKT and growth in cell lines with intact PTEN. Thus P-REX2a is a component of the PI3K pathway that can antagonize PTEN in cancer cells.
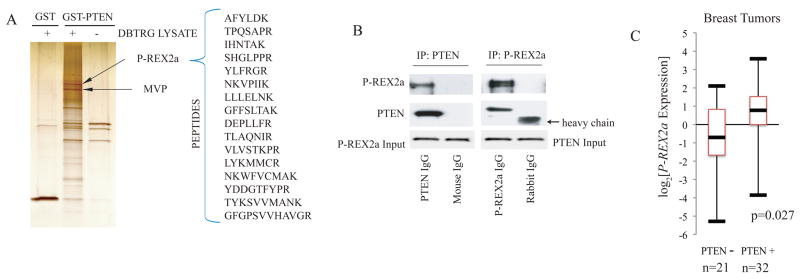
The PTEN (phosphatase and tensin homolog on chromosome ten) gene is frequently lost in cancers, and germline PTEN mutations are linked to inherited cancer predisposition syndromes (1). The PTEN protein dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PIP3), the critical lipid second messenger generated by phosphoinositide 3-kinase (PI3K) upon stimulation of cells by external mitogens (2, 3). Inactivation of PTEN leads to accumulation of PIP3, and as a consequence, increases activity of the kinase AKT, which promotes cellular survival, cell cycle progression, and growth, thereby contributing to oncogenesis. Studies of human tumors have revealed alterations in multiple components of the PTEN-PI3K-AKT axis, all of which result in increased signaling (4). There is mounting evidence that post-translational modifications, including oxidation, phosphorylation, and ubiquitinylation, regulate PTEN (5, 6). To further elucidate cellular regulatory mechanisms, we identified PTEN interacting proteins by affinity purification. We postulated that a PTEN-mutant cell line would be a rich source for interacting proteins without competing endogenous PTEN and selected DBTRG-05MG, a human glioblatoma cell line, since it grows rapidly in culture and lacks detectable PTEN due to an in frame deletion of codons 274 to 342 (7). PTEN-binding proteins were purified from cytoplasmic extracts on an affinity column with PTEN as a glutathione-S-transferase (GST) fusion protein and sequenced by mass spectrometry (Fig. 1A) (8). Associated proteins included PTEN-binding protein, Major Vault Protein (MVP), and a second protein named P-REX2a (9).
Fig. 1. P-REX2a as a PTEN-binding protein.
(A) Silver stain of 1M salt elutions of affinity purified PTEN binding proteins. GST and GST-PTEN columns were incubated with (left, middle) and without (right) DBTRG-05MG cytoplasmic extract and proteins eluted with high salt were separated and identified by mass spectrometry. P-REX2a and MVP are indicated with arrows. (B) Co-immunoprecipitation of endogenous PTEN and P-REX2a. Immunoprecipitions were performed as per Materials and Methods and proteins were detected by immunoblotting. (C) Box plot of P-REX2a expression in PTEN positive and negative breast tumors. Upper and lower bars represent the maximum and minimum expression respectively. The box delineates the first to third quartiles of expression and the central bar represents the median. P-REX2a levels are significantly associated with PTEN status (p=0.027) by two-tailed t-test.
P-REX2a is a guanine nucleotide exchange factor (GEF) for the RAC guanosine triphosphatase (GTPase) which was discovered in a search for proteins with sequence similarity to the leukocyte-specific RAC GEF, P-REX1 (10, 11). P-REX1 GEF activity is critical for RAC-mediated formation of reactive oxygen species in response to PIP3 and Gβγ signaling in neutrophils (12). P-REX1 expression is increased in metastatic prostate cancers and has been shown to mediate a RAC dependent metastatic and invasive phenotype in prostate cancer cell lines (13). P-REX2a is a widely expressed paralog of P-REX1 and contains an N-terminal Dbl homology and pleckstrin homology (DHPH) domain [which confers GEF activity], pairs of PDZ and DEP domains, and a C-terminus with weak similarity to inositol 4-polyphosphate phosphatase.
To demonstrate an endogenous interaction, we co-immunoprecipitated P-REX2a and PTEN from HEK293 extracts using either a polyclonal antibody to P-REX2a or a monoclonal antibody to PTEN (Fig. 1B). We used Flag-tagged PTEN truncation mutants co-expressed with V5-tagged P-REX2a in HEK293 cells to map the binding domain of PTEN. Deletion of the tail and PDZ-binding domain of PTEN (PTENΔTail, aal–353) disrupted binding to PREX2a (Fig. S1). On the other hand, the C2 and tail domains alone (C2TΔPDZ) were able bind to PREX2a, indicating that the tail of PTEN, not including the PDZ-binding domain, is required for binding to P-REX2a. In in vitro experiments, PTEN bound the DHPH domain, an interaction which was mediated by the PH domain (Fig. S2). When overexpressed, PTEN and P-REX2a also displayed subcellular co-localization both diffusely throughout the cytoplasm and at peripheral foci in U87 cells (Fig. S3). In stimulation experiments, only those cells exposed to either PDGF or insulin after starvation displayed peripheral foci of co-localization. Starved and EGF stimulated cells retained diffuse cytoplasmic localization only (Fig. S4).
The P-REX2a gene is located on chromosome 8q13, a region of frequent amplification in breast, prostate, and colorectal cancers (14–16) which has also been linked to aggressive cancer phenotypes and metastatic progression (17, 18). We investigated P-REX2a expression by qRT-PCR in a breast tumor data set thoroughly annotated for PI3K pathway alterations. P-REX2a showed a significant two-tailed association with PTEN status (p=0.027) and the median P-REX2a expression was 3 fold greater in tumors that retained PTEN than in those that did not (Fig. 1C). Additionally, gene expression data sets from other cancer databases demonstrate increased expression of P-REX2a in various tumors including breast and prostate compared to that in normal tissues (Fig. S5). Mutations in P-REX2a were not found in a breast tumor mutation survey (19), however, our analysis of publicly available databases yielded numerous somatic mutations in P-REX2a in other tumors including those of the colon, pancreas and lung, making it one of the most commonly mutated GEF’s in cancer (Fig. S6). We thus suspected that P-REX2a might be a PTEN-regulating factor that is co-opted in tumors to stimulate PI3K signaling.
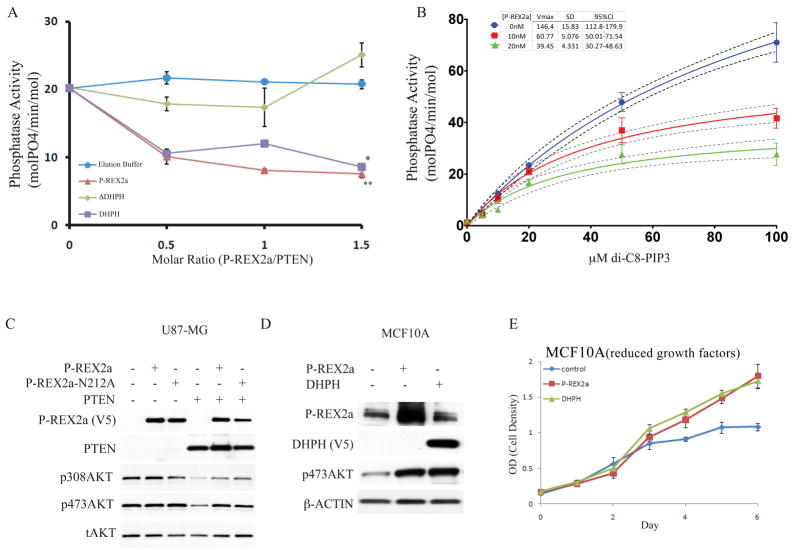
Affinity purified full-length P-REX2a (hereafter referred to as P-REX2a) protein (Fig. S7) decreased PTEN lipid phosphatase activity at various molar ratios of P-REX2a to PTEN (Fig. 2A) using a soluble lipid substrate di-C8-PIP3. At equimolar quantities of P-REX2a, PTEN’s phosphatase activity was decreased by 61.6% (SD= +/− 2.2%). A decrease of 43.2% (SD= +/− 2.8%) in phosphatase activity was recorded with the DHPH domain of P-REX2a (DHPH). Both P-REX2a and DHPH significantly inhibited PTEN activity (p<0.005 for P-REX2a, p<0.05 for DHPH, ANOVA). Conversely, addition of a P-REX2a lacking the DHPH domain (ΔDHPH) showed only modest effects on PTEN activity and was not significantly different than buffer alone (p>0.05, ANOVA).
Fig. 2. Inhibition of PTEN phosphatase activity by P-REX2a.
(A) Full-length P-REX2a, or a deletion of the DHPH domain (ΔDHPH) or the DHPH domain alone were added in the indicated molar volumes to 40nM PTEN, purified from HEK293 cells, and phosphatase activity of PTEN was assayed with 20μM di-C8-PIP3. This is a representative experiment, error bars indicate standard deviation (n=3), **p<0.005, *p<0.05 by ANOVA. (B) P-REX2a (10nM or 20nM) was added to the reactions and phosphate released was measured over a range of di-C8-PIP3 using 40nM PTEN. Regression line (solid) to Michaelis-Menten kinetics is shown along with 95% confidence interval (dotted lines). Vmax values are shown in table along with standard deviation and 95% confidence interval. Representative experiment shown and error bars represent +/− standard deviation (n=3). (C) Effect of P-REX2a and a GEF dead point mutant of P-REX2a, N212A, on phosphorylation of AKT in presence and absence of PTEN. (D) Effect of expression of P-REX2a alone in MCF10A cells on abundance of p473AKT. The phosphorylation status of T308AKT was not detectable under normal growth conditions in MCF10A cells. (E) Effect of P-REX2a and DHPH on proliferation of MCF10A cells grown in reduced growth factors (0.1% serum).
We measured activity of 40 nM PTEN over a range of substrate (di-C8-PIP3) concentrations from 5μM to 100μM and used non-linear regression analysis (GraphPad Prism 5.0) to best fit the data to the Michaelis-Menten equation (Fig. 2B). Increasing amounts of P-REX2a resulted in decreases in the Vmax of PTEN, indicative of non- competitive inhibition. We calculated the dissociation constant for inhibitor binding Ki between P-REX2a and PTEN to be 18.97nM (SD: +/− 1.769nM, 95% CI =15.41–22.52) (GraphPad Prism 5.0). This high affinity was calculated in the presence of soluble lipid substrate and may be altered at the physiological membrane where PTEN has been shown to be a highly efficient interfacial phosphatase (20). We found that co-expression of P- REX2a, but not ΔDHPH, decreased the phosphatase activity of PTEN immunoprecipitated from these cells (Fig. S8). Using a point mutant modeled after P-REX1, we further showed that PTEN phosphatase inhibition was independent on the GEF activity of P-REX2a (Fig. S9) (21).
Expression of PTEN in PTEN-deficient U87-MG cells decreased the amount of phosphorylated AKT (pAKT) (Figs. 2C, S10-12). Overexpression of P-REX2a had no effect on pAKT alone, but when expressed with PTEN, it restored phosphorylation of AKT at both S473 and T308 (Fig. 2C). This restoration was independent of P-REX2a’s GEF activity but dependent on both the tail (Fig. S10) and catalytic activity of PTEN (Fig. S11). We observed a similar rescue of AKT phosphorylation in a second PTEN-deficient glioblastoma cell line (Fig. S10). In platelet derived growth factor (PDGF) stimulated U87-MG cells (Fig. S12) overexpression of PTEN expression alone reduced amounts of phosphorylation of threonine-308 AKT, serine-473 AKT, and glycogen synthetase kinase 3 β(GSK3β). P-REX2a co-expression with PTEN restored levels of phosphorylated AKT and GSK3β to baseline. Among other agonists investigated, P-REX2a was able to also rescue PTEN suppression of insulin signaling but was ineffectual for those cells stimulated with epidermal growth factor (EGF) (Fig. S13). Interestingly, throughout these experiments, we observed that wildtype P-REX2a expression increased levels of simultaneously overexpressed PTEN (Fig. 2C, S11-12). This may be a direct result of PTEN inactivation as previous studies have shown PTEN stability and activity are inversely correlated (6).
We explored the effect of overexpression of P-REX2a on endogenous wild type PTEN in an immortalized human mammary cell line, MCF10A. Cells engineered to express either P-REX2a or DHPH exhibited increased amounts of p473AKT and enhanced proliferation in tissue culture (Figs. 2, D and E). The presence of growth factors was required to observe this difference in proliferation. P-REX2a may not be sufficient to impart growth factor-independent proliferation because effects of PTEN inhibition would still require PIP3 generation to allow for PI3K activation.
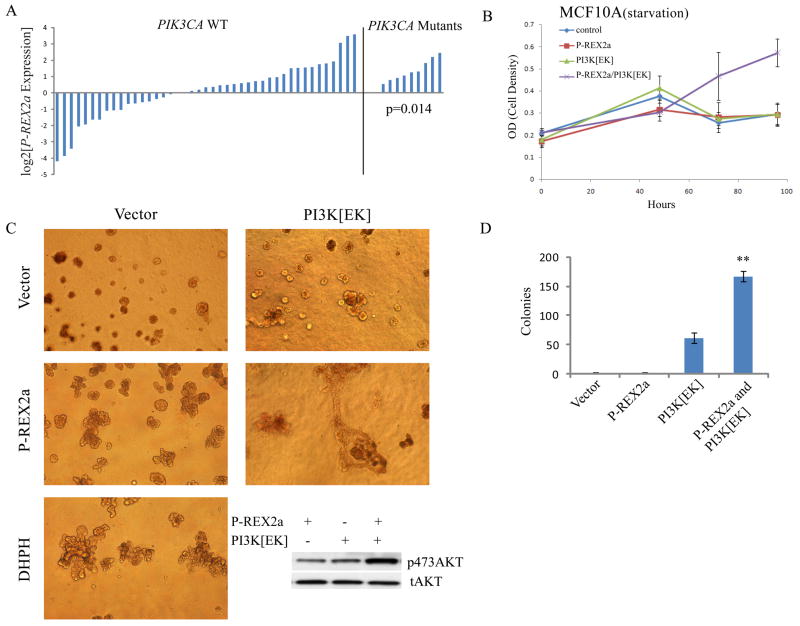
Analysis of P-REX2a mRNA in a cohort of PTEN-expressing breast tumors showed a significant association between increased P-REX2a expression levels and activating mutations in PIK3CA (p=0.014, t-test) (Fig. 3A). Expression of either P-REX2a or PI3Kα E545K constitutively active mutant (PI3K[EK]) alone did not allow growth in the absence of all growth factors, but over-expression of PI3K[EK] and P-REX2a together allowed for growth factor-independent cellular proliferation (Fig. 3B). In three-dimensional matrigel culture, MCF10A cells normally form single acinar cell clusters; however expression of either P-REX2a or DHPH resulted in multiacinar epithelial structures, similar to published results of cells expressing activated AKT, ERRB2 or PI3K (Fig. 3C) (22, 23). Expression of PI3K[EK] with P-REX2a led to formation of large, branched and highly dysmorphic structures and evidence of matrigel invasion. This effect was also observed in soft agar assays in which P-REX2a and PI3K[EK] expression together yielded approximately three times as many transformed colonies as were seen with PI3K[EK] alone (p<0.01, t-test) (Fig. 3D). Amounts of phosphorylated AKT increased as well (Fig. 3C, inset).
Fig. 3. P-REX2a and PI3K mutants cooperate.
(A) P-REX2a expression levels in PTEN wild-type breast tumors without PIK3CA PI3K activating mutations (right) (p=0.014; two tailed t-test). (B) Proliferation in the absence of all growth factors and serum in MCF10A cells expressing P-REX2a and the PI3K mutant E545K (PI3K[EK]). (C) Microscopy of branched multilobulated mammospheres in MCF10A cells expressing P-REX2a or DHPH as indicated when grown in matrigel. Amount of pAKT shown (inset). All phase contrast images are displayed at 100x magnification. (D) Colony formation by MCF10A cells grown in soft agar. Cells expressing P-REX2a and PI3K[EK] as indicated (**p<0.01; t-test).
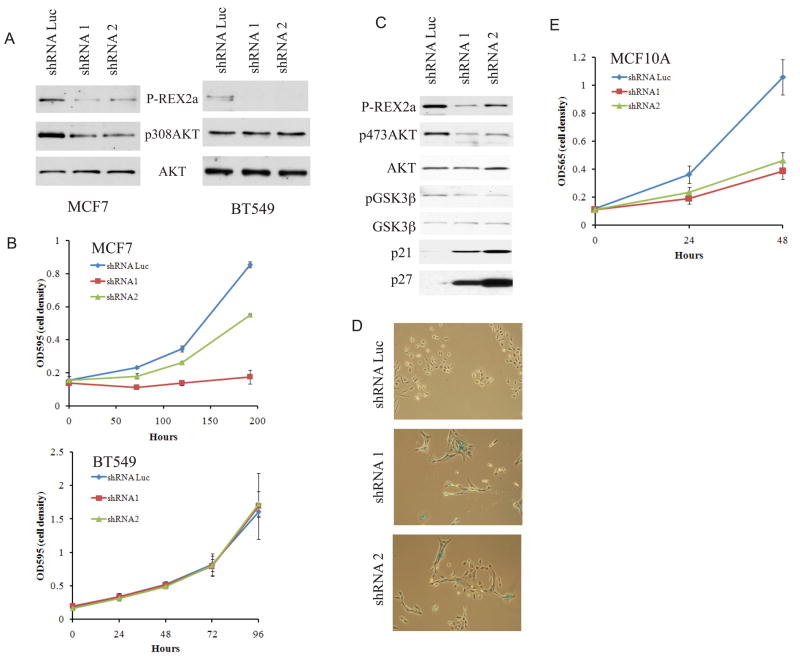
To test whether endogenous P-REX2a influenced the PI3K pathway in tumor cells, we depleted P-REX2a with shRNA in a PTEN-wild type cancer cell line harboring an E545K PIK3CA mutation, MCF7, and a PTEN-deficient cell line, BT549. MCF7 cells displayed decreased amounts of pAKT and reduced cell proliferation after introduction of P-REX2a shRNAs, whereas amounts of pAKT and extent of proliferation were not perturbed in BT549 cells (Figs. 4, A and B). Thus the effects of depletion of PREX2a on the PI3K pathway and cell proliferation are dependent on the presence of PTEN. Depletion of P-REX2a in MCF10A cells decreased amounts of pAKT and pGSK3β (Fig. 4C) and a reduced proliferation rate (Fig. 4E). Cells depleted of P-REX2a appeared to be larger, which was confirmed by forward scatter on flow cytometry (Fig. S14). Staining of these cells with X-gal at pH 6.0, an assay for cell senescence, resulted in marked increase in blue staining cells in both shRNA knockdown cell lines compared to control cells (Fig. 4D, S14). Increased amounts of the senescence-associated cell cycle inhibitors p21 and p27 were also detected by immunblot (Fig. 4C). Senescence similarly occurs in human fibroblasts after inhibition of the PI3K pathway by either LY294002 or overexpression of PTEN (24).
Fig. 4. Diminished phosphorylation of AKT and proliferation after depletion of P-REX2a.
Phosphorylation of AKT (A) and proliferation (B) after depletion of P-REX2a in PTEN wild type MCF7 cells or PTEN-null BT549 cells. (C) Effect of P-REX2a depletion in MCF10A cells on pAKT and pGSK3β and amounts of the p21 and p27 cell cycle inhibitors. (D) X-gal staining of indicated cell lines. Phase contrast images displayed at 40x magnification. (E) MCF10A proliferation after P-REX2a depletion.
Within the PI3K pathway (Fig. S15), P-REX2a is poised to amplify PI3K signaling by decreasing catabolism of PIP3. As an activator of the PI3K pathway PREX2 may contribute to numerous pathological or physiological processes such as tumorigenesis, diabetes, and aging. Its place in the pathway as a coordinator of RAC and PTEN makes it an intriguing target for further analysis of signal transduction and therapeutic intervention.
Supplementary Material
Acknowledgments
We thank W. Zhang and the Herbert Irving Comprehensive Cancer Center Proteomics Shared Resource, M. Li, W. Gu, V. Seshan, S. Voronov, G. DiPaolo, R. Baer, J. Settleman, K. Kinzler, A. Hall, N. Leslie, C. P. Downes, and the members of the Parsons Laboratory for technical assistance or critical reading. B.F. is supported by the NIH Medical Scientist Training Program. Supported by NCI CA097403, the Avon Foundation, Octoberwoman Foundation, and the Adele Meyer Fund.
Footnotes
References and Notes
- 1.Eng C. Hum Mutat. 2003;22:183. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 2.Maehama T, Dixon JE. J Biol Chem. 1998;273:13375. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 3.Cantley LC. Science. 2002;296:1655. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 4.Cully M, You H, Levine AJ, Mak TW. Nat Rev Cancer. 2006;6:184. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 5.Baker SJ. Cell. 2007;128:25. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Leslie NR, Downes CP. Biochem J. 2004;382:1. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, et al. Science. 1997;275:1943. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 8.Materials and methods are available as supporting material on Science Online.
- 9.Yu Z, et al. J Biol Chem. 2002;277:40247. doi: 10.1074/jbc.M207608200. [DOI] [PubMed] [Google Scholar]
- 10.Rosenfeldt H, Vazquez-Prado J, Gutkind JS. FEBS Lett. 2004;572:167. doi: 10.1016/j.febslet.2004.06.097. [DOI] [PubMed] [Google Scholar]
- 11.Donald S, et al. FEBS Lett. 2004;572:172. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 12.Welch HC, et al. Cell. 2002;108:809. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 13.Qin J, et al. Oncogene. 2009;28:1853. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fejzo MS, Godfrey T, Chen C, Waldman F, Gray JW. Genes Chromosomes Cancer. 1998;22:105. doi: 10.1002/(sici)1098-2264(199806)22:2<105::aid-gcc4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, et al. Prostate. 2007;67:692. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 16.Vogelstein B, et al. N Engl J Med. 1988;319:525. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 17.Adler AS, et al. Nat Genet. 2006;38:421. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieche I, Lidereau R. Genes Chromosomes Cancer. 1995;14:227. doi: 10.1002/gcc.2870140402. [DOI] [PubMed] [Google Scholar]
- 19.Sjoblom T, et al. Science. 2006;314:268. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 20.McConnachie G, Pass I, Walker SM, Downes CP. Biochem J. 2003;371:947. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill K, et al. J Biol Chem. 2005;280:4166. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- 22.Isakoff SJ, et al. Cancer Res. 2005;65:10992. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 23.Debnath J, Brugge JS. Nat Rev Cancer. 2005;5:675. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 24.Courtois-Cox S, et al. Cancer Cell. 2006;10:459. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.