Abstract
Objective
To compare gene expression profiles between ankylosing spondylitis (AS) and undifferentiated spondyloarthritis (USpA) patients with inflammatory low back pain.
Methods
Peripheral blood mononuclear cells (PBMC) from AS, USpA and healthy subjects were screened with genome-wide microarrays, followed by validation with Real-time PCR.
Results
Microarray profiling and Real-time PCR assays showed that any differences between AS and healthy subjects were only minor. In contrast, 20 genes were strikingly more highly expressed in USpA. The “regulator of G-protein signaling 1” (RGS1) was identified as the most useful biomarker distinguishing especially USpA patients and to a lesser extent AS patients, from control subjects (p= 2.3×10-7 and 6.7×10-3 respectively). All these findings were verified with an independent cohort of patients, which also included rheumatoid arthritis and patients with mechanical low back pain. The Receiver-operator-characteristics (ROC) area under the curve (AUC) values in the first and second cohorts of USpA patients were 0.98 and 0.93 respectively (p=1×10-4). To evaluate the possible derivation of RGS1, we cultured a monocyte-derived cell line with a panel of cytokines and chemokines. RGS1 was significantly induced either by TNF-α or by IL-17.
Conclusion
(1) The PBMC of USpA carry strikingly more highly expressed genes compared to AS or healthy subjects. (2) TNF-α and IL-17 – inducible RGS1 is a potential biomarker for USpA patients and to a lesser extent for AS patients, with inflammatory low back pain.
Spondyloarthritis (SpA) is a family of diseases consisting of the following members: ankylosing spondylitis (AS), undifferentiated spondyloarthritis (USpA), reactive arthritis (ReA), arthritis associated with inflammatory bowel diseases (IBD), and arthritis associated with psoriasis (PsA) (1). The most common are AS and USpA (2, 3). The diagnosis of ReA, PsA and arthritis associated with IBD is frequently made straightforward by the corresponding extra-articular features. However, the diagnosis of AS or USpA can be difficult, especially when the predominant clinical feature is low back pain, and not peripheral extremity inflammation. This is because low back pain alone is a very common primary complaint, contributing to 3% of annual medical visits in the U.S (4). However, only 5% of the chronic back pain seen in general practice is associated with SpA (5). Whereas the back pain associated with AS and USpA is designated as “inflammatory” back pain, non-SpA-associated back pain is commonly designated as “mechanical” back pain (6, 7). Currently, diagnosis of AS and USpA relies on clinical and imaging parameters that, in combination provide an estimate of the degree of probability of whether a patient has SpA (8). The degree of confidence is often not 100%, especially with USpA. This is because the most specific clinical parameter for diagnosing AS in general practice is plain-radiological sacroiliitis. Whereas plain-radiographic sacroiliitis is always positive in AS, it is negative in USpA. Frequently, years of followup are required before some of the USpA patients develop positive sacroiliitis. Both longitudinal and cross-sectional studies indicate that in some patients, USpA is an early form of AS (3, 9). Currently, there is no single blood-derived biomarker which by itself is highly sensitive and specific for distinguishing AS and USpA from patients with “mechanical” back pain (10). Although HLA-B27, TNF-α and perhaps IL-17 are pathogenic factors, as biomarkers when used alone they have either poor sensitivity or poor specificity (11). However, since some of the plain-radiographic sacroiliitis-negative patients show bone edema of the sacroiliac joints by MRI (12), it is possible that such local processes can lead to release of blood-derived protein- or nucleotide- based biomarkers useful in diagnosis of SpA.
In the first part of the study, we used whole-genome 20,589 probe gene expression microarrays each containing 16,283 curated RefSeq genes to screen for genes differentially expressed in AS and USpA relative to healthy subjects. The ultimate purpose of this entire study is to identify a small number of genes whose expression profile might serve as a cost-effective set of surrogate biomarkers for AS and USpA. This particular paper will not follow an alternate strategy which is to identify a diagnostic microarray gene expression pattern.
From the microarray data, we selected 25 most promising individual candidate genes, based on the degree of differential expression and the degree of statistical significance. These were then subjected to a validation test using Real-time PCR, still using the same cohort of AS and USpA patients. At least 6 biomarkers were validated to show potential diagnostic value when assessed by the area under the curve (AUC) of the Receiver-operator-characteristics (ROC). Biomarkers with ROC AUC 0.8 to 1.0 are usually considered as being useful in clinical practice (13). In the third part of our study, we submitted these 6 candidates to an even more stringent level of screening by using an independent cohort of AS and USpA patients as well as healthy subjects. In addition, we also compared the results to those of rheumatoid arthritis (RA) and also patients with mechanical low back pain. From these exhaustive studies, we discovered that the PBMC of USpA contained many more high expression genes compared to AS and healthy subjects. In addition, we identified “regulator of G-protein signaling 1” (RGS1) as the most promising candidate biomarker for USpA and to a lesser extent also for AS patients. In the human peripheral blood mononuclear cells (PBMC), RGS1 has been reported to be constitutively expressed only in monocytes (14). There are as yet no reports on which arthritis-inducing factors might drive the expression of RGS1 in monocytes. To study this, we stimulated in culture a monocyte-derived cell line with a panel of 25 cytokines and chemokines. We discovered that the most effective for generating RGS1 were TNF-α and IL-17. Thus, RGS1 might be a surrogate of the effects of these 2 arthritis-causing cytokines.
Materials and Methods
Research subjects
The cohorts of subjects providing samples for microarray as well as PCR studies were recruited from Beijing and Guangzhou in China, while the second cohort providing samples for PCR assays was collected from Taichung in Taiwan. All AS and USpA patients also fulfilled the Calin criteria for inflammatory back pain (6). Patients from the first cohort were also selected for their severity of disease activity. Patients from the second cohort were selected without regard to their disease activity or severity. The common selection criterion was clinical predominance of back pain. From Taichung, we also collected a cohort of rheumatoid arthritis (RA) patients, and a cohort of patients with mechanical low back pain. None of the patients in either cohort had psoriasis, inflammatory bowel diseases or history of precedent infections. The study and consent forms have been approved by the respective ethical committees, and signed by all participants.
Microarray studies
Peripheral venous blood samples were collected into CPT tubes (BD Diagnostics). PBMC were separated, and RNA extracted with Trizol®. Labeling of RNA was performed using the Illumina TotalPrep® RNA Amplification Kit (Applied Biosystems Ambion, Austin, TX). cRNA was hybridized to Sentrix Human Ref-8_v2 Beadchips followed by staining according to instructions from the manufacturer (Illumina, San Diego, CA).
Data were normalized together by the quantile normalization method (15). Data from AS and USpA were then separately compared to healthy subjects. These microarray results were submitted to 2 consecutive statistical screening processes to eliminate unlikely candidates. In the first screening, we eliminated genes which were less than two-fold different between patients and healthy subjects. In the second screening, the microarray values were log transformed, and genes with low variances among all subjects were again eliminated from subsequent analysis.
Real-time PCR assays
Total RNA was reverse-transcribed to cDNA by using QuantiTec Whole Transcriptome Amplification Kit (QIAGEN, Valencia, CA). RPLP0 (ribosomal protein, large, P0, NM_001002) and GAPDH were used as the housekeeping reference genes for PBMC and cell lines respectively. Relative levels of target gene transcripts were assayed in triplicate using quantitative real-time PCR in the 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The reactions were performed in 384-well plates (Applied Biosystems) in a volume of 10μl containing SYBR Green PCR Master Mix (Applied Biosystems), 500nM of each primer, and about 10ng of the cDNA template.
With the exception of three pairs of primers, all primers including RGS1 primer pair #1 were purchased from GeneGlobe of QIAGEN. Sequences of primers which were synthesized separately were RPLP0: 5′-CCACGCTGCTGAACATGCT-3′ and 5′-TCGAACACCTGCTGGATGAC-3′ (16); and RGS1 primer pair #2: 5′-TAGTCTTCACAAGCCAGCCA-3′ and 5′GGAAAAACTTCTTGCCAACC-3′ (qPrimerDepot, NIH); RGS-1 primer pair #3: 5′- ATTGAGTTCTGGCTGGCTTG-3′ and 5′-GATTCTCGAGTGCGGAAGTC-3′ (derived from Primer 3 v.0.4.0). Efficiencies of all primers exceeded 90%. Cycling conditions are in the footnote of Table 2. The relative amount of the target gene in each sample was calculated by the 2-ΔΔCt method.
Table 2.
Microarray and Real-time PCR data of those genes which were differentially expressed to statistically significant degree
| Microarray Results | Realtime PCR Results | ||||
|---|---|---|---|---|---|
| Symbol | Accession # | Fold-change in USpA | Fold-change in AS | Fold-change in USpA | Fold-change in AS |
| OLR1 | NM_002543.2 | 33.7 * | 270.1 * | ||
| FOSB | NM_006732.1 | 26.2 *** | 10.8 ** | ||
| RGS1 | NM_002922.3 | 17.3 *** | 3.2 | 52.7 *** | 5.4 * |
| NR4A2 | NM_006186.2 | 17.0 *** | 3.5 | 49.7 *** | |
| CXCL2 | NM_002089.1 | 17.0 *** | 159.5 *** | 12.3 | |
| CCL20 | NM_004591.1 | 16.9 * | |||
| CCL3L3 | NM_001001437.2 | 14.8 *** | 13.1 * | ||
| ATF3 | NM_001030287.1 | 14.3 *** | 24.6 *** | ||
| CCL3L1 | NM_021006.4 | 13.2 ** | 14.4 * | ||
| HBEGF | NM_001945.1 | 12.8 *** | 2.8 | 27.4 *** | |
| IL-1B | NM_000576.2 | 11.6 *** | 6.9 * | ||
| IL-1A | NM_000575.3 | 11.4 * | 597.2 *** | ||
| CD83 | NM_004233.2 | 11.2 *** | 21.7 ** | ||
| NR4A3 | NM_173199.1 | 10.4 *** | 2.6 | ||
| OSM | NM_020530.3 | 10.3 *** | 4.5 * | ||
| PAI | NM_002575.1 | 9.8 *** | 33.5 *** | 3.5 | |
| IL8 | NM_000584.2 | 8.9 ** | 11.5 ** | ||
| SNF1LK | NM_173354.2 | 8.8 *** | 5.9 * | ||
| CCL3 | NM_002983.1 | 8.6 ** | 15.1 ** | ||
| NR4A1 | NM_173158.1 | 6.1 * | 8.1 | ||
| SOCS3 | NM_003955.3 | 4.7 *** | 2.7 | 3.4 * | |
| IL6 | NM_000600.1 | 4.5 | 35.4 *** | ||
| IL1RN | NM_173842.1 | 3.8 ** | |||
| DEFA1 | NM_004084.2 | -5.0 ** | -7.9 * | -6.9 | |
| CAMP | NM_004345.3 | -6.3 ** | -75.4 | ||
Cells with no numbers indicate that mean values are not different from control subjects
Fold-changes refer to comparison with healthy controls
p< 1 × 10 -5
p< 1 × 10 −4
p< 1 × 10 −2
Absence of asterisk next to numericals = p<0.05
All comparisons are to the healthy cohort.
OLR1 = oxidized low density lipoprotein receptor 1; FOSB = FBJ murine osteosarcoma viral oncogene homolog B; RGS1 = regulator of G-protein signaling 1; NR4A2 = nuclear receptor subfamily 4 group A member 2; CXCL2 = chemokine C-X-C motif ligand 2; CCL20 = chemokine C-C motif ligand 20; CCL3L3 = chemokine C-C motif ligand 3-like 3; ATF3 = activating transcription factor 3; CCL3L1 = chemokine C-C motif ligand 3-like 1; HBEGF = heparin-binding EGF-like growth factor; IL-1B = interleukin 1 beta; IL-1A = interleukin 1 alpha; NR4A3 = nuclear receptor subfamily 4 group A member 3; OSM = oncostatin M; PAI = placental plasminogen activator inhibitor; IL8 = interleukin 8; SNF1LK = salt-inducible kinase 1; CCL3 = chemokine C-C motif ligand 3; NR4A1 = nuclear receptor subfamily 4 group A member 1; SOCS3 = suppressor of cytokine signaling 3; IL6 = interleukin 6; IL1RN = interleukin 1 receptor antagonist; DEFA1 = defensin alpha 1; CAMP = cathelicidin antimicrobial peptide.
Cycling conditions for PCR assays: 95°C for 15 minutes, followed by amplification for 40 cycles of 94 °C for 15 seconds, 55 °C for 30 seconds and 72 °C for 30 seconds.
Statistics
Data analysis utilized the following software: Microsoft Excel, Prism (Graphpad), SPSS and MedCalc (Mariakerke, Belgium), Multiexperiment Viewer (Dana-Farber Cancer Institute, Boston), BRB Array Tools (Biometric Research Branch, NCI), Prediction Analysis with Microarray (PAM), and the 7900HT Fast Real-Time PCR System Software SDS 2.1 (Applied Biosystems).
Expression values of each gene were expressed as mean ± standard deviation. To compare expression levels of the same gene between two groups of subjects, after verifying that the distributions of values were non-parametric, they were submitted to the Mann-Whitney U test. All p values in microarray and in PCR results were corrected by Bonferroni factor for multiple testing (17). The correction factors were 23,000 for microarray and 35 for Real-time PCR data. P values of <0.05 were considered as being statistically significant. In our study, genes which were statistically significant by Mann-Whitney U test also carried false discovery rates of less than 0.05.
Culture of cells with cytokines
U937 and Jurkat cells were cultured with cytokines at the optimum concentrations recommended by the manufacturers (18).
Results
Demographics of subjects studied
The demographics and clinical parameters of the first cohort of AS and USpA patients are shown in the first cohort columns Table 1. All AS and USpA patients complained of back pain, with only a very few also complaining of pain in the peripheral extremities. All AS and USpA patients were on NSAIDs and some on sulfasalazine. None were on other DMARDs, corticosteroids or biologics. The AS and USpA patients of the first cohort were recruited from two separate institutes. As control subjects, we recruited from the same institutes 13 male and 7 female subjects of mean age 35 ± 9.1 years.
Table 1.
Demographics and clinical parameters of first and second cohorts of SpA patients.
| First cohort | Second cohort | |||
|---|---|---|---|---|
| USpA | AS | USpA | AS | |
| Number of patients | 28 | 21 | 18 | 23 |
| Age (yr) | 26.4 ± 7.6 | 28.8 ± 8.7 | 39.6 ± 7.3 | 31.8 ± 6.9 |
| Gender | 16M/12F | 21M/0F | 7M/11F | 19M/4F |
| HLA-B27 | 26/28 | 21/21 | 21/22 | |
| Duration (months) | 48.8 ± 47.3 | 116.6 ± 78.2 | ||
| Patients with spinal pain | 28/28 | 21/21 | 18/18 | 23/23 |
| Patients with swollen joints or heels | 4 | 3 | 4 | 5 |
| Patients with hip involvement | 4/28 | 13/21 | ||
| Back pain score | 4.9 ± 1.9 | 6.4 ± 1.5 | 5.2 ± 2.8 | 6.4 ± 1.6 |
| Morning stiffness (minutes) | 17.5 ± 24.6 | 42.9 ± 37.9 | ||
| BAS-G | 5.4 ± 2.0 | 7.6 ± 1.6 | 5.1 ± 1.4 | 4.9 ± 2.1 |
| BASFI | 1.7 ± 1.4 | 7.6 ± 1.6 | 2.5 ± 1.4 | 3.2 ± 1.9 |
| BASDAI | 4.3 ± 1.5 | 5.8 ± 1.1 | 3.9 ± 1.8 | 5.4 ± 1.4 |
| ESR (mm/lst hr) | 22.3 ± 16.5 | 37.3 ± 23.9 | 39.6 ± 7.3 | 25.5 ± 19.8 |
| CRP (mg/l) | 7.2 ± 11.9 | 3.2 ± 2.4 | 1.7 ± 0.9 | 1.4 ± 1.3 |
| Tragus-to-wall (cm) | 10.5 ± 0.7 | 15.2 ± 7.3 | ||
| Occiput-to-wall (cm) | 1.9 ± 4.8 | |||
| Schober (cm) | 4.3 ± 0.9 | 3.3 ± 1.7 | 3.9 ± 2.7 | |
| Lateral flexion of lumbar spine (cm) | 18.5 ± 3.5 | 9.8 ± 5.4 | 12.0 ± 6.1 | |
| Chest expansion (cm) | 4.4 ± 1.1 | 2.1 ± 5.4 | 4.1 ± 1.7 | |
| X-rays with positive sacroiliitis | 0/28 | 21/21 | 0/18 | 23/23 |
Cells with no values indicate that those parameters have not been measured. Upper limit of normal of CRP was 0.8 mg/dl for first cohort and 0.3 mg/dl for second cohort.
All AS patients fulfilled the 1984 Modified New York criteria for AS (38). All USpA patients fulfilled the ESSG criteria for SpA, but did not have features for other subtypes of SpA and did not fulfill the classification criteria for AS (3, 39). BASDAI, BASFI and BAS-G are referenced in (40-42). Scores were on scales of 0 to 10.
A second cohort of AS and USpA patients was also collected from a third institute to validate the results of the first cohort. The demographics and clinical parameters of the second cohort of patients are also shown in Table 1. The medications they were taking were similar to the first cohort. From the same institute were recruited 26 healthy controls, 13 male and 13 female subjects, with age 27 ± 9 years. We also recruited a cohort of 12 female rheumatoid arthritis (RA) patients of age 43.3 ± 12.6 years. These patients fulfilled the 1988 classification criteria for RA (19). The mean RA DAS28 score was 4.8 ± 0.8 (20); ESR was 27.5 ± 15.7 mm/lst hr; CRP was 0.5 ± 0.4 mg/l. All RA patients were on NSAIDs and methotrexate. None of the patients in the either cohorts were on corticosteroids or biologics. Lastly, we also carefully selected 3 male and 5 female patients diagnosed as having chronic “mechanical low back pain”. Diagnosis was made jointly by an orthopedic and a rheumatology specialist. The mean age was 52 ± 13 years. The diagnosis was based on the clinical parameters as well as either MRI or nerve conduction studies (21).
An overview of gene expression patterns among AS, USpA and control groups
Hierarchical clustering was used to obtain an overview of gene expression among control, AS and USpA subjects. In the heat map shown in Figure 1, high expression genes were marked in red. The pattern of expression in the USpA group was globally distinct, with a relatively higher number of highly expressed genes compared to either the control or the AS groups.
Figure 1.
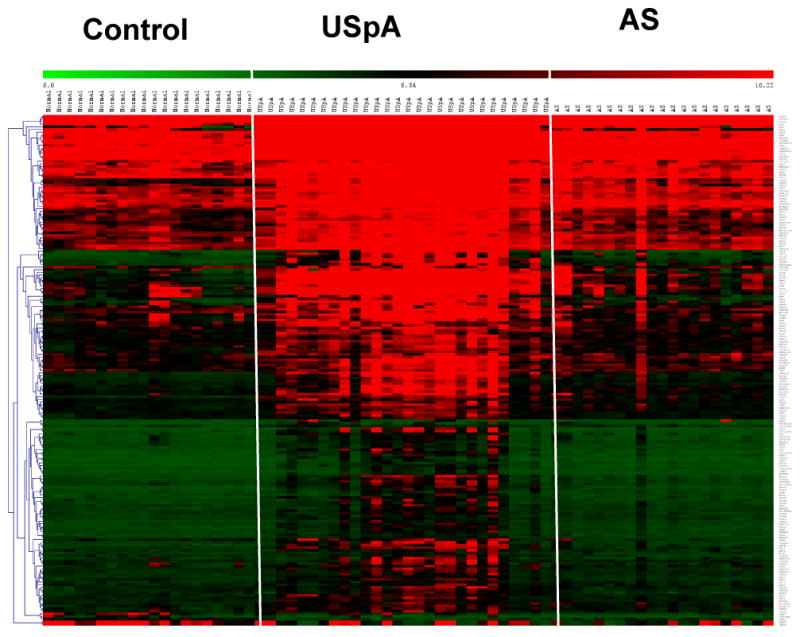
Heat map of microarray results. For clarity, the three groups of subjects were separated by white margins. The genes were hierarchically clustered by average group linkage. Each column represents one subject, and each row one gene. The red, green and black colors indicate high, low and mid expression levels respectively.
When the actual microarray values were analyzed, only 4 genes were significantly more highly expressed in AS compared to control subjects: RGS1, NR4A2, HBEGF and SOCS3. The fold-change in AS versus controls was subtle varying from 2.7 to 3.5 fold. Only two genes were under-expressed in AS: DEFA1 and CAMP. The fold-change of mean microarray values as well as statistical significance of these 6 differentially expressed genes is shown in Table 2. In Table 2, empty cells indicate that values corresponding to those cells did not reach statistical significance.
In contrast, in the USpA group, 38 genes were more than 2 fold over-expressed compared to the control group, all with corrected p values <0.05. The 4 genes over-expressed in AS were also over-expressed in USpA. In USpA, as many as 20 genes were more than 6-fold higher than healthy subjects. The list of these 20 most highly differentially expressed USpA genes and their p values are listed in the “microarray column” of Table 2. Nine of those genes appeared to be related to inflammatory processes: CXCL2, CCL20, CCL3L3, CCL3L1, CCL3, IL-1A, IL-1B, IL-8 and IL-6. Out of the 20,589 probes in the microarray, only two genes were exceptional in being over-expressed to more than 3-fold in both AS and USpA. They were RGS1 and NR4A2.
Verifying gene expression diagnostic potential using Real-time PCR
The commonly accepted “gold standard” of gene expression measurements is Real-time PCR. For validation by Real-time PCR, we selected from the microarray data the 6 genes which were differentially expressed in AS, and the 20 most highly expressed genes in USpA. In addition, IL-1RN, IL-6 and SOCS3 were also selected in spite of their lower statistical values, because of their frequently alleged potential in SpA (Table 2).
Genes differentially expressed in USpA but not selected for PCR were: CARD3, MTD118, CXCR4, IRAK3, CTL4, PLAUR, IL1RL2, MAPK6, COX-2, CXCL16, SOD2, CD278, ICAM1, SOCS1 and RGS2. Their microarray values were all less than 4.5 folds higher than control.
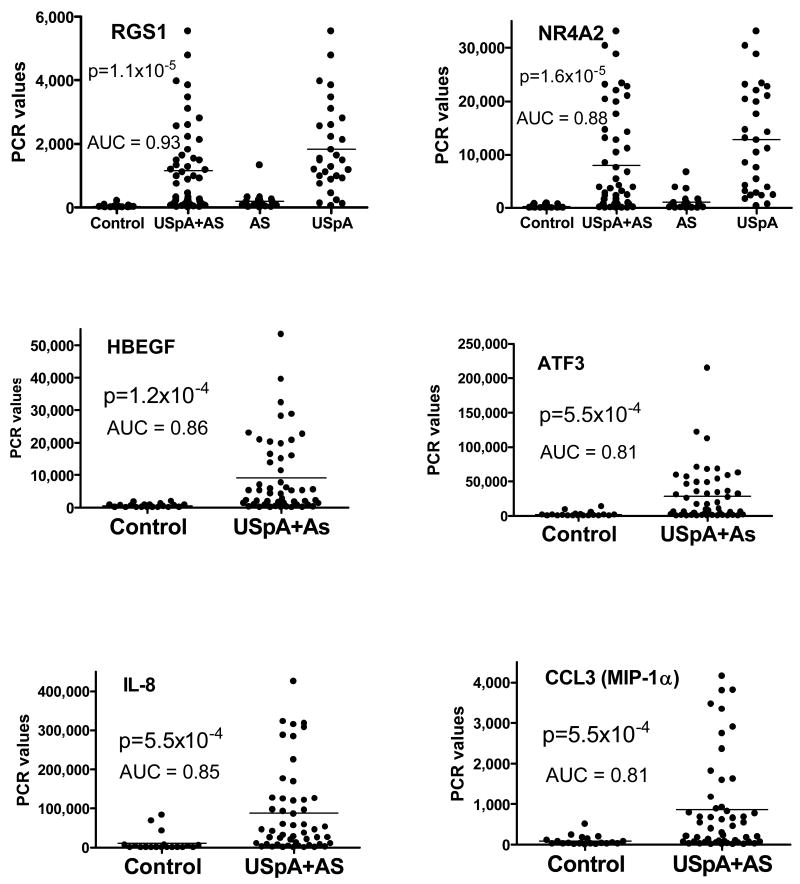
Confirming a high degree of accuracy in our microarray procedures, our results showed a high degree of correlation between the mean values of microarray and Real-time PCR data (r=0.75, p<1×10-4). Most of the genes differentially expressed in microarray were also differentially expressed in Real-time PCR. An exception was NR4A2, which was differentially expressed in AS by microarray but not by Real-time PCR. Also confirming the microarray results, other than CCL20, the mean values of all the highly expressed genes were statistically higher in USpA compared to AS (corrected p values = 1.6×10-2 – 7×10-7). Mean values of all Real-time PCR and their degrees of statistical significance are shown in the “Real-time PCR results column” in Table 2. For both USpA and AS, the gene with the highest degree of statistical significance was RGS1 (p= 2.3×10 -7 and 6.7×10-3 respectively). We also ranked the diagnostic potential of each gene by their ROC AUC. RGS1 was also of the highest diagnostic potential with ROC AUC for USpA and AS being 0.99 (95% CI 0.91-0.99) and 0.84 (95% CI 0.69-0.94) respectively (p<1×10-4) (Figure 2).
Figure 2.
Real-time PCR values of genes differentially expressed between the AS and USpA and control subjects in the first cohort. ROC AUC and p values are in comparison between the combined AS and USpA group and the control subjects. Each dot represents result from one individual. Bars indicate the mean values. For RGS1 and NR4A2, the AS and the USpA groups are also shown separately.
To parallel what is commonly practiced in patient care in trying to distinguish inflammatory from mechanical low back pain, we combined the USpA and AS into one group designated as “USpA+AS”. The diagnostic potential of all the genes in diagnosing inflammatory low back pain were then ranked by their ROC AUC values. When these calculations were applied to our combined USpA and AS group, only 6 genes showed diagnostic potential. The values of individual subjects are shown in Figure 2. Of these RGS1 still showed the highest promise as a diagnostic biomarker with a very high ROC AUC value of >0.9 (p<1×10-4).
The PCR primers used to measure RGS1 in Figure 2 were RGS1 primer pair #1 which amplified a segment spanning exons 3 and 4 of the RGS1 gene. To ensure that the PCR values in Figure 2 were not artifacts of this particular pair of primers, the samples of AS, USpA and control subjects were assayed again with two different pairs of primers. One pair, RGS1 primer pair #2 amplified a segment overlapping with that of the RGS1 primer pair #1. The third pair, RGS1 primer pair #3 amplified a segment spanning exons 4 and 5 of the RGS1 gene. The correlation coefficient between values derived from the RGS1 primer pairs #1 and #2 was high at 0.88 (p=1×10-6). That for values derived from primer pairs #1 and #3 was also high at 0.89 (p=1×10-6)
RGS1 is a member of the RGS family. To ensure that the results of RGS1 in USpA were unique to the RGS1 member alone, we measured the expression of the following homologues of RGS1 with our USpA and control samples: RGS -2, -3, -4, -5, and -8. Transcripts of RGS -4, -5 and -8 were undetectable. Expression of RGS -2 and -3 in USpA patients were not different from control subjects (p>0.05). These results confirmed the specificity of RGS1 in the RGS family.
Using correlation matrix to test if gene expression correlated with clinical parameters
The PCR values of all genes shown in Table 2 were higher with the USpA patients compared to the AS patients (p<1×10-4). The relative expression was not due to increased disease activity in USpA relative to AS patients. As a group, these USpA patients were different from the AS patients in having lower values of BASDAI, BASFI and BAS-G (all p<1×10-4), shorter duration of disease (p<3×10-4), better chest expansion and lateral flexion of the lumbar spine, and less radiological sacroiliitis (all p<1×10-4). A matrix correlation analysis showed that there was no positive correlation of RGS1 or any other genes with ESR, CRP, or disease activity indices or peripheral arthritis in either AS or USpA.
Validation testing of candidate biomarkers in an independent cohort
We then collected an independent cohort of AS and USpA patients and healthy control subjects from a different geographical area (Table 1). Patient diagnosis was carried out by different rheumatologists. RNA was extracted in a Real-time PCR laboratory instead of the microarray laboratory. Samples were assayed at least 3-6 months after completion of the study of the first cohort. Also included in this cohort was a group of patients with RA, and a carefully selected group of patients with mechanical low back pain (see section on patient demographics).
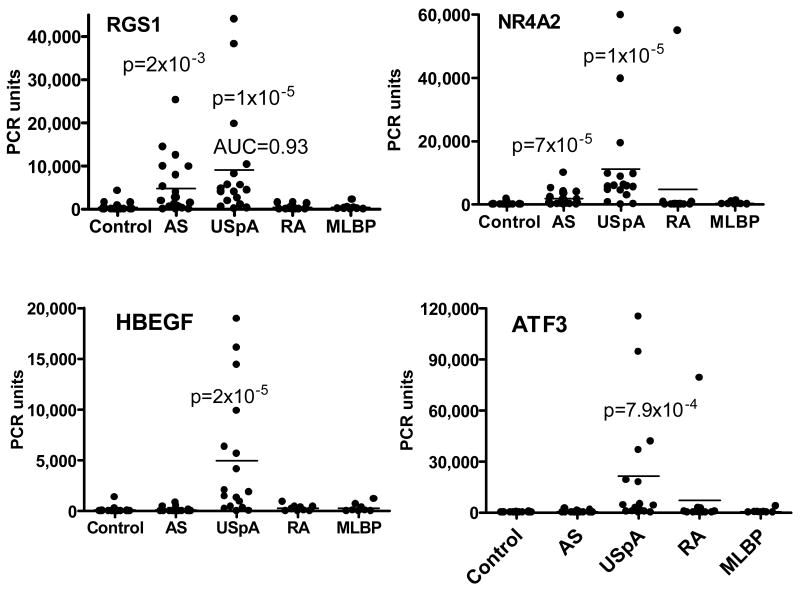
Samples from this second cohort were tested for Real-time PCR expression of those genes shown in Figure 2, because those genes showed the highest diagnostic potential in the first cohort. To avoid fortuitous omissions, we also tested several genes of only borderline significance in the first cohort. These were CAMP, DEFA1, CXCL2, IL-1α and IL-6. In this second cohort, no differences were detected between either the USpA or the AS groups compared to the control groups for IL-1α, IL-6, CCL3, CAMP and DEFA1. IL-8 and CXCL2 were of poor diagnostic values with ROC AUC <0.6 (Results not shown). As in the first cohort, NR4A2, ATF3 and HBEGF were 6 – 40 folds higher in the USpA group compared to the AS group (P= 5×10-4 – 1×10 -5) (Figure 3).
Figure 3.
Real-time PCR values of genes differentially expressed between the patients and control subjects in the second cohort. ROC AUC and p values are in comparison to healthy control subjects. Each dot represents result from one individual. MLBP = mechanical low back pain. When patients with mechanical low back pain were compared to control subjects, there was no difference in any of the genes tested with the exception of a borderline increase in CXCXL2 (p=0.05).
As in the first cohort, the most useful candidate biomarkers appeared to be RGS1. The mean values of RGS1 in the USpA and AS patients were 20.9 and 11.1 fold higher than those of healthy subjects (p=4×10-6 and 2×10-4, respectively), and were also higher in comparison to both the RA and the mechanical low back pain groups (p= 1×10-2 to 1×10-4). Levels of RGS1 in the latter two groups were not different from those in healthy subjects. Individual PCR values are shown in Figure 3. Even more helpful as a biomarker, for this second cohort, RGS1 appeared to be equally highly expressed in both USpA and AS patients. There were no statistically significant differences between USpA and AS in their mean PCR values or their ROC AUC values. This was in contrast to the first cohort, in which RGS1 was more useful for USpA compared to AS. We also pooled the results of AS and USpA into a single combined group. When this entire group of combined AS and USpA was compared to the mechanical low back pain group, the following biomarker values were observed for RGS1: ROC AUC 0.86, sensitivity 84.2, specificity 87.5, +LR 6.7, PPV 97 and NPV 53.8. Threshold values for these calculations were based on automatic selection by the Medcalc software. The predictive values were based on the assumption that the prevalence of AS and USpA in patients of chronic low back pain was 5.0% (5). The prevalence of SpA among the Chinese population has been reported to be 0.93% (22). When the values of SpA were compared to the general population based on that particular prevalence, the diagnostic values were similar to those for distinguishing from mechanical low back pain.
The RGS1 values shown in Figure 3 were derived from the PCR RGS1 primer pair #1. We also measured the control and the SpA samples with RGS1 primer pair #3. There is a high degree of correlation between results derived from the two different pairs of primers (r=0.85, p=1×10-6).
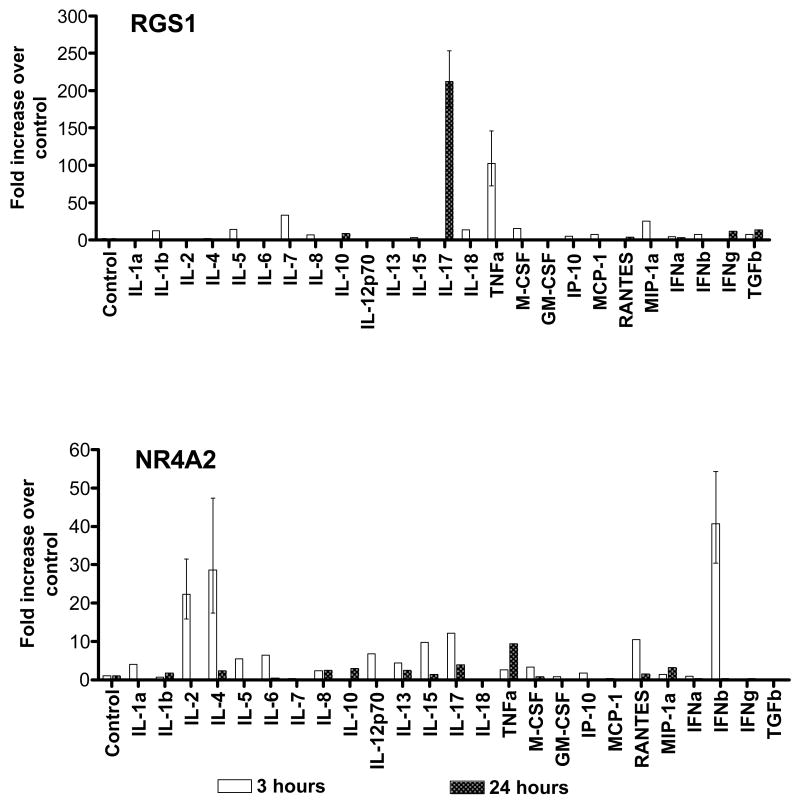
Evaluating cytokine control of RGS1
U937 cells were separately cultured with a panel of 25 cytokines and chemokines, and the expression of RGS1 was assayed 3 and 24 hours later. RGS1 transcription was strikingly induced by culture with TNF-α as well as IL-17 (Figure 4 upper panel). As a control for specificity, we also assayed for NR4A2 in the same samples. In contrast to RGS1, NR4A2 was induced by multiple other cytokines (Figure 4 lower panel). As negative control, we also assayed for RGS1 in Jurkat cells. Similar to a report published previously, RGS1 was undetectable in Jurkat cells (23).
Figure 4.
Effect of cytokines and chemokines on expression of RGS1 and NR4A2 in U937 cells. Error bars represent range of values. Error bars are provided only for those highly induced genes.
Discussion
There is a large degree of uncertainty among many clinicians as how to diagnose early AS. That is probably why the diagnosis of AS is frequently delayed for several years (24). Part of the reason is because unlike diseases such as RA, the blood-derived biomarkers currently used for diagnosing AS in clinical practice have low sensitivities and specificities. Numerous papers have been published in attempts to identify more useful blood-derived biomarkers for AS such as MMP3 (10). The present paper is distinct in several regards. First, we based our search on a preliminary screening for differentially expressed genes with genome-wide microarrays. In our analysis, gene selection was driven by statistical significance avoiding the biases of biology-based candidate gene selection approaches. Second, for the microarray study, we tested AS as well as USpA patients within the same study. These patients were recruited by several different investigators, but adhering to the same classification criteria. Third, we verified our microarray results with Real-time PCR. Fourth, we validated the reliability of the most promising candidate RGS1 by comparing its results to homologous members of the same family of genes. Fifth, for the most promising candidate, we validated the results of the first PCR assays by comparing them to two completely different set of PCR primers. The use of these three pairs of primers also suggested that the particular isoform of RGS1 which was expressed in our cohorts was Sp1 (EMBL-EBI Alternative Splicing Database). Sixth, we retested the most promising candidates in an independent cohort of AS and USpA patients and healthy controls, which were recruited by a different clinical investigator from a different geographical location, and also with RNA purified at a different laboratory. Seventh, we compared the AS and USpA results to a cohort of RA patients, and also to a very carefully selected cohort of mechanical low back pain patients.
With each successive experimental step, false candidates were eliminated. Using these procedures, we arrived at three conclusions. The first conclusion is that overall gene expression is higher in USpA compared to AS patients, suggesting that early axial SpA is associated with a more systemic inflammatory process. This is an advantage to clinicians because biomarkers are more in demand in the early stage of SpA rather than the late stage. The second conclusion is that the most promising candidate for both USpA and AS is RGS1. The third conclusion is that, like the other genes observed in this study, RGS1 is probably more highly expressed in USpA compared to AS.
The discovery of RGS1 as an AS and USpA biomarker is unanticipated. Since the expression of RGS1 in our first cohort was much lower in AS compared to USpA, our discovery would have been unlikely without genome-wide screening of both AS and USpA patients. RGS1 is a member of a family of “Regulators of G-protein signaling” (RGS) proteins. These were initially characterized as inhibitors of signaling cascades initiated by G-protein-coupled receptors (GPCRs). RGS proteins are so designated because they can down regulate the signaling initiated by engagement of ligands with GPCRs (25, 26). However, it is now understood that in addition to G proteins RGS proteins also interact with a large network of lipids, ions and other proteins, suggesting that RGS proteins have a diverse array of regulatory functions (27). Among the various RGS proteins, RGS1 is expressed mainly in hematopoetic cells. There is only limited information on the biology RGS1 (28-30). It is not clear from the limited information which particular RGS1 activity is related to SpA disease processes. Nevertheless, although the exact biology of RGS1 is still uncertain, it has already been discovered by random SNP screening to be candidate genes for both Type 1 diabetes and Celiac disease (31, 32), and an independent prognostic marker of disease survival in melanomas (33).
Little is known about the arthritis-related factors which might enhance RGS1 expression (29). We tested a panel of 25 cytokines and chemokines on a monocyte-derived human cell line, and discovered that the two strongest activators of RGS1 expression were TNF-α and IL-17. The kinetics of expression of TNFα and IL-17 were quite different, perhaps reflecting differences in pathways of activation similar to what we observed with HLA-B27 gene (18). Perhaps not fortuitous, TNF-α and IL-17 are currently being considered as key modulators of SpA processes (34, 35).
In conclusion, our genome-wide profiling has identified RGS1 as a candidate biomarker especially with USpA. For any new candidate biomarkers to be used in clinical practice, the candidates will have to undergo the much more vigorous processes of multicenter multi-ethnicity standardization and testing, with well-characterized healthy subjects and also other disease groups (36). Comparison to MRI changes of the sacroiliac joints and the most recent classification criteria for early axial spondyloarthritis will also be essential (37). Our present results serve to contribute a candidate which can be considered for such selection processes. In addition, they might contribute to future discoveries of other more clinical laboratory – friendly and disease-relevant biomarkers, useful in both diagnosis and disease management.
Acknowledgments
The authors gratefully acknowledge Dr. Chin-Hsiu Liu (Taiwan) for assisting in the Real-time PCR experiments, and Ms Shu-Yuan Chang for collection of clinical data.
Grant supporters: J Gu: grants (30325019 and 30872328) from the National Natural Sciences Foundation of China, clinical subject (2007) of Ministry of public health of China and grants (2002Z3-E4021 and 2005A30801005) from Foundation of Guangzhou and Guangdong province of China; YL Wei and D Yu: Nora Eccles Treadwell Foundation; JCC Wei and MS Jan: National Science Council, Taiwan, NSC 93-2320-B-040-030; F Huang: grant A30771983 from the National Natural Sciences Foundation of China. M.B. Frank:P20 RR016478-077281, P20 RR016478-087197, P20 RR016478-096180, P20 RR020143-047652, U19 AI062629-040006, P20 RR015577-086790 and RR015577-096179 from NIH.
Footnotes
Author Contribution: Dr. Yu had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study design. Yu, Frank, Centola.
Acquisition of data: Gu, Huang, Wei, Wei, Jan, Frank
Analysis and interpretation of data: Yu, Frank, Wei.
Manuscript preparation: Yu, Frank, Centola, Wei, Gu.
Statistical analysis: Yu, Frank, Wei.
References
- 1.Sieper J, Rudwaleit M, Khan MA, Braun J. Concepts and epidemiology of spondyloarthritis. Best Pract Res Clin Rheumatol. 2006;20(3):401–17. doi: 10.1016/j.berh.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Collantes E, Zarco P, Munoz E, Juanola X, Mulero J, Fernandez-Sueiro JL, et al. Disease pattern of spondyloarthropathies in Spain: description of the first national registry (REGISPONSER) extended report. Rheumatology (Oxford) 2007;46(8):1309–15. doi: 10.1093/rheumatology/kem084. [DOI] [PubMed] [Google Scholar]
- 3.Zochling J, Brandt J, Braun J. The current concept of spondyloarthritis with special emphasis on undifferentiated spondyloarthritis. Rheumatology (Oxford) 2005;44(12):1483–91. doi: 10.1093/rheumatology/kei047. [DOI] [PubMed] [Google Scholar]
- 4.Licciardone JC. The epidemiology and medical management of low back pain during ambulatory medical care visits in the United States. Osteopath Med Prim Care. 2008;2:11. doi: 10.1186/1750-4732-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underwood MR, Dawes P. Inflammatory back pain in primary care. Br J Rheumatol. 1995;34(11):1074–7. doi: 10.1093/rheumatology/34.11.1074. [DOI] [PubMed] [Google Scholar]
- 6.Calin A, Porta J, Fries JF, Schurman DJ. Clinical history as a screening test for ankylosing spondylitis. JAMA. 1977;237(24):2613–4. [PubMed] [Google Scholar]
- 7.Rudwaleit M, Metter A, Listing J, Sieper J, Braun J. Inflammatory back pain in ankylosing spondylitis: a reassessment of the clinical history for application as classification and diagnostic criteria. Arthritis Rheum. 2006;54(2):569–78. doi: 10.1002/art.21619. [DOI] [PubMed] [Google Scholar]
- 8.Rudwaleit M, Feldtkeller E, Sieper J. Easy assessment of axial spondyloarthritis (early ankylosing spondylitis) at the bedside. Ann Rheum Dis. 2006;65(9):1251–2. doi: 10.1136/ard.2005.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudwaleit M, Haibel H, Baraliakos X, Listing J, Marker-Hermann E, Zeidler H, et al. The early disease stage in axial spondylarthritis: results from the German Spondyloarthritis Inception Cohort. Arthritis Rheum. 2009;60(3):717–27. doi: 10.1002/art.24483. [DOI] [PubMed] [Google Scholar]
- 10.Chen CH, Yu DTY, Chou CT. Biomarkers in spondyloarthropathies. In: Lopez-Larrea C, editor. Molecular Mechanisms of spondyloarthropathies. Austin, Texas: Landes Bioscience; 2008. [Google Scholar]
- 11.Brown MA. Human leucocyte antigen-B27 and ankylosing spondylitis. Intern Med J. 2007;37(11):739–40. doi: 10.1111/j.1445-5994.2007.01491.x. [DOI] [PubMed] [Google Scholar]
- 12.Bennett AN, McGonagle D, O'Connor P, Hensor EM, Sivera F, Coates LC, et al. Severity of baseline magnetic resonance imaging-evident sacroiliitis and HLA-B27 status in early inflammatory back pain predict radiographically evident ankylosing spondylitis at eight years. Arthritis Rheum. 2008;58(11):3413–8. doi: 10.1002/art.24024. [DOI] [PubMed] [Google Scholar]
- 13.Rao G. What is an ROC curve? J Fam Pract. 2003;52(9):695. [PubMed] [Google Scholar]
- 14.Denecke B, Meyerdierks A, Bottger EC. RGS1 is expressed in monocytes and acts as a GTPase-activating protein for G-protein-coupled chemoattractant receptors. J Biol Chem. 1999;274(38):26860–8. doi: 10.1074/jbc.274.38.26860. [DOI] [PubMed] [Google Scholar]
- 15.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 16.Liu PT, Krutzik SR, Kim J, Modlin RL. Cutting edge: all-trans retinoic acid down-regulates TLR2 expression and function. J Immunol. 2005;174(5):2467–70. doi: 10.4049/jimmunol.174.5.2467. [DOI] [PubMed] [Google Scholar]
- 17.Verhoeven KJF, Simonsen K, McIntyre LM. Implemeting false dicovery rate method will generate less false negatives but more false positives. Oikos. 2005;108:643–64. [Google Scholar]
- 18.Zhao L, Fong Y, Granfors K, Gu J, Yu D. Identification of cytokines that might enhance the promoter activity of HLA-B27. J Rheumatol. 2008;35(5):862–8. [PubMed] [Google Scholar]
- 19.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 20.Prevoo ML, van't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38(1):44–8. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 21.Vroomen PC, de Krom MC, Wilmink JT, Kester AD, Knottnerus JA. Diagnostic value of history and physical examination in patients suspected of lumbosacral nerve root compression. J Neurol Neurosurg Psychiatry. 2002;72(5):630–4. doi: 10.1136/jnnp.72.5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng SC, Liao Z, Yu DT, Chan ES, Zhao L, Gu J. Epidemiology of spondyloarthritis in the People's Republic of China: review of the literature and commentary. Semin Arthritis Rheum. 2007;37(1):39–47. doi: 10.1016/j.semarthrit.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Moratz C, Kang VH, Druey KM, Shi CS, Scheschonka A, Murphy PM, et al. Regulator of G protein signaling 1 (RGS1) markedly impairs Gi alpha signaling responses of B lymphocytes. J Immunol. 2000;164(4):1829–38. doi: 10.4049/jimmunol.164.4.1829. [DOI] [PubMed] [Google Scholar]
- 24.Dincer U, Cakar E, Kiralp MZ, Dursun H. Diagnosis delay in patients with ankylosing spondylitis: possible reasons and proposals for new diagnostic criteria. Clin Rheumatol. 2008;27(4):457–62. doi: 10.1007/s10067-007-0727-6. [DOI] [PubMed] [Google Scholar]
- 25.De Vries L, Gist Farquhar M. RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol. 1999;9(4):138–44. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 26.Hendriks-Balk MC, Peters SL, Michel MC, Alewijnse AE. Regulation of G protein-coupled receptor signalling: focus on the cardiovascular system and regulator of G protein signalling proteins. Eur J Pharmacol. 2008;585(2-3):278–91. doi: 10.1016/j.ejphar.2008.02.088. [DOI] [PubMed] [Google Scholar]
- 27.Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116(3):473–95. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lattin J, Zidar DA, Schroder K, Kellie S, Hume DA, Sweet MJ. G-protein-coupled receptor expression, function, and signaling in macrophages. J Leukoc Biol. 2007;82(1):16–32. doi: 10.1189/jlb.0107051. [DOI] [PubMed] [Google Scholar]
- 29.Riekenberg S, Farhat K, Debarry J, Heine H, Jung G, Wiesmuller KH, et al. Regulators of G-protein signalling are modulated by bacterial lipopeptides and lipopolysaccharide. Febs J. 2009;276(3):649–59. doi: 10.1111/j.1742-4658.2008.06813.x. [DOI] [PubMed] [Google Scholar]
- 30.Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH. Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: implications for G protein-coupled receptor signaling. J Immunol. 2004;172(9):5175–84. doi: 10.4049/jimmunol.172.9.5175. [DOI] [PubMed] [Google Scholar]
- 31.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–77. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangel J, Nosrati M, Leong SP, Haqq C, Miller JR, 3rd, Sagebiel RW, et al. Novel role for RGS1 in melanoma progression. Am J Surg Pathol. 2008;32(8):1207–12. doi: 10.1097/PAS.0b013e31816fd53c. [DOI] [PubMed] [Google Scholar]
- 34.Braun J, Sieper J. Ankylosing spondylitis. Lancet. 2007;369(9570):1379–90. doi: 10.1016/S0140-6736(07)60635-7. [DOI] [PubMed] [Google Scholar]
- 35.Jandus C, Bioley G, Rivals JP, Dudler J, Speiser D, Romero P. Increased numbers of circulating polyfunctional Th17 memory cells in patients with seronegative spondylarthritides. Arthritis Rheum. 2008;58(8):2307–17. doi: 10.1002/art.23655. [DOI] [PubMed] [Google Scholar]
- 36.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6(7):565–71. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 37.Rudwaleit M, van der Heijde D, Landewe R, Listing J, Akkoc N, Brandt J, et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis. 2009;68(6):777–83. doi: 10.1136/ard.2009.108233. [DOI] [PubMed] [Google Scholar]
- 38.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27(4):361–8. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 39.Dougados M, van der Linden S, Juhlin R, Huitfeldt B, Amor B, Calin A, et al. The European Spondylarthropathy Study Group preliminary criteria for the classification of spondylarthropathy. Arthritis Rheum. 1991;34(10):1218–27. doi: 10.1002/art.1780341003. [DOI] [PubMed] [Google Scholar]
- 40.Calin A, Garrett S, Whitelock H, Kennedy LG, O'Hea J, Mallorie P, et al. A new approach to defining functional ability in ankylosing spondylitis: the development of the Bath Ankylosing Spondylitis Functional Index. J Rheumatol. 1994;21(12):2281–5. [PubMed] [Google Scholar]
- 41.Garrett S, Jenkinson T, Kennedy LG, Whitelock H, Gaisford P, Calin A. A new approach to defining disease status in ankylosing spondylitis: the Bath Ankylosing Spondylitis Disease Activity Index. J Rheumatol. 1994;21(12):2286–91. [PubMed] [Google Scholar]
- 42.Jones SD, Steiner A, Garrett SL, Calin A. The Bath Ankylosing Spondylitis Patient Global Score (BAS-G) Br J Rheumatol. 1996;35(1):66–71. doi: 10.1093/rheumatology/35.1.66. [DOI] [PubMed] [Google Scholar]