Abstract
To elucidate the molecular basis of the specialization of cortical architectures, we searched for genes differentially expressed among neocortical areas of Old World monkeys by restriction landmark cDNA scanning . We found that mRNA of SLIT1, an axon guidance molecule, was enriched in the prefrontal cortex but with developmentally related changes. In situ hybridization analysis revealed that SLIT1 mRNA was mainly distributed in the middle layers of most cortical areas, robustly in the prefrontal cortex and faintly in primary sensory areas. The lowest expression was in the primary visual area. Analyses of other SLIT (SLIT2 and SLIT3) mRNAs showed preferential expression in the prefrontal cortex with a distinct laminar pattern. By contrast, the receptor Roundabout (ROBO1 and ROBO2) mRNAs were widely distributed throughout the cortex. Perinatally, SLIT1 mRNA was abundantly expressed in the cortex with modest area specificity. Downregulation of expression initially occurred in early sensory areas around postnatal day 60 and followed in the association areas. The prefrontal area–enriched SLIT1 mRNA expression results from a relatively greater attenuation of this expression in the other areas. These results suggest that its role is altered postnatally and that this is particularly important for prefrontal connectivity in the Old World monkey cortex.
Keywords: area-specific gene expression, axon guidance molecule, in situ hybridization, postnatal development, prefrontal cortex
Introduction
One of the most outstanding features of the primate cerebral cortex is the marked enlargement of the association areas, particularly the prefrontal cortex (Fuster 2002). Prefrontal cortex plays essential roles in various cognitive functions, such as planning, reasoning, attention, and decision making. Such functions emerge both from local connections of neurons with various intrinsic physiological properties and from connectional interactions with other brain structures. Recent studies have shown that the synaptic dynamics and local connectivity of the prefrontal cortex are specialized to provide sustained neuronal firing, as might be appropriate for supporting working memory processes (Goldman-Rakic 1995; Wang 2001; Gonzalez-Burgos et al. 2004). It is also reported that the pyramidal neurons of the prefrontal cortex have more complex dendritic structures, with more spines than neurons in other cortical areas. This is interpreted as related to the ability to integrate a large number of inputs (Elston 2003; Elston et al. 2006). Identification of molecules that are specifically expressed in the prefrontal cortex will be useful for understanding the mechanisms underlying the functional specialization of neurons in this area.
We have been interested in the molecular basis of the differentiation of cortical architecture across areas and have identified several genes that are differentially expressed in monkey neocortical areas (Tochitani et al. 2001; Watakabe, Sugai, et al. 2001; Komatsu et al. 2005; Watakabe, Komatsu, et al. 2006; Watakabe et al. 2008; Takahata et al. 2009; Takaji et al. 2009). Among the screened genes, Retinol-binding protein 4 (RBP4) and Paraneoplastic antigen-like 5 (PNMA5) are highly expressed in the sensory and prefrontal association areas but are only poorly expressed in V1 (Komatsu et al. 2005; Takaji et al. 2009). The area-specific expression pattern of these genes was not observed in rodents. These findings suggest that a unique set of genes in primates may endow cortical neurons with the properties required for associative functions.
In a new round of screening, we found that SLIT1 mRNA is highly expressed in the prefrontal cortex of monkeys. SLIT is a family of axon guidance molecules, which is well conserved in various species including invertebrates (Brose and Tessier-Lavigne 2000). In Drosophila, SLIT is synthesized by midline glia and functions as a chemorepellent factor for preventing commissural axons from recrossing the midline (Rothberg et al. 1990; Kidd et al. 1999). The chemorepellent effect of SLIT is mediated by Roundabout (ROBO), a family of transmembrane receptor proteins (Kidd et al. 1998). Only one Slit gene is known in invertebrates, whereas 3 Slit genes, Slit1–3, have been identified in vertebrates (Brose et al. 1999). To date, 4 mammalian Robo homologues have been identified (Robo1–4) (Kidd et al. 1998; Sundaresan et al. 1998; Yuan et al. 1999; Huminiecki et al. 2002). All SLITs have been shown to bind to all ROBOs with comparable affinity (Brose et al. 1999; Li et al. 1999; Sabatier et al. 2004). In mammals, SLIT/ROBO signaling is known to play essential roles in the development of the nervous system, including midline crossing, as observed in Drosophila (Nguyen-Ba-Charvet and Chedotal 2002; Chedotal 2007), and disruption of Slit or Robo genes in mammals results in misrouting of major projection tracts (Bagri et al. 2002; Plump et al. 2002; Andrews et al. 2006; Lopez-Bendito et al. 2007). All Slit genes are also expressed in the postnatal stage (Marillat et al. 2002; Whitford et al. 2002), but the role of SLITs after the axon guidance event remains to be determined.
To obtain an insight into the role of SLIT genes in cortical organization, we performed detailed in situ hybridization (ISH) analysis of SLIT and ROBO genes in various cortical areas of macaque monkeys. Furthermore, we investigated SLIT1 mRNA expression in the newborn cortex. Although there were already modest area differences at birth, our results demonstrate that the area- and laminar-specific downregulation of SLIT1 mRNA occurs during postnatal periods and leads to its prefrontal-enriched pattern in the macaque neocortex.
Materials and Methods
Experimental Animals
For restriction landmark cDNA scanning (RLCS) and semiquantitative reverse transcription (RT)-polymerase chain reaction (PCR) analyses, postmortem brain tissues of African green monkeys (Cercopithecus aethiops) were obtained from the Japan Poliomyelitis Research Institute, as previously described (Watakabe et al. 2008; Takahata et al. 2009; Takaji et al. 2009). For histological analysis, brain tissues were obtained from 10 macaque monkeys. Three macaques (Macaca fuscata) whose body weights (BWs) were 2.9 kg (monkey A), 4.7 kg (monkey B), and 4.8 kg (monkey C) were considered to correspond to juvenile to young adult monkeys. We generally use the term “young adult monkeys” in the text but specify the macaque monkey used for each figure and table in the legend. Seven infant macaques (Macaca fascicularis, postnatal day (P) 1 and P2, n = 1; P30 and P60, n = 2; M. fuscata, P95, n = 1) were also used. All the experiments described here were performed in accordance with the guidelines for animal experiments of the Okazaki National Research Institute, Japan, and the National Institutes of Health, United States.
RLCS Analyses
Differential gene expressions among 4 cortical areas shown in Figure 1A (and see below) were identified by RLCS as previously described (Suzuki et al. 1996; Shintani et al. 2004). Total RNAs were extracted separately from the cortical areas as previously described (Watakabe, Fujita, et al. 2001; Komatsu et al. 2005). Poly (A)+ RNA was purified using BioMag Oligo (dT)20 (PerSeptive Biosystems, Framingham, MA). Double-stranded cDNAs were synthesized by reverse transcription. They were digested with XmaI as the first restriction enzyme and with HinfI as the second restriction enzyme for 2D gel electrophoresis. Clones corresponding to differential spots were selected by restriction fragment length polymorphism (RFLP) analysis. Thirty colonies containing the inserts derived from a spot were subjected to colony PCR, and the amplified fragments were digested using a 4-base cutter, AluI. The sequence of the most frequent insert as revealed by RFLP was determined by the dideoxy chain–termination method. On the basis of this sequence, we designed the PCR primers (see Supplementary Table S1), and the consistency of the area difference was confirmed between 2 individuals by semiquantitative RT-PCR.
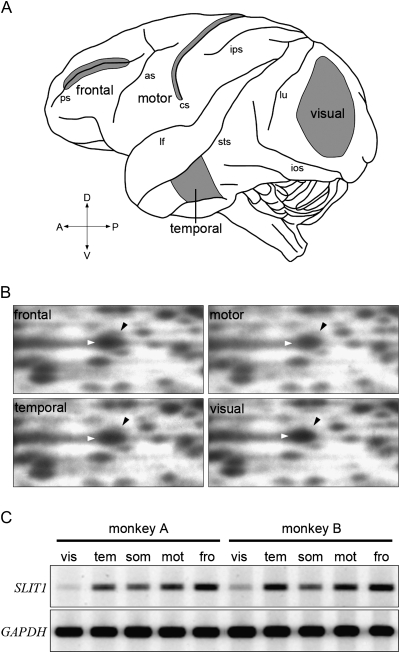
Figure 1.
RLCS profiles for 4 different cortical areas. (A) Lateral surface view of a monkey brain. Anterior is to the left and posterior is to the right. Major sulci are indicated by lower case letters: ps, principal sulcus; as, arcuate sulcus; cs, central sulcus; ips, intraparietal sulcus; sts, superior temporal sulcus; ios, inferior occipital sulcus, and lu, lunate sulcus. Samples for RLCS analyses were taken from the 4 cortical areas in gray; frontal, temporal, motor, and visual areas. (B) A cDNA spot for SLIT1 exhibited differential intensity in each of the RLCS gels for the 4 distinct areas of the neocortex depicted in panel A. Each black arrowhead indicates the spot corresponding to SLIT1 cDNA, which is located in the upper right position of the larger spot that is observed in all areas (white arrowheads). Although the spots shown by the white arrowheads were observed in all the areas, the signal intensity of SLIT1 spot in the visual area was lowest. (C) Semiquantitative RT-PCR demonstrated that the SLIT1 mRNA expression level was the highest in the frontal area and lowest in the visual area. Essentially the same result was obtained from 2 different individuals. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as the control for constitutive expression. vis, visual; tem, temporal; mot, motor; fro, frontal; and som, somatosensory areas.
Synthesis of RNA Probes
All the PCR primers for monkeys were designed on the basis of the human sequence database. The cDNA fragments were obtained by RT-PCR using the primers listed in Supplementary Table S1 and subcloned into the pBlueScriptII vector. We used VGLUT1 as the marker of glutamatergic excitatory neurons (Takamori et al. 2000; Fujiyama et al. 2001; Komatsu et al. 2005) and GAD67, a gamma-aminobutyric acid (GABA)-synthesizing enzyme, for identifying GABAergic inhibitory neurons (Fitzpatrick et al. 1987; Komatsu et al. 2005). We used more than 2 types of probe per gene for SLIT1, SLIT2, ROBO1, ROBO2, and VGLUT1 and confirmed that each probe exhibits the same patterns of signal distribution (data not shown). After the initial confirmation, the multiple probes were mixed together to enhance ISH signals. We also confirmed that the sense probes detected no apparent signals above the background level. The digoxygenin (DIG)- and fluorescein isothiocyanate (FITC)-labeled riboprobes were produced using these plasmids as templates for in vitro transcription. The concentration of all the riboprobes used in this study was adjusted to 0.1 μg/μL, and they were stored at −30 °C.
Tissue Preparation
For histological analysis, macaque monkeys were deeply anesthetized with Nembutal (60 mg/kg, BW intraperitoneally, i.p. injection) following ketamine pretreatment (16 mg/kg BW, intramuscularly, i.m. injection) and perfused through the heart with warmed 0.9% NaCl containing 2 U/mL heparin followed by ice-cold 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.4). After the removal from the skull, the brains were postfixed in 4% PFA for 4–6 h at room temperature and then cryoprotected in 30% sucrose in 0.1 M PB at 4 °C. The tissue blocks were frozen on dry ice and stored at −80°C until use. The frozen blocks were cut into 20- to 35-μm-thick sections using a sliding microtome (ROM-380, Yamato-kohki, Saitama, Japan). The sections were maintained in a cryoprotectant solution (30% glycerol, 30% ethylene glycol, and 40% 0.1 M phosphate-buffered saline [PBS], pH 7.4) at −30 °C until use.
In Situ Hybridization
Single-color ISH was carried out as previously described (Liang et al. 2000). Briefly, free-floating sections (35-μm thick) were soaked overnight in 4% PFA in 0.1 M PB (pH 7.4) at 4 °C, treated with 0.5–5.0 μg/mL proteinase K for 30 min at 37 °C, acetylated, and then incubated in hybridization buffer containing 0.5–1.0 μg/mL DIG-labeled riboprobes at 60–65 °C. The sections were sequentially treated with 2× 16.65 mM NaCL, 16.65 mM trisodium citrate dihydrate (SSC)/50% formamide/0.1% N-lauroylsarcosine for 15 min at 50 °C, twice; 30 min at 37 °C in RNase buffer (10 mM Tris–HCl, pH 8.0, 1 mM ethylenediaminetetraacetic acid [EDTA], 500 mM NaCl) containing 20 μg/mL RNase A (Sigma–Aldrich, Saint Louis, MO); 15 min at 37 °C in 2× SSC/0.1% N-lauroylsarcosine, twice; and 15 min at 37 °C in 0.2× SSC/0.1% N-lauroylsarcosine, twice. The hybridized probes were detected with an alkaline phosphatase (AP)-conjugated anti-DIG antibody using a DIG nucleic acid detection kit (Roche Diagnostics, Basel, Switzerland).
Fluorescence double ISH was also performed as described previously (Komatsu et al. 2005; Watakabe et al. 2007). Sections of 20-μm thickness were used for double ISH. DIG- and FITC-labeled riboprobes were used to detect the expressions of 2 genes. The prehybridization and washing steps were carried out under the same conditions as the single ISH. For the detection of the FITC probes, the sections were incubated with an anti-FITC antibody conjugated with horseradish peroxidase (Roche Diagnostics; 1:2000–1:8000 accordingly in 1% blocking buffer) for 2–5 h at room temperature. After washing in TNT solution (0.1 M Tris–HCl, pH 7.5, 0.15 M NaCl, 0.1% Tween 20) 3 times for 15 min, the sections were treated with 1:50 diluted TSA-Plus dinitrophenyl hapten (DNP) reagents, the kit for tyramide signal amplification (Yang et al. 1999), for 30 min in accordance with the manufacturer's instructions (Perkin–Elmer, Norwalk, CT), and the FITC signals were converted to DNP signals. After washing in TNT 3 times for 15 min, the sections were incubated overnight at 4 °C with an anti-DNP antibody conjugated with Alexa 488 (1:500, Molecular Probes, Eugene, OR) in 1% blocking buffer for the fluorescence detection of the DNP signals. At this step, an anti-DIG antibody conjugated with AP (1:1,000, Roche Diagnostics) was included in the 1% blocking buffer for the detection of the DIG probes. The sections were washed 3 times in TNT, twice in TS 8.0 (0.1 M Tris–HCl, pH 8.0, 0.1 M NaCl, and 50 mM MgCl2) for 10 min, and AP activity was detected using an 2-hydroxy-3-naphtoic acid-2'-phenylanilide phosphate fluorescence detection set (Roche Diagnostics). The incubation of this substrate was carried out for 20–40 min and stopped by washing in PBS containing 10 mM EDTA. The sections were then counterstained with Hoechst 30442 (Molecular Probes) diluted in PBS to 1:1000 for 5 min.
The images for the ISH were obtained by 2 methods: First, the low-magnification photographs shown in Figures 2A and 5A were captured using a flat-headed image scanner GTX-800 (EPSON, Tokyo, Japan). Second, the images of all magnified views of the cortex were obtained using a digital color camera DP70 (Olympus, Tokyo, Japan) attached to a BX-51 microscope (Olympus). The brightness and contrast of images were adjusted with Adobe Photoshop CS2 (Adobe Systems, San Jose, CA). Although the sections for ISH shrank, the scale bars in the figures are not adjusted for such shrinkage.
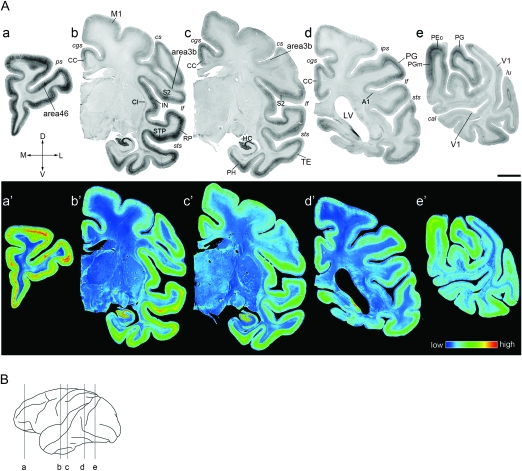
Figure 2.
ISH Analysis of SLIT1 in Macaque Brain. (A) Coronal sections of a macaque monkey brain were obtained from the positions corresponding to a–e in the brain diagram shown in (B). The representative 6 cortical areas (area 46, TE, PG, M1, area 3b, and V1) are magnified in Figure 3. Pseudocolor representations of the same sections in a–e are shown in a′–e′. SLIT1 mRNA expression was observed in the entire cerebral cortex at variable levels. Note that the most intense signal was observed in the frontal pole section (a,a′). These images are derived from monkey (B). A1, primary auditory area (core region); M1, primary motor area; V1, primary visual area; CC, cingulate cortex; Cl, claustrum; HC, hippocampus; IN, insular cortex; LV, lateral ventricle; PEc, PE caudal part; PGm, PG medial part; PH, parahippocampal area; RP, rostral parabelt area; STP, superior temporal polysensory area; S2, secondary somatosensory area; cal, calcarine sulcus; cgs, cingulate sulcus; cs, central sulcus; ips, inferior parietal sulcus; lf, lateral fissure; lu, lunate sulcus; ps, principal sulcus; sts, superior temporal sulcus. Orientation of each section is indicated: (D) dorsal; V, ventral; L, lateral; and M, medial. Scale bar = 5 mm. (B) Lateral view of the macaque neocortex. The lines indicate the planes sliced for the sections shown in (A).
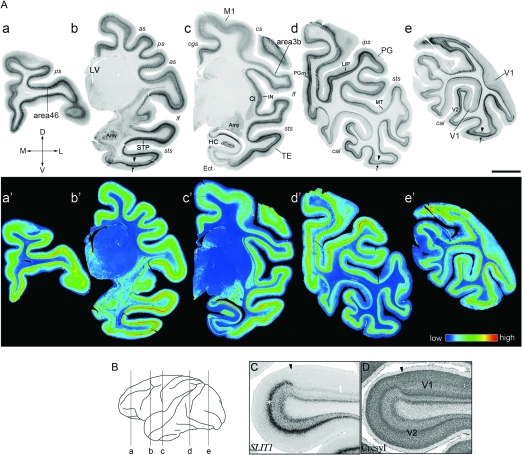
Figure 5.
ISH Analysis of SLIT1 in Infant Brain. (A) Coronal sections of the P2 macaque brain were obtained from the positions corresponding to the brain diagram as a–e in (B). The representative 6 cortical areas (area 46, TE, PG, M1, area 3b, and V1) are magnified in Figure 6. Arrows and arrowheads in b, d, and e indicate the ISH signals at layers IV and VI, respectively. These laminar signals were observed throughout the neocortex. Pseudocolor representation of the same images in a–e is shown in a′–e′. as, arcuate sulcus; Ect, entorhinal cortex; LIP, lateral intraparietal area. Other abbreviations are the same as those in Figure 2. The orientation of each section is indicated: D, dorsal; V, ventral; L, lateral; and M, medial. Scale bar = 5 mm. (B) Lateral view of macaque neocortex. The lines indicate the planes sliced for the sections shown in (A). (C) ISH and (D) cresyl violet-stained sections around the V1/V2 border of P2 monkey. Arrowheads indicate the border. Note that the signal intensity observed in the middle layer (*) in V2 markedly decreased in V1.
Cortical Areas Examined in Old World Monkey Brain
The cortical areas were identified using a brain atlas (Paxinos et al. 1999). For RLCS analyses, we compared the gene expressions among 4 cortical areas. Because the tissue was obtained from relatively large areas, they were labeled as frontal, temporal, motor, and visual (Fig. 1A) instead of area 46, TE, M1, and V1, which constitute only a part of these samples. The exact locations of these areas are as follows: frontal, both banks of the principal sulcus (ps); temporal, the region ventral to the superior temporal sulcus (sts); motor, the anterior bank of the central sulcus (cs); visual, the region posterior to the lunate sulcus (lu), and superior to the inferior occipital sulcus (ios). In addition to the above areas, the somatosensory area corresponding to the posterior bank of cs was analyzed for semiquantitative RT-PCR.
Although we investigated the expressions of SLIT1 and its related genes in various brain regions, we limited our description to selected cortical areas. To clarify how the area-specific expression pattern was formed (see next section), we selected 6 cortical areas including 2 primary sensory areas (visual, V1; somatosensory, area 3b), the primary motor area, M1, and 3 association areas (prefrontal, area 46; parietal, PG; and temporal, TE). Area 3b is a part of the primary somatosensory area, which consists of different 4 subareas: areas 1, 2, 3a, and 3b (Burton and Sinclair 1996). The above areas were selected because they have well-known functional differences and represent hierarchically different positions in the cortical circuit. Other areas selected for single ISH, secondary (V2) and third (V3) visual areas were also described. For double ISH, we focused on area 46, TE, and M1. Throughout the text, we use the terms V1, V2, V3, and M1 instead of areas 17, 18, 19, and 4, respectively.
Quantification of In Situ Hybridization Signals
We used 4 types of quantification methods to estimate mRNA expression levels. For each quantification method, images were taken using the same lamp voltage, condenser aperture, and exposure time.
First, to demonstrate the gross distribution of SLIT1 mRNA in the macaque brain, the relative optical density (ROD) of ISH sections was pseudocolored (Figs. 2A and 5A). The gray level of each pixel in ISH sections was initially converted into ROD. A stronger labeling corresponded to darker regions coded as red. All the processing was performed using an image analyzer (Image Pro Plus, Media Cybernetics, Silver Spring, MD).
Second, to estimate the relative amount of SLIT1 mRNA in each cortical area and layer of different ages (Figs. 3B and 6B), we introduced “normalized staining intensity (NSI)” in accordance with a method previously described (Meberg and Routtenberg 1991; Higo et al. 1999). Briefly, we calculated normalized ROD from ISH sections (ISH RODsignal – ISH RODbackground). In this method, the mean gray level in a region of interest (ROI) was converted into ROD using Image Pro Plus. The measurement was performed in each layer of a 200-μm-wide column from layers II–VI (ISH RODsignal). Three ROIs were set in each cortical layer. Background RODs were sampled in each ROI from the cell-free space in each layer (ISH RODbackground). We also calculated normalized ROD from adjacent cresyl violet–stained sections (cresyl RODsignal − cresyl RODbackground). This value represents the cell density in each area. To compensate for differences in cell density between areas and layers, we determined the ratio as follows:
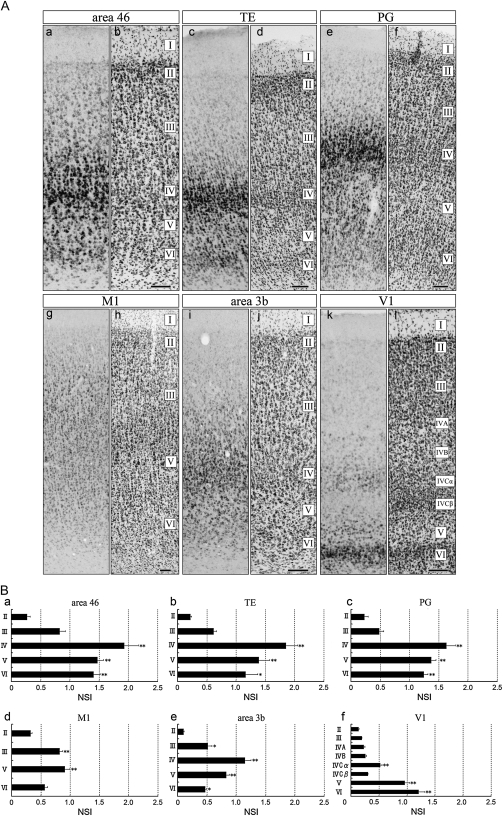
Figure 3.
Layer Distribution of SLIT1 mRNA in Macaque Cortex. (A) Area 46 (a,b), TE (c,d), PG (e,f), M1 (g,h), area 3b (i,j), and V1 (k,l) of macaque cortex. (a,c,e,g,i,k) Coronal sections for ISH. (b,d,f,h,j,l) Coronal sections for cresyl violet staining. In all areas, clear hybridization signals for SLIT1 mRNA were observed in layers II–VI. These images are derived from monkey C. Scale bar = 100 μm. (B) To quantify the hybridization signal level, NSI (SLIT1 OD/cresyl OD ratios) in each of 6 cortical areas (a, area 46; b, TE; c, PG; d, M1; e, area 3b, and f, V1) was calculated (see Materials and Methods for details). The ratios were higher in the middle layers than in layers II or VI except for V1. Three cortices were used for calculation. Asterisks indicate a significant difference from the lowest value in each area (*P < 0.05; **P < 0.01, one-way analysis of variance (ANOVA), n = 9). Data are expressed as mean ± standard error of the mean (SEM). WM, white matter.
Figure 6.
Layer Distribution of SLIT1 mRNA in Infant Cortex. (A) Area 46 (a,b), TE (c,d), PG (e,f), M1 (g,h), area 3b (i,j), and V1 (k,l) of the infant macaque cortex. (a,c,e,g,i,k) Coronal sections for ISH. (b,d,f,h,j,l) Coronal sections for cresyl violet staining. Scale bar = 100 μm. (B) To quantify the hybridization signal level, NSI (SLIT1 OD/cresyl OD ratios) in each of 6 cortical areas was calculated. The ratios were higher in deep layers than in surface layers. The signal intensity of layer VI in V1 was the highest among the layers examined. For this calculation, 3 cortices were used for area 46 (P1, P2, and P30). Two cortices were used for other areas (P1 and P2 for PG; P1 and P30 for TE, M1, area 3b and V1). Asterisks indicate significant difference from the lowest value in each area (*P < 0.05; **P < 0.01, one-way ANOVA, n = 9 for area 46 and n = 6 for other areas). Data are expressed as mean ± SEM. WM, white matter.
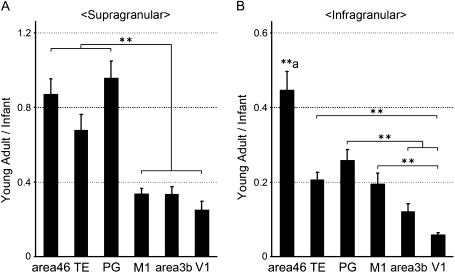
Although SLIT1 mRNA signals were observed in layer I, signals were too weak and sparse to estimate the correct staining intensity. Thus, we omitted layer I from this analysis. For this quantification, we used 3 infants and 3 young adults. The SLIT1 mRNA expression patterns were similar for the samples from P1, P2, and P30 monkeys, which enabled us to compile data from these individuals for statistical analyses (infant, Fig. 6B). On the basis of these data, we determined the ratio of NSI in young adults to that in infants (young adult/infant) for supra and infragranular layers of each area (Fig. 7). The ratio was useful for confirming how the laminar distribution of SLIT1 mRNA was altered postnatally among cortical areas.
Figure 7.
Alteration of SLIT1 mRNA Distribution between Infant and Young Adult Cortices. (A) Postnatal alteration of SLIT1 mRNA expression in supragranular layers. Each bar represents the ratio of SLIT1 mRNA expression levels between infants (P1, P2, and P30 monkeys) and young adults (monkeys A, B, and C) in supragranular layers (layers II and III) of the 6 cortical areas (young adult/infant). The asterisk indicates significant difference between the ratios in the association areas (area 46, TE, and PG) and those in the primary areas (M1, area 3b, and V1, **P < 0.01, 1-way ANOVA). (B) Postnatal alteration of SLIT1 mRNA expression in infragranular layers. Each bar represents the ratio in infragranular layers (layers V and VI). Asterisks indicate significant difference in each pair (**P < 0.01, 1-way ANOVA). (**a) The ratio in area 46 was significantly higher than those in the remaining areas (P < 0.01). Data are expressed as mean ± SEM.
Third, to compare the laminar patterns of different ISH signals (Fig. 4), signals were extracted from the background component by converting the 8-bit gray-scale images into binary images. The threshold used here was set to the standard deviation beyond the average intensity of each cortical section. Then, we calculated averaged values with respect to each row to obtain the line profile in ROIs (100-μm bin, height of the cortex through layer I to white matter) using Image Pro Plus.
Figure 4.
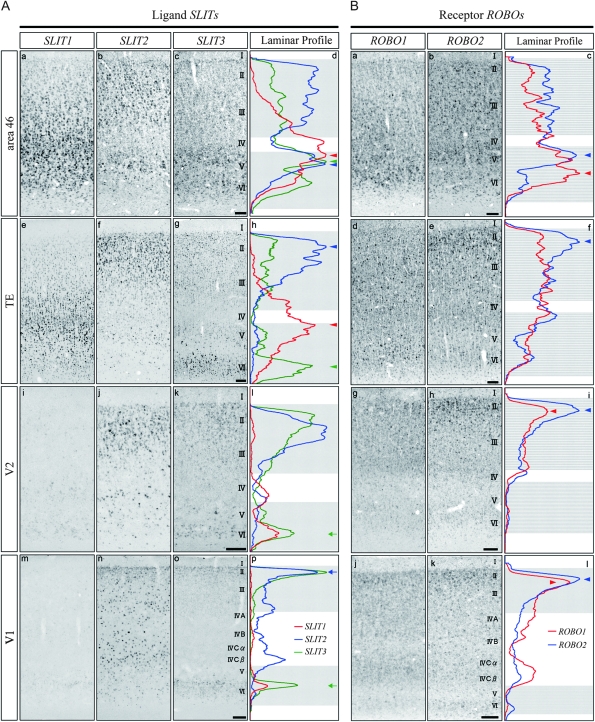
Differential Laminar Patterns of SLIT and ROBO mRNAs in Macaque Cortex. (A) ISH of SLIT1, SLIT2, and SLIT3 mRNAs in area 46 (a–c), TE (e–g), V2 (i–k), and V1 (m–o), respectively. The laminar profiles of the ISH signals are also shown. Red, blue, and green lines indicate the profiles of SLIT1, SLIT2, and SLIT3 mRNAs, respectively (d,h,l,p). The ISH signals of SLIT mRNAs were strongest in area 46 and weakest in V1. Their laminar patterns showed complementarity in TE. Arrowheads indicate the peaks of ISH signals for SLIT mRNAs in the association areas. Arrows indicate the peaks of the signals in early sensory areas (see in more detail in text). These images are derived from monkey A. (B) ISH of ROBO1 and ROBO2 mRNAs in area 46 (a,b), TE (d,e), V2 (g,h), and V1 (j,k). The laminar profiles of the ISH signals are also shown. Red and blue lines indicate the profiles of ROBO1 and ROBO2 mRNAs (c,f,i,l), respectively. Arrowheads indicate the peaks of ISH signals for ROBO mRNAs. These images are derived from monkey C. Scale bar = 100 μm.
Fourth, for double ISH experiments, cells double positive for the 2 genes of interest were manually counted (Tables 1 and 2) as previously described (Takahata et al. 2006; Watakabe, Ohsawa, et al. 2006) with minor modification. Briefly, the images captured in 3 channels (ISH signals in the red and green channels and Hoechst nuclear staining in the blue channel) were layered into a single file. Next, Hoechst signals were plotted on the blank layer to define the center position of the nucleus. By displaying this plotted layer, the cells positive for red or green ISH fluorescent signals were separately plotted onto other blank layers manually as dots for later counting. After plotting the positive cells for each of the 2 ISH images, the double-positive cells were identified as the overlapping dots. After plotting the single- and double-positive cells, the numbers of these dots were counted using Image Pro Plus. Counting was performed in squared ROIs (100 × 100 μm2) set in each layer. At least 5 ROIs were set in each layer. Note that our cell counts provide the ratio and not the absolute numbers of the positive cells. Although stereological method is not used, the risk of miscalculation is made minimal by providing the ratio (Saper 1996).
Table 1.
Percentages of SLIT1+ neurons among excitatory and inhibitory neurons in macaque cortex
| Area | Layer | SLIT1/VGLUT1 (%) | SLIT1/GAD67 (%) |
| Area 46 | II | 48 ± 3.0 | 50 ± 13.1 |
| III | 82 ± 3.8 | 66 ± 6.1 | |
| IV | 76 ± 5.0 | 41 ± 13.8 | |
| V | 65 ± 4.0 | 43 ± 6.9 | |
| VI | 78 ± 3.0 | 47 ± 15.4 | |
| TE | II | 46 ± 6.0 | 36 ± 11.3 |
| III | 75 ± 3.2 | 51 ± 7.7 | |
| IV | 75 ± 3.8 | 45 ± 8.6 | |
| V | 83 ± 3.7 | 26 ± 5.7 | |
| VI | 82 ± 3.6 | 31 ± 16.3 | |
| M1 | II | 50 ± 5.9 | 64 ± 12.8 |
| III | 84 ± 4.7 | 51 ± 7.3 | |
| IV | 67 ± 4.8 | 31 ± 10.9 | |
| V | 67 ± 6.1 | 36 ± 17.4 |
The ratios of SLIT1-positive neurons among either VGLUT1- or GAD67-positive neurons were calculated for area 46, TE, and M1. These data are derived from monkey A. Data are expressed as mean ± SEM.
Table 2.
Percentages of double-positive neurons to either SLIT1- or ROBO-single positive neurons in macaque cortex
| Area | Layer |
SLIT1/ROBO1 |
SLIT1/ROBO2 |
||
| Doublea/SLIT1+ (%) | Doublea/ROBO1+ (%) | Doubleb/SLIT2+ (%) | Doubleb/ROBO2+ (%) | ||
| Area 46 | II | 84 ± 2.1 | 78 ± 3.1 | 73 ± 3.7 | 82 ± 3.5 |
| III | 90 ± 2.6 | 90 ± 4.1 | 74 ± 3.6 | 80 ± 6.8 | |
| V | 91 ± 3.1 | 82 ± 4.2 | 73 ± 5.9 | 82 ± 5.1 | |
| VI | 97 ± 2.3 | 88 ± 4.5 | 89 ± 3.6 | 90 ± 3.0 | |
| TE | II | 87 ± 4.3 | 69 ± 3.1 | 90 ± 3.4 | 73 ± 2.4 |
| III | 88 ± 1.1 | 82 ± 3.7 | 62 ± 3.5 | 77 ± 3.6 | |
| V | 83 ± 3.3 | 74 ± 5.4 | 45 ± 2.9 | 74 ± 3.9 | |
| VI | 88 ± 3.1 | 86 ± 2.4 | 74 ± 3.4 | 90 ± 2.1 | |
| M1 | II | 88 ± 3.1 | 71 ± 4.0 | 92 ± 3.3 | 63 ± 1.6 |
| III | 82 ± 5.3 | 65 ± 4.3 | 70 ± 7.0 | 58 ± 3.8 | |
| V | 78 ± 5.9 | 64 ± 7.0 | 61 ± 9.2 | 39 ± 3.4 | |
| VI | 76 ± 4.2 | 60 ± 3.4 | 53 ± 3.8 | 47 ± 6.0 | |
The ratios of double-positive neurons to either SLIT1- or ROBO-single positive neurons were calculated for area 46, TE, and M1. These data are derived from monkey A. Data are expressed as mean ± SEM.
SLIT1 and ROBO1 mRNAs double-positive neurons.
SLIT1 and ROBO2 mRNAs double-positive neurons.
Results
Identification of SLIT1 cDNA by RLCS Analyses
To screen area-specific molecules in the monkey cortex, we carried out screening by RLCS as previously reported (Watakabe et al. 2008; Takahata et al. 2009; Takaji et al. 2009). In this analysis, mRNAs were purified from 4 distinct cortical areas (frontal, motor, temporal, and visual, Fig. 1A), converted to cDNA by reverse transcription, and then digested with a pair of restriction enzymes for 2D electrophoresis. As a result, we found a spot that was very faint in the visual area compared with the other 3 areas (Fig. 1B). Spot cloning and sequence analyses showed that the cDNA corresponding to this spot was homologous to the 3′ UTR (untranslated region, 6408–6622 bp) of the human Slit homologue 1 (SLIT1) gene (Genbank; NM_003061). To confirm the area specificity of SLIT1 mRNA expression, we performed semiquantitative RT-PCR. The results showed the same tendency of area difference as that for RLCS, that is, the SLIT1 mRNA expression was the highest in the frontal area, with intermediate levels in the temporal, somatosensory and motor areas, and the lowest in the visual area (Fig. 1C).
Laminar and Area Distribution of SLIT1 mRNA in Cerebral Cortex
To investigate the expression pattern of SLIT1 in detail, we performed ISH using a macaque cerebral hemisphere. Consistent with the RLCS analysis, the expression level of SLIT1 mRNA was generally high in the sensory and prefrontal association areas and low in the primary sensory areas. The upper panel of Figure 2A is a series of images showing the gross distribution of SLIT1 mRNA in the macaque cerebral hemisphere. The lower panel of Figure 2A shows the pseudocolor representation of the same images quantified for ROD. As shown, the SLIT1 mRNA signals were abundant in the frontal cortex (Fig. 2A,a), the dorsal bank of the sts corresponding to the superior temporal polysensory area (STP, Fig. 2A,b) and the hippocampus (HC, Fig. 2A,b,c). Moderate to intense expression was observed in parietal fields, PG (Fig. 2A,d,e), PG medial part (PGm, Fig. 2A,e), PE caudal part (PEc, Fig. 2A,e), the parahippocamal area (PH, Fig. 2A,c), the insular cortex lying within the lateral fissure (IN, Fig. 2A,b), the claustrum present under the insular cortex (Cl, Fig. 2A,b), the limbic cortices, such as the middle and posterior cingulate cortex (CC, Fig. 2A,b,c,d), and the entorhinal cortex (data not shown). In contrast, the expression level was very low in the primary sensory areas, such as the somatosensory (area 3b, Fig. 2A,b,c) and the core auditory (A1, Fig. 2A,d) areas. Compared with area 3b, the secondary somatosensory area showed stronger signals (S2, Fig. 2A,b,c). Among the auditory cortex, the rostral parabelt (RP) area showed obviously stronger signals than the core auditory area (Fig. 2A,b). The lowest expression level was observed in the primary visual area (V1, Fig. 2A,e). Thus, the result of our histological analysis was consistent with those of RLCS and RT-PCR analyses and showed the association area-enriched pattern over primary areas.
We next examined the laminar distribution patterns of SLIT1 mRNA shown in the high-magnification images of Figure 3A. In all the areas examined, SLIT1 mRNA was widely expressed in layers II–VI, with only weak SLIT1 mRNA signals in a scattered population of layer I (data not shown). There were, however, conspicuous laminar differences in intensity across areas. In Figure 3B, we quantified ISH signals by measuring the ROD of each layer. To estimate the differences in expression level of SLIT1 mRNA across areas and layers, we calculated each NSI of 6 representative cortical areas: NSI is the ROD of the ISH signal divided by the cresyl violet staining intensity, which normalizes for cell density that varies markedly across layers and areas (see Materials and Methods). Despite the large difference in the overall signal level, the laminar distribution patterns of SLIT1 mRNA were similar among area 46, TE, PG, M1, and area 3b, in the sense that the peak of signal intensity was observed in the middle layers (layers III–V), which gradually decreased toward the upper and deeper layers (Fig. 3A,a,c,e,g,i). Except for area 3b and V1, the differences between the middle layers and outer layers (layers II or VI) were statistically significant (*P < 0.05; **P < 0.01, Fig. 3B). At cellular resolution, the large pyramidal neurons in the upper part of layer V of area 46 showed the most intense hybridization signals (Fig. 3A,a). Accordingly, the NSI of layer V in area 46 was the highest among the layers investigated (Fig. 3B,a). Intense signals were also observed in the upper part of layer V of TE and PG. Compared with the other neocortical areas, the SLIT1 mRNA distribution pattern in V1 showed a unique profile: In layers II–V, the number of SLIT1-mRNA-positive cells was low, and the mRNA level per cell was also considerably low. The cells that exhibited weak to moderate signals were concentrated in layer VI (Fig. 3A,k). The signal intensity changed gradually across area borders and did not exhibit a sharp transition.
SLIT1 mRNA Is Mainly Expressed in Excitatory Neurons in the Cortex
In considering the roles of SLIT1 in the neocortical circuit, it is important to determine the types of cell that express it. To confirm this point, we performed fluorescence double ISH using VGLUT1 and GAD67 probes, which label excitatory and inhibitory neurons, respectively.
For this analysis, we chose 3 areas: area 46, TE, and M1. Representative photographs of the double ISH in area 46 are shown in Supplementary Figure S1. In area 46, SLIT1 mRNA colocalized with both VGLUT1 and GAD67 mRNAs, indicating that both excitatory and inhibitory neurons expressed SLIT1 mRNA. Similar results were also obtained in TE and M1 (Table 1). In our counting, 65–84% of the excitatory neurons expressed SLIT1 mRNA in layers III–VI of these areas. Even in layer II, which showed weak SLIT1 mRNA signals, approximately one-half of the excitatory neurons expressed SLIT1 mRNA. The ratios in each layer did not show any significant difference across areas (data not shown). Thus, a majority of excitatory neurons expressed SLIT1 mRNA despite a large difference in staining intensity. As for the inhibitory neurons, 26–66% of the cells expressed SLIT1 mRNA irrespective of the area and layer (Table 1).
Distinct Laminar Expression of SLIT and ROBO Genes in Cortex
Because SLIT1 and other SLITs have been shown to interact with all ROBOs (Brose et al. 1999; Li et al. 1999; Sabatier et al. 2004), we examined the expressions of SLIT2, SLIT3, ROBO1, and ROBO2 mRNA and compared them with that of SLIT1 mRNA (Fig. 4).
In both area 46 and TE, the expression levels of all SLIT mRNAs were high, but the laminar distributions were different. In area 46, the SLIT2 mRNA expression was high in layers II, III, and V (Fig. 4A,b). The SLIT3 mRNA expression was high in layers V and IV and was low in layers II and III (Fig. 4A,c). All SLIT mRNAs were highly expressed in layer V in area 46 (Fig. 4A,d, arrowheads). Although the expression levels of all SLIT mRNAs were also high in TE, those of SLIT2 and SLIT3 mRNA were markedly low in layer V, which resulted in a complementary laminar pattern of SLIT mRNAs in TE (Fig. 4A,h, arrowheads).
In contrast to these association areas, the expression levels of all SLIT mRNAs were generally low in the visual areas (Fig. 4A, V1 and V2). For example, although not as conspicuous as that of SLIT1, the SLIT3 mRNA expression level in layer VI of sensory areas was much lower than those of association areas (Fig. 4A,l,p, green arrows). The signal intensity of SLIT3 mRNA in the supragranular layers was also much lower in V1 than in V2. At first glance, SLIT2 mRNA seemed to be abundantly expressed in V1. However, most of the scattered strong signals of SLIT2 mRNA in layers III–VI were in GAD67-mRNA-positive inhibitory neurons (data not shown). The excitatory population that expressed weak SLIT2 mRNA signals was restricted to the upper part of the supragranular layers of V1 (Fig. 4A,p, blue arrow). This is in stark contrast to the pattern in the association areas.
In comparison with the expression of SLIT mRNAs, ROBO mRNAs were widely expressed in neuronal populations. In area 46, the intensity of ISH signals of ROBO mRNAs was the highest among the cortical areas. The profile curves indicated that the signals were intense in the infragranular layers in this area (Fig. 4B,c), which was different in other areas. Except area 46, both ROBO1 and ROBO2 mRNAs showed dense signals in layer II (Fig. 4B,f,i,l, arrowheads), and the signal intensity gradually decreased toward the deeper layers. Some scattered cells in layers II–VI, particularly in early sensory areas, expressed higher levels of ROBO1 and ROBO2 mRNAs than the surrounding cells.
Considering the widespread distributions of ROBO mRNAs, it is likely that they are coexpressed with SLIT1 mRNA. In Table 2, we summarized the results of cell counting in area 46, TE and M1. In area 46 and TE, 83–97% of SLIT1 positive cells expressed ROBO1 mRNA (see also Supplementary Fig. S2). Compared with the result of coexpression of SLIT1 and ROBO1 mRNAs, the ratio of SLIT1+/ROBO2+ cells among SLIT1+ cells in the 3 areas was generally low except in layer II. Nonetheless, the mean ratio was more than 65% in the areas examined.
In summary, although SLIT1 mRNA exhibited the most obvious area specificity among the family and receptor mRNAs investigated, all these genes were generally more abundantly expressed in the sensory and prefrontal association areas than in early sensory areas, and each exhibited a unique laminar profile. At the single cell level, SLIT1 and ROBO mRNAs were colocalized in the majority of cortical neurons in the sensory and prefrontal association areas.
Postnatal Alteration of SLIT1 mRNA Expression in Cortex
In our previous studies, the area-specific mRNA expressions of RBP4 and OCC1 genes were found to be established during postnatal development (Tochitani et al. 2003; Komatsu et al. 2005). This led us to examine the expression pattern of SLIT1 mRNA in macaque monkeys at early postnatal stages.
The upper panel of Figure 5A shows the distribution of SLIT1 mRNA in coronal sections of P2 monkey brain. The lower panel of this figure shows the pseudocolor representation quantified for ROD. As these data show, the area difference in SLIT1 mRNA expression in the newborn monkey was generally similar to that in young adult monkeys. Intense hybridization signals were observed in the frontal cortex (Fig. 5A,a), lateral intraparietal area (LIP, Fig. 5A,d), the STS bank containing STP (Fig. 5A,b), parietal association areas, PG and PGm (Fig. 5A,d). Moderately strong signals were observed in M1, area 3b, amygdala complex (Amy, Fig. 5A,b), Cl (Fig. 5A,c), and IN (Fig. 5A,c). In contrast, the SLIT1 mRNA expression level was low in the middle temporal visual area (MT, Fig. 5A,d) and lowest in V1 (Fig. 5A,e).
Despite the overall similarity of the area specificity, we observed clear differences in the laminar distribution of SLIT1 mRNA between the infant (P1, P2, and P30, Fig. 6A) and the young adult cortices. One key feature of the expression pattern in the infant cortex is the high SLIT1 mRNA expression level in deep layers. As shown in the magnified images, SLIT1 mRNA was concentrated in layer IV, abundantly expressed in layers V and VI, but was scarce in layers II and III. This expression pattern was prevalent in the association areas such as area 46, TE, and PG, but the deep-layer preference was common across areas (Fig. 6A). In order to compare the laminar profiles of different areas quantitatively, we calculated the NSI of each layer of 6 different areas, which takes into account local neuronal density (Fig. 6B). As Figure 6B shows, the normalized laminar profiles of area 46, TE and PG were very similar (Fig. 6B,a,b,c). The NSIs of M1 and area 3b were lower than those of these association areas, but the area difference was not as large as that in the more aged monkeys (see Fig. 3B). The SLIT1 mRNA signal abruptly diminished at the V1/V2 border (Fig. 5C, arrowhead). This is mainly due to the lack of SLIT1 mRNA expression in layer IV (Fig. 5C and Fig. 6A,k). Unlike in young adults, however, strong signals of SLIT1 mRNA were detected in layer VI of infant V1 (Fig. 6A,k). The NSI was relatively high in layer V as well (Fig. 6B,f).
Comparing the NSIs in the infants (Fig. 6B) with those in older monkeys (Fig. 3B), it appears that a large reduction of expression in layers IV–VI occurred postnatally in all the areas examined, but the change in laminar profiles was different across areas. To investigate how the area-specific expression is formed in detail, we calculated the young adult/infant ratios of NSI for 6 areas, separately for supra and infragranular layers (Fig. 7). This analysis revealed a differential reduction of SLIT1 mRNA signals in the supra and infragranular layers across various areas. In the supragranular layers, the young adult/infant ratios of the 3 association areas (area 46, TE, and PG) were close to 1 and significantly higher than those of the remaining areas (M1, area 3b, and V1, P < 0.01, Fig. 7A). The ratios suggest that the supragranular expression of SLIT1 mRNA in the association areas did not change much postnatally, whereas those in M1, area 3b, and V1 significantly decreased.
We also observed large area differences in the young adult/infant ratios in the infragranular layers (Fig. 7B). In this case, the ratio in area 46 was significantly higher than those in all the other areas examined, including TE and PG (P < 0.01). The ratios in area 3b and V1 in the infragranular layers were even lower than those in M1, TE, and PG. These graphs demonstrate the area/layer-specific decrease in SLIT1 mRNA expression level during the postnatal period that consequently led to the characteristic expression pattern in the macaque cortex.
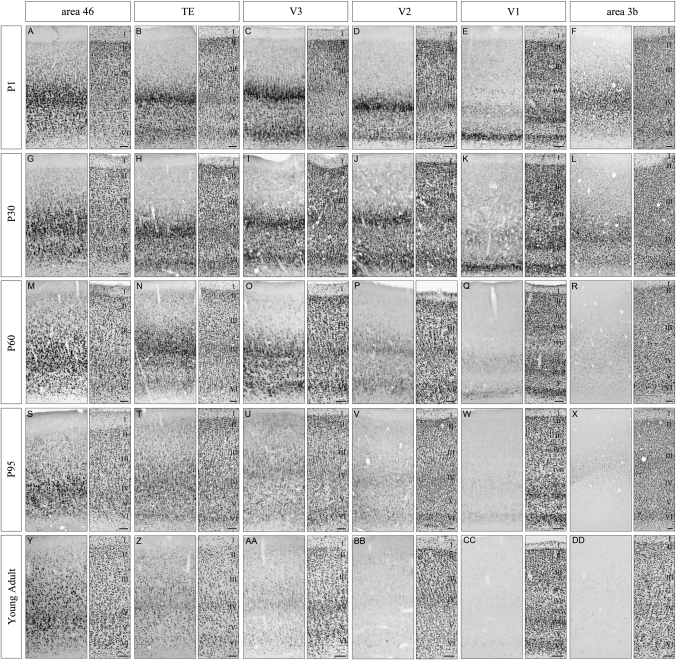
To further examine when and how such a decrease in expression level occurs, we investigated the SLIT1 mRNA expression in the cortices of P60 and P95 monkeys. Figure 8 shows the SLIT1 mRNA expression pattern in 6 cortical areas (area 3b, V1, V2, V3, TE, and area 46) in 5 different postnatal ages (P1, P30, P60, P95 and young adult). As mentioned above, the expression patterns in these 6 areas were almost the same between P1 and P30 monkeys (Fig. 8A–L). At P60, although the levels of SLIT1 mRNA expression in area 46, TE and V3 remained high, the expression levels in V2, V1 and area 3b were lower than those in P30 (Fig. 8M–R). At P95, we observed only faint SLIT1 mRNA signals in V2, V1 and area 3b (Fig. 8V–X), whereas difference in the expression level between area 46 and TE was relatively smaller in P95 than in young adult (Fig. 8S,T). Thus, the alteration of SLIT1 mRNA expression started at approximately P60–P95 with a different time course in each cortical area. Therefore, the characteristic expression pattern of SLIT1 mRNA was established by gradual downregulation in the postnatal periods (P60–P95 and older ages), although some area differences were already observed at birth.
Figure 8.
Postnatal Alteration of SLIT1 mRNA Expression in Various Cortical Areas. The expression of SLIT1 mRNA in 6 cortical areas (area 3b, V1, V2, V3, TE, and area 46) at 5 postnatal ages are shown. Coronal sections for ISH of SLIT1 mRNA (left panels) and the adjacent sections for cresyl violet staining (right panels) of the macaque neocortex are shown. (A–F) P1, (G–L) P30, (M–R) P60, (S–X) P95, and (Y–DD) young adult (monkey C). Scale bar = 100 μm.
Discussion
In this study, we characterized the SLIT1 gene as showing a prefrontal-enriched expression in the monkey cortex. Although SLIT1 is known to function as an axon guidance cue in developing CNS, our expression analysis suggests a novel role of SLIT1 and SLIT/ROBO signaling in the sensory and particularly prefrontal association areas of the postnatal brain. By comparing the SLIT1 mRNA expression between the newborn and young adult monkeys, we found that the high degree of area specificity is established by downregulation of the expression in an area- and laminar-specific manner during postnatal development.
Enriched Expression of SLIT1 mRNA in Prefrontal Cortex
Lines of evidence suggest that the prefrontal-enriched expression of SLIT1 mRNA is a common feature between Old World monkeys and humans. Northern blot analysis indicated that the SLIT1 mRNA expression level is high in the frontal lobe and low in the occipital lobe of humans (Itoh et al. 1998). In aged human brains, DNA microarray analysis revealed that the SLIT1 mRNA expression level is specifically low in V1, whereas it is relatively high in the prefrontal cortex (area 10 and area 11) and HC (Liang et al. 2007). In contrast, SLIT1 mRNA expression in the rat cortex did not show such an area difference: The frontal area showed a moderately strong signal, and there was no marked reduction in primary areas (data not shown). Slit1 mRNA signals were mainly located in layers Va and VI throughout the neocortex in rats (data not shown), a laminar distribution that is different from that of monkeys. We speculate that the distinct area-specific expression of SLIT1 mRNA in Old World monkeys and humans is observed because of a high degree of functional specification of these cortices.
We have reported that the RBP4 and PNMA5 genes are also highly expressed in the sensory and prefrontal association areas of macaque monkeys and their distribution is similar to each other (Komatsu et al. 2005; Takaji et al. 2009). The mRNA expression levels of SLIT1 and these genes increase gradually from the primary sensory to sensory association areas. In primates, pyramidal neurons of cortical areas involved in more integrative processing have more complex and widespread dendrites, with more abundant spines, than those in areas involved in the early stages of sensory processing. Elston and his colleagues reported that, in the prefrontal cortex (area 10), TE and V1, the average total numbers of spines in the basal dendrites of layer III neurons are 8766, 7260, and 643, respectively (Elston et al. 2001; Elston 2007). Interestingly, these numbers can be seen as correlating with the expression level of SLIT1 mRNA in layer III. It is thus possible that the association area-enriched genes contribute to a greater capacity for integrating inputs.
Despite the similarity in area expression, the laminar profile of SLIT1 mRNA was different from those of RBP4 and PNMA5 mRNAs. These genes are both widely expressed in the excitatory neurons of layers II, III, and V in the sensory and prefrontal association areas, but are restricted to the upper part of the supragranular layers in early sensory areas. In contrast, intense ISH signals of SLIT1 mRNA were observed in the middle layers, leading to a “bell-shaped” laminar distribution in most cortical areas. Because the cortical middle layers mainly receive 2 types of projection, that is, feedforward corticocortical and thalamocortical projections (Rockland and Pandya 1979; Felleman and Van Essen 1991; Jones 1998), SLIT1 may thus play a role in how neurons receive and/or process these inputs. The difference in the laminar expression pattern between SLIT1 and RBP4/PNMA5 genes may reflect different roles of these genes in the association cortex.
To understand the role of SLIT1 in primates, it would be useful to examine the expression patterns of other SLITs and ROBOs because all SLITs interact with all ROBOs (Brose et al. 1999; Li et al. 1999; Sabatier et al. 2004). By ISH, we found that all SLIT mRNAs were abundant in the association areas, although SLIT1 mRNA showed the highest area specificity. All SLIT genes exhibited different laminar expression patterns and their expressions altogether covered layers II–VI of the association areas. In particular, the laminar distributions of SLIT mRNAs were strikingly complementary in TE. This observation suggests that they play different roles in the macaque cortex.
Interestingly, the SLIT2 mRNA expression in excitatory neurons was similar to that of RBP4-type genes. Our double ISH analysis showed that SLIT2 and RBP4 mRNAs were colocalized in the cortical neurons (data not shown). Thus, SLIT2, RBP4, and PNMA5, whose expressions show the RBP4-like expression in the cortex, may all exert influence over a similar type of cortical neuron.
Compared with SLIT mRNAs, ROBO1 and ROBO2 mRNAs were more widely distributed across layers and areas, and their expressions were mostly overlapping. These data suggest that most cortical neurons could potentially respond to SLIT signaling. Given that cortical neurons, particularly in the association areas, simultaneously express SLIT and ROBO genes, SLITs could act in either an autocrine or a paracrine manner. Further studies are required to understand how SLITs and ROBOs interact in specific aspects of monkey cortical physiology.
Alteration of SLIT1 mRNA Expressions in Cortex during Postnatal Periods
We have demonstrated that the prefrontal-enriched SLIT1 mRNA expression pattern was established postnatally. From the late prenatal (data not shown) to early postnatal stages, the SLIT1 mRNA expression level was intense in deeper layers, particularly in layers IV and VI throughout the cortex (except for V1 where the expression level in layer IV is very low). Neurons in these 2 layers receive and send thalamocortical and corticothalamic connections, respectively. In macaque monkeys, the thalamocortical projections are known to be established by E151 (Darian-Smith et al. 1990). The primate brain, however, undergoes significant changes during postnatal development as well. During the perinatal period, axonal elongation (LaMantia and Rakic 1990) and synaptogenesis (Rakic et al. 1986; Bourgeois and Rakic 1993; Bourgeois et al. 1994) occur vigorously in the monkey cortex. Corticocortical connections are being formed until P60 (Batardiere et al. 2002; Kennedy and Burkhatler 2003; Kennedy et al. 2007). Subsequently, synaptic elimination continues until puberty (Bourgeois et al. 1994; Levitt 2003). Selective elimination of neural connections plays a major role in the establishment of mature circuits. In this regard, the postnatal decrease in SLIT1 mRNA signals may reflect rearrangement of cortical circuitry. Our results indicate that SLIT1 mRNA expression in the infragranular layers is downregulated to a greater degree than that in the superficial layers. It may be noted that these layers exhibit a large decrease in the staining of nonphosphorylated Neurofilament (NEFM), as observed in the marmoset cortex (Burman et al. 2007).
Note that the time course of the decreased expression level was markedly different across areas. From P30 to P95, the level of SLIT1 mRNA expression dropped sharply in V1, but decreased slowly in TE. This time course appears to be coincident with the time course of cortical maturation, that is, the association areas mature after sensory areas (Brody et al. 1987; Conde et al. 1996; Gogtay et al. 2004; Guillery 2005). In this regard, although area 3b is located more anteriorly than V3, the downregulation of the SLIT1 mRNA expression in area 3b occurred faster than that in V3. This is difficult to be explained only by the caudal to rostral gradient. We also found that MT was lightly stained at the newborn stage, which may be consistent with the early maturation of this area as shown in humans and marmosets (Annese et al. 2005; Bourne and Rosa 2006). Among the cortical areas, the prefrontal cortex is considered to be one of the last areas to mature in humans and nonhuman primates (Goldman and Galkin 1978; Chugani et al. 1987; Chiron et al. 1992; Huttenlocher and Dabholkar 1997). We thus can speculate that the prefrontal cortex may require a larger amount of SLIT1 than primary or early sensory areas, in accord with longer periods of developmental remodeling.
Functional Consideration of Role of SLIT1 in Postnatal Cortex
Slit homologues have been found in a wide variety of taxonomic groups (Holmes et al. 1998; Itoh et al. 1998; Li et al. 1999; Halloran et al. 2000; Hao et al. 2001; Vargesson et al. 2001; Marillat et al. 2002). They appear to function as diffusible, repellent guidance molecules for axons in the CNS, and this function is well conserved evolutionally (Andrews et al. 2007; Chedotal 2007). Although the role of SLITs/ROBOs in axon guidance during development may be as expected, their roles in postnatal monkey brains, where there are no major guidance events, is as yet unclarified. One plausible possibility is that SLIT1 plays a role in maintaining certain neuronal morphology and circuits (Nestler 2000; Murai and Pasquale 2002; Bahi and Dreyer 2005; Lin et al. 2009). Recent studies have demonstrated the dynamic mobility of neuronal processes in the mature brain (Holtmaat et al. 2005; Stettler et al. 2006). In this respect, we note that SLIT1 mRNA was preferentially expressed in regions known to show high neuronal integration and/or plasticity, such as the prefrontal cortex and HC. Previous studies revealed that some “axon” guidance molecules also regulate dendritic morphology and spine growth (Ethell et al. 2001; Morita et al. 2006; Yoshida et al. 2008). For example, semaphorin 3A (Sema3A) promotes dendritic branching and spine maturation in cultured cortical neurons, and its deficiency causes a lower density of cortical spines (Morita et al. 2006). SLIT1 is also reported to promote dendritic growth and branching of both pyramidal and nonpyramidal neurons in the developing rat cortex (Whitford et al. 2002).
Clinical evidence suggests a link between spine morphology and mental retardation. In idiopathic mental retardation patients, an immature or deformed morphology of spines is widely observed in the cortex (Purpura 1974; Fiala et al. 2002). Interestingly, SLIT-ROBO specific-GAP3 (SRGAP3), which is one of the components of the SLIT/ROBO signaling pathway, is reported to have a putative role in mental retardation (Endris et al. 2002). Thus, our results raise the possibility that the preferential expression of SLIT1 in the prefrontal cortex contributes to the maintenance of the highly branched dendrites, greater spine density and its morphology in these areas that have long been known as important for higher cognitive functions. This possibility needs to be tested in future studies.
Supplementary Material
Supplementary materials can be found at: http://www.cercor.oxfordjournals.org/.
Funding
Grants-in-Aid for Scientific Research on Priority Areas (A, 17024055 to T.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Material
Acknowledgments
We thank Drs Junichi Yuasa-Kawada and Masaharu Noda of the National Institute for Basic Biology and Shingo Akiyoshi of the JFCR Cancer Institute for help with RLCS. We thank Dr Fumiko Ono of the Corporation for Production and Research of Laboratory Primates; Dr Keiji Terao of the Tsukuba Primate Research Center, National Institute of Infectious Diseases; and Drs Hitoshi Horie, Shinobu Abe and Sou Hashizume of the Japan Poliomyelitis Research Institute for supplying monkey tissues. We are also grateful to Dr Kathleen S. Rockland for valuable discussions and critical reading of this manuscript. The authors declare that no competing interests exist. Conflict Of Interest: None declared.
References
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Andrews WD, Barber M, Parnavelas JG. Slit–Robo interactions during cortical development. J Anat. 2007;211:188–198. doi: 10.1111/j.1469-7580.2007.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annese J, Gazzaniga MS, Toga AW. Localization of the human cortical visual area MT based on computer aided histological analysis. Cereb Cortex. 2005;15:1044–1053. doi: 10.1093/cercor/bhh205. [DOI] [PubMed] [Google Scholar]
- Bagri A, Marin O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, Tessier-Lavigne M. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron. 2002;33:233–248. doi: 10.1016/s0896-6273(02)00561-5. [DOI] [PubMed] [Google Scholar]
- Bahi A, Dreyer JL. Cocaine-induced expression changes of axon guidance molecules in the adult rat brain. Mol Cell Neurosci. 2005;28:275–291. doi: 10.1016/j.mcn.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Batardiere A, Barone P, Knoblauch K, Giroud P, Berland M, Dumas AM, Kennedy H. Early specification of the hierarchical organization of visual cortical areas in the macaque monkey. Cereb Cortex. 2002;12:453–465. doi: 10.1093/cercor/12.5.453. [DOI] [PubMed] [Google Scholar]
- Bourgeois JP, Goldman-Rakic PS, Rakic P. Synaptogenesis in the prefrontal cortex of rhesus monkeys. Cereb Cortex. 1994;4:78–96. doi: 10.1093/cercor/4.1.78. [DOI] [PubMed] [Google Scholar]
- Bourgeois JP, Rakic P. Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J Neurosci. 1993;13:2801–2820. doi: 10.1523/JNEUROSCI.13-07-02801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JA, Rosa MG. Hierarchical development of the primate visual cortex, as revealed by neurofilament immunoreactivity: early maturation of the middle temporal area (MT) Cereb Cortex. 2006;16:405–414. doi: 10.1093/cercor/bhi119. [DOI] [PubMed] [Google Scholar]
- Brody BA, Kinney HC, Kloman AS, Gilles FH. Sequence of central nervous system myelination in human infancy. I. An autopsy study of myelination. J Neuropathol Exp Neurol. 1987;46:283–301. doi: 10.1097/00005072-198705000-00005. [DOI] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr Opin Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Lui LL, Rosa MG, Bourne JA. Development of non-phosphorylated neurofilament protein expression in neurones of the New World monkey dorsolateral frontal cortex. Eur J Neurosci. 2007;25:1767–1779. doi: 10.1111/j.1460-9568.2007.05442.x. [DOI] [PubMed] [Google Scholar]
- Burton H, Sinclair R. Somatosensory cortex and tactile perceptions. In: Carterette E, editor. Pain and touch. 2nd ed. London: Academic Press; 1996. pp. 105–177. [Google Scholar]
- Chedotal A. Slits and their receptors. Adv Exp Med Biol. 2007;621:65–80. doi: 10.1007/978-0-387-76715-4_5. [DOI] [PubMed] [Google Scholar]
- Chiron C, Raynaud C, Maziere B, Zilbovicius M, Laflamme L, Masure MC, Dulac O, Bourguignon M, Syrota A. Changes in regional cerebral blood flow during brain maturation in children and adolescents. J Nucl Med. 1992;33:696–703. [PubMed] [Google Scholar]
- Chugani HT, Phelps ME, Mazziotta JC. Positron emission tomography study of human brain functional development. Ann Neurol. 1987;22:487–497. doi: 10.1002/ana.410220408. [DOI] [PubMed] [Google Scholar]
- Conde F, Lund JS, Lewis DA. The hierarchical development of monkey visual cortical regions as revealed by the maturation of parvalbumin-immunoreactive neurons. Brain Res Dev Brain Res. 1996;96:261–276. doi: 10.1016/0165-3806(96)00126-5. [DOI] [PubMed] [Google Scholar]
- Darian-Smith C, Darian-Smith I, Cheema SS. Thalamic projections to sensorimotor cortex in the newborn macaque. J Comp Neurol. 1990;299:47–63. doi: 10.1002/cne.902990104. [DOI] [PubMed] [Google Scholar]
- Elston GN. Cortex, cognition and the cell: new insights into the pyramidal neuron and prefrontal function. Cereb Cortex. 2003;13:1124–1138. doi: 10.1093/cercor/bhg093. [DOI] [PubMed] [Google Scholar]
- Elston GN. Specializations in pyramidal cell structure during primate evolution. In: Preuss TM, editor. Evolution of nervous systems. London: Academic Press; 2007. pp. 191–242. [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J. The pyramidal cell in cognition: a comparative study in human and monkey. J Neurosci. 2001;21:RC163. doi: 10.1523/JNEUROSCI.21-17-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, Elston A, Zietsch B, Defelipe J, Manger P, Casagrande V, Kaas JH. Specializations of the granular prefrontal cortex of primates: implications for cognitive processing. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:26–35. doi: 10.1002/ar.a.20278. [DOI] [PubMed] [Google Scholar]
- Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher ER, Tariverdian G, Kirsch S, Karch D, et al. The novel Rho-GTPase activating gene MEGAP/srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci U S A. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D, Lund JS, Schmechel DE, Towles AC. Distribution of GABAergic neurons and axon terminals in the macaque striate cortex. J Comp Neurol. 1987;264:73–91. doi: 10.1002/cne.902640107. [DOI] [PubMed] [Google Scholar]
- Fujiyama F, Furuta T, Kaneko T. Immunocytochemical localization of candidates for vesicular glutamate transporters in the rat cerebral cortex. J Comp Neurol. 2001;435:379–387. doi: 10.1002/cne.1037. [DOI] [PubMed] [Google Scholar]
- Fuster JM. Frontal lobe and cognitive development. J Neurocytol. 2002;31:373–385. doi: 10.1023/a:1024190429920. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, 3rd, Herman DH, Clasen LS, Toga AW, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman PS, Galkin TW. Prenatal removal of frontal association cortex in the fetal rhesus monkey: anatomical and functional consequences in postnatal life. Brain Res. 1978;152:451–485. doi: 10.1016/0006-8993(78)91103-4. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14:477–485. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Krimer LS, Urban NN, Barrionuevo G, Lewis DA. Synaptic efficacy during repetitive activation of excitatory inputs in primate dorsolateral prefrontal cortex. Cereb Cortex. 2004;14:530–542. doi: 10.1093/cercor/bhh015. [DOI] [PubMed] [Google Scholar]
- Guillery RW. Is postnatal neocortical maturation hierarchical? Trends Neurosci. 2005;28:512–517. doi: 10.1016/j.tins.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Hao JC, Yu TW, Fujisawa K, Culotti JG, Gengyo-Ando K, Mitani S, Moulder G, Barstead R, Tessier-Lavigne M, Bargmann CI. C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron. 2001;32:25–38. doi: 10.1016/s0896-6273(01)00448-2. [DOI] [PubMed] [Google Scholar]
- Higo N, Oishi T, Yamashita A, Matsuda K, Hayashi M. Quantitative non-radioactive in situ hybridization study of GAP-43 and SCG10 mRNAs in the cerebral cortex of adult and infant macaque monkeys. Cereb Cortex. 1999;9:317–331. doi: 10.1093/cercor/9.4.317. [DOI] [PubMed] [Google Scholar]
- Holmes GP, Negus K, Burridge L, Raman S, Algar E, Yamada T, Little MH. Distinct but overlapping expression patterns of two vertebrate slit homologs implies functional roles in CNS development and organogenesis. Mech Dev. 1998;79:57–72. doi: 10.1016/s0925-4773(98)00174-9. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Huminiecki L, Gorn M, Suchting S, Poulsom R, Bicknell R. Magic roundabout is a new member of the roundabout receptor family that is endothelial specific and expressed at sites of active angiogenesis. Genomics. 2002;79:547–552. doi: 10.1006/geno.2002.6745. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Itoh A, Miyabayashi T, Ohno M, Sakano S. Cloning and expressions of three mammalian homologues of Drosophila slit suggest possible roles for Slit in the formation and maintenance of the nervous system. Brain Res Mol Brain Res. 1998;62:175–186. doi: 10.1016/s0169-328x(98)00224-1. [DOI] [PubMed] [Google Scholar]
- Jones EG. Viewpoint: the core and matrix of thalamic organization. Neuroscience. 1998;85:331–345. doi: 10.1016/s0306-4522(97)00581-2. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Burkhatler A. Ontogenesis of cortical connectivity. In: Chalupa LM, Werner JS, editors. The visual neurosciences. Cambridge (MA): MIT Press; 2003. pp. 146–159. [Google Scholar]
- Kennedy H, Douglas R, Knoblauch K, Dehay C. Self-organization and pattern formation in primate cortical networks. In: Bock G, Goode J, editors. Cortical development: genes and genetic abnormalities. Vol. 288. Hobeken (NJ): Wiley Interscience; 2007. pp. 178–194. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Retinol-binding protein gene is highly expressed in higher-order association areas of the primate neocortex. Cereb Cortex. 2005;15:96–108. doi: 10.1093/cercor/bhh112. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Rakic P. Axon overproduction and elimination in the corpus callosum of the developing rhesus monkey. J Neurosci. 1990;10:2156–2175. doi: 10.1523/JNEUROSCI.10-07-02156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P. Structural and functional maturation of the developing primate brain. J Pediatr. 2003;143:S35–S45. doi: 10.1067/s0022-3476(03)00400-1. [DOI] [PubMed] [Google Scholar]
- Li HS, Chen JH, Wu W, Fagaly T, Zhou L, Yuan W, Dupuis S, Jiang ZH, Nash W, Gick C, et al. Vertebrate slit, a secreted ligand for the transmembrane protein roundabout, is a repellent for olfactory bulb axons. Cell. 1999;96:807–818. doi: 10.1016/s0092-8674(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genom. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Hatanaka Y, Saito H, Yamamori T, Hashikawa T. Differential expression of gamma-aminobutyric acid type B receptor-1a and -1b mRNA variants in GABA and non-GABAergic neurons of the rat brain. J Comp Neurol. 2000;416:475–495. [PubMed] [Google Scholar]
- Lin L, Lesnick TG, Maraganore DM, Isacson O. Axon guidance and synaptic maintenance: preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009;32:142–149. doi: 10.1016/j.tins.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G, Flames N, Ma L, Fouquet C, Di Meglio T, Chedotal A, Tessier-Lavigne M, Marin O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J Neurosci. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V, Cases O, Nguyen-Ba-Charvet KT, Tessier-Lavigne M, Sotelo C, Chedotal A. Spatiotemporal expression patterns of slit and robo genes in the rat brain. J Comp Neurol. 2002;442:130–155. doi: 10.1002/cne.10068. [DOI] [PubMed] [Google Scholar]
- Meberg PJ, Routtenberg A. Selective expression of protein F1/(GAP-43) mRNA in pyramidal but not granule cells of the hippocampus. Neuroscience. 1991;45:721–733. doi: 10.1016/0306-4522(91)90284-u. [DOI] [PubMed] [Google Scholar]
- Morita A, Yamashita N, Sasaki Y, Uchida Y, Nakajima O, Nakamura F, Yagi T, Taniguchi M, Usui H, Katoh-Semba R, et al. Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signaling. J Neurosci. 2006;26:2971–2980. doi: 10.1523/JNEUROSCI.5453-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai KK, Pasquale EB. Can Eph receptors stimulate the mind? Neuron. 2002;33:159–162. doi: 10.1016/s0896-6273(02)00565-2. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Genes and addiction. Nat Genet. 2000;26:277–281. doi: 10.1038/81570. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet KT, Chedotal A. Role of slit proteins in the vertebrate brain. J Physiol Paris. 2002;96:91–98. doi: 10.1016/s0928-4257(01)00084-5. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huang X, Toga AW. The rhesus monkey brain in stereotaxic coordinates. San Diego (CA): Academic Press; 1999. 408 p. [Google Scholar]
- Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Purpura DP. Dendritic spine “dysgenesis” and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Rockland KS, Pandya DN. Laminar origins and terminations of cortical connections of the occipital lobe in the rhesus monkey. Brain Res. 1979;179:3–20. doi: 10.1016/0006-8993(79)90485-2. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. Slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Sabatier C, Plump AS, Le M, Brose K, Tamada A, Murakami F, Lee EY, Tessier-Lavigne M. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/s0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- Saper CB. Any way you cut it: a new journal policy for the use of unbiased counting methods. J Comp Neurol. 1996;364:5. doi: 10.1002/(SICI)1096-9861(19960101)364:1<5::AID-CNE1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Shintani T, Kato A, Yuasa-Kawada J, Sakuta H, Takahashi M, Suzuki R, Ohkawara T, Takahashi H, Noda M. Large-scale identification and characterization of genes with asymmetric expression patterns in the developing chick retina. J Neurobiol. 2004;59:34–47. doi: 10.1002/neu.10338. [DOI] [PubMed] [Google Scholar]
- Stettler DD, Yamahachi H, Li W, Denk W, Gilbert CD. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron. 2006;49:877–887. doi: 10.1016/j.neuron.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Roberts I, Bateman A, Bankier A, Sheppard M, Hobbs C, Xiong J, Minna J, Latif F, Lerman M, et al. The DUTT1 gene, a novel NCAM family member is expressed in developing murine neural tissues and has an unusually broad pattern of expression. Mol Cell Neurosci. 1998;11:29–35. doi: 10.1006/mcne.1998.0672. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yaoi T, Kawai J, Hara A, Kuwajima G, Wantanabe S. Restriction landmark cDNA scanning (RLCS): a novel cDNA display system using two-dimensional gel electrophoresis. Nucleic Acids Res. 1996;24:289–294. doi: 10.1093/nar/24.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Activity-dependent expression of occ1 in excitatory neurons is a characteristic feature of the primate visual cortex. Cereb Cortex. 2006;16:929–940. doi: 10.1093/cercor/bhj034. [DOI] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Differential expression patterns of occ1-related genes in adult monkey visual cortex. Cereb Cortex. 2009;19:1937–1951. doi: 10.1093/cercor/bhn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaji M, Komatsu Y, Watakabe A, Hashikawa T, Yamamori T. Paraneoplastic antigen-like 5 gene (PNMA5) is preferentially expressed in the association areas in a primate specific manner. Cereb Cortex. 2009;19:2865–2879. doi: 10.1093/cercor/bhp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- Tochitani S, Hashikawa T, Yamamori T. Expression of occ1 mRNA in the visual cortex during postnatal development in macaques. Neurosci Lett. 2003;337:114–116. doi: 10.1016/s0304-3940(02)01311-3. [DOI] [PubMed] [Google Scholar]
- Tochitani S, Liang F, Watakabe A, Hashikawa T, Yamamori T. The occ1 gene is preferentially expressed in the primary visual cortex in an activity-dependent manner: a pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex. Eur J Neurosci. 2001;13:297–307. doi: 10.1046/j.0953-816x.2000.01390.x. [DOI] [PubMed] [Google Scholar]
- Vargesson N, Luria V, Messina I, Erskine L, Laufer E. Expression patterns of Slit and Robo family members during vertebrate limb development. Mech Dev. 2001;106:175–180. doi: 10.1016/s0925-4773(01)00430-0. [DOI] [PubMed] [Google Scholar]
- Wang XJ. Synaptic reverberation underlying mnemonic persistent activity. Trends Neurosci. 2001;24:455–463. doi: 10.1016/s0166-2236(00)01868-3. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Fujita H, Hayashi M, Yamamori T. Growth/differentiation factor 7 is preferentially expressed in the primary motor area of the monkey neocortex. J Neurochem. 2001;76:1455–1464. doi: 10.1046/j.1471-4159.2001.00177.x. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Ichinohe N, Ohsawa S, Hashikawa T, Komatsu Y, Rockland KS, Yamamori T. Comparative analysis of layer-specific genes in mammalian neocortex. Cereb Cortex. 2007;17:1918–1933. doi: 10.1093/cercor/bhl102. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Nawa H, Yamamori T. Gene expression profiling of primate neocortex: molecular neuroanatomy of cortical areas. Genes Brain Behav. 2006;5(Suppl. 1):38–43. doi: 10.1111/j.1601-183X.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Sadakane O, Shimegi S, Takahata T, Higo N, Tochitani S, Hashikawa T, Naito T, Osaki H, et al. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb Cortex. 2008;19:1915–1928. doi: 10.1093/cercor/bhn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Ohsawa S, Hashikawa T, Yamamori T. Binding and complementary expression patterns of semaphorin 3E and plexin D1 in the mature neocortices of mice and monkeys. J Comp Neurol. 2006;499:258–273. doi: 10.1002/cne.21106. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Sugai T, Nakaya N, Wakabayashi K, Takahashi H, Yamamori T, Nawa H. Similarity and variation in gene expression among human cerebral cortical subregions revealed by DNA macroarrays: technical consideration of RNA expression profiling from postmortem samples. Brain Res Mol Brain Res. 2001;88:74–82. doi: 10.1016/s0169-328x(01)00019-5. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chedotal A, Ghosh A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/s0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Yang H, Wanner IB, Roper SD, Chaudhari N. An optimized method for in situ hybridization with signal amplification that allows the detection of rare mRNAs. J Histochem Cytochem. 1999;47:431–446. doi: 10.1177/002215549904700402. [DOI] [PubMed] [Google Scholar]
- Yoshida J, Kubo T, Yamashita T. Inhibition of branching and spine maturation by repulsive guidance molecule in cultured cortical neurons. Biochem Biophys Res Commun. 2008;372:725–729. doi: 10.1016/j.bbrc.2008.05.124. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Cox LA, Dasika GK, Lee EY. Cloning and functional studies of a novel gene aberrantly expressed in RB-deficient embryos. Dev Biol. 1999;207:62–75. doi: 10.1006/dbio.1998.9141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.