Abstract
Background
A novel gene, rat pHyde, has been cloned by us recently. The rat pHyde was shown by the same group to have growth inhibitory effects on human prostate cancer through the induction of apoptosis.
Methods
In this report, a human homologue, hpHyde of the rat pHyde, was cloned by cDNA libraries screening. The database search and in situ hybridization were used to map the genomic loci of hpHyde in human chromosome. The anti-prostate cancer effects of pHyde in conjunction with chemotherapy agent were analyzed by in vitro and in vivo assays using adenoviral vector expressing pHyde (AdRSVpHyde) in combination with DNA damaging chemotherapeutic agent, cisplatin, and docetaxel, respectively.
Results
Database search and FISH analysis consistently indicated that hpHyde gene localizes at human chromosome 2q14. Protein sequence analysis suggests that hpHyde may be a plasma membrane protein. hpHyde is differentially expressed in various normal human tissues and organs, suggesting that hpHyde may play roles in development and differentiation. Growth suppression and induction of apoptosis were additively greater in DU145 human prostate cancer cells treated with AdRSVpHyde and cisplatin than either agent alone both in vitro and in vivo. Moreover, AdRSVpHyde and docetaxel also have a similar additively inhibitory effect on DU145 cell growth.
Conclusions
A novel gene hpHyde, the human homologue of rat pHyde, has been cloned and its genomic location in the human chromosome has been identified. Our results support the potential use of pHyde for prostate cancer gene therapy coupled with chemotherapy to improve therapeutic index.
Keywords: pHyde, cisplatin, apoptosis, prostate cancer, adenovirus
Introduction
Prostate cancer is the most frequently diagnosed malignancy and the second leading cause of cancer deaths in American men today with an estimated 218,890 new cases of prostate cancer and 27,050 deaths estimated in year 2007.1 Like many carcinomas, prostate cancer formation is a multi-step process involving tumor initiation, promotion, transformation, and progression.2,3 This process is driven by multiple factors including chromosomal instability, spontaneous mutations, and carcinogen-induced genetic and epigenetic changes. A better understanding of the molecular mechanisms responsible for prostate cancer may ultimately lead to new effective therapies. Therefore, identification and characterization of the critical genes that are associated with and involved in prostate carcinogenesis, are important. Particularly, identification and characterization of novel genes that successfully inhibit or destroy prostate cancer cells may have potential utility in clinical therapeutic treatment.
A novel gene, rat pHyde, was cloned using a cDNA competition hybridization technique from two cDNA libraries derived from the metastatic (AT-3) and the nonmetastatic (AT-1) rat prostatic cancer cell lines. Our previous study has shown that rat pHyde is able to suppress human prostate cancer through induction of apoptosis.4 In this study, the human homologue of rat pHyde gene, the human pHyde gene (hpHyde), has been isolated and the sequencing has been completed. We have determined the chromosomal localization of hpHyde genomic gene, predicted the physiological function of hpHyde protein based on protein structure analysis, and assessed differential expression of hpHyde gene in various organs and tissues. To further investigate whether pHyde prostate cancer gene therapy can be used to improve clinical outcome of prostate cancer chemotherapy, we examined the potential additive effects of adenoviral-mediated pHyde gene expression coupled with chemotherapeutic agents, cisplatin or docetaxel, respectively, in human prostate cancer in vitro and in the nude mice models.
Materials and Methods
Cloning and DNA sequencing
A full-length cDNA gene of human homologue of rat pHyde gene, the human pHyde gene (hpHyde), was isolated by screening human prostate cDNA libraries (Invitrogen, Carlsbad, CA) using rat pHyde cDNA4 as a probe by following the methods as described previously.5 The sequencing of hpHyde cDNA gene was carried out in the Molecular Resource Center of The University of Tennessee Health Science Center. DNA sequencing was performed using Big Dye Reaction Mix (Applied Biosystems, Foster City, CA) at 1/2 X strength with 500 ng of double-stranded plasmid and 3.2 pmol of the relevant primer in a total volume of 20 μL. The reactions were passed through Centi-Sep 8 columns (Princeton Separations, Adelphia, NJ) to remove the unincorporated nucleotides and other reaction components, dried, and resuspended in 12 μL of formamide. Following a 5 min denaturation at 95 °C, the extension products were analyzed on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequencing of full-length of hpHyde cDNA gene was carried out by the “sequencing walk-through” strategy; that is, those sequences of hpHyde cDNA determined by initial sequencing were used to choose and design the new primers to determine the unknown region of the gene. The complete cDNA sequence was confirmed by a twice “sequencing walk-through” using two different sets of primers from both directions.
Database search
The sequence for the hpHyde cDNA (1884 nucleotides, GenBank accession number AY082673) was submitted to BLAST against nr (non-redundant) database. The new BLAST Human Genome facility was used to visualize in silico chromosome localizations of the hpHyde gene, as well as identify other genes with partial sequence homology with hpHyde, which would elucidate the function of hpHyde gene product. As a rule of thumb, the scores above 250 and expect value (e-value) of 0 (or as close as 0) were considered highly significant sequence similarity.6 Predicted genes with significant hits of high similarity were analyzed by a confirmatory pairwise BLAST of the hpHyde protein sequence with the predicted gene.6 If the result showed a 100% identity, it indicates that the predicted gene (annotated by Human Genome Project with unknown function) was presumably the hpHyde genomic gene. The chromosomal location, e.g., contig and megabase from centromere or telomere, of the predicted gene was identified. This information would also be depicted using the Human Genome BLAST facility with an expect value of 0.1, which utilizes the MapViewer resource6 to graphically delineate the regions of similarity between the hpHyde cDNA sequence and the contig backbone, including a clear suggestion of the possible intron/exon structure. Furthermore, using an expect value of 0.01, other similar genes of interest would be identified.
Fluorescent in situ hybridization (FISH)
To confirm the in silico prediction results, FISH analysis was used to confirm the physical chromosomal localization of hpHyde gene. First, by using hpHyde cDNA as a probe to screen RPCI-11 human BAC genomic libraries (Roswell Park Cancer Institute, Buffalo, NY), a matched clone, BAC RPCI-11-17N4, was identified. A genomic sequence of hpHyde was then amplified by PCR from that clone using primers specific to hpHyde cDNA sequences. The resulted PCR product was labeled with biotin (by standard nick-translation incorporation of biotin-14-dATP hapten, Gibco BRL, Gaithersburg, MD) and in situ hybridized with normal human metaphase chromosome spread, which was prepared as following: cytogenetic slides with metaphase chromosomes were prepared as previously described7 from normal male lymphoblasts using a 1.5 hour colcemid treatment followed by 75 mM KCl hypotonic treatment. Hybridization of the biotin-labeled probe to the metaphase targets was allowed to proceed overnight. After washing, the FISH signals were detected with fluorescein isothiocyanate (FITC). DAPI (4′,6-diamidino-2-phenylinidole)-stained images and inverted DAPI-stained images were both used to make it easier to locate FISH signals and delineate the chromosomes better, respectively.
Protein structure analysis
hpHyde protein sequence was analyzed by multiple database for functional domains/motifs, sorting signal and structure features, including Pubmed (http://www.ncbi.nlm.nih.gov/), Prosite (http://www.expasy.ch/sport/prosite.html), pfam (http://genome.wust.edu/Pfam/ and http://www.sanger.ac.uk/Pfam/), Psort (http://psort.nibb.ac.jp/), SOSUI transmembrane protein prediction site (http://sosui.proteome.bio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_submit.html), and PredictProtein (http://www.embl-heidelberg.de/predictprotein/).
Cell lines and tissue culture conditions
Human prostate cancer cell line DU145 was purchased from American Type Culture Collection (ATCC, Rockville, MD) and grown in RPMI-1640 medium (Cellgro, Herndon, VA) containing 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) at 37°C and 5% CO2. Human embryonic kidney cell line 293 was purchased from ATCC and grown in D-MEM medium (Cellgro) containing 10% heat inactivated fetal bovine serum at 37°C and 5% CO2.
Viral construction, amplification, titration and transduction
The generation of adenoviral vector expressing pHyde (AdRSVpHyde)4 and adenoviral vector expressing bacterial ß-galactosidase (AdRSVlacZ)8 was previously described. Viral vectors were propagated in 293 cells and were purified by twice CsCl2 gradient ultracentrifugation. The viral titration and transduction were performed by methods described by Graham and Prevec.9
Generation of antibody to rat pHyde protein
The rabbit anti-rat pHyde antibody was generated (custom-made) by Research Genetics, Inc. (Huntsville, AL). Briefly, a synthetic peptide, that consists of 17 amino acids (N terminal-NFIRDVLQPYIRKDENK-C terminal) corresponding to an antigenic region of the pHyde protein sequence deduced from rat pHyde cDNA sequence in the open reading frame, was coupled with a carrier protein (Keyhole hemolymbet) and used as the immunogen to raise rabbit polyclonal antibody against rat pHyde protein.
Cell-surface immunofluorescence staining for pHyde
Cells were grown on coverslips and either untreated or transduced with virus (AdRSVpHyde or AdRSVlacZ) at multiplicity of infection (moi) of 200 for 48 h. The cells were fixed with 3.5% formaldehyde in PBS for 30 min at room temperature. After 3 times washing with PBS, the cells were processed to immunofluorescence staining under non-permeabilization condition as described previously.10 The cells were incubated with 10% normal goat serum (NGS) in PBS for 20 min for blocking, then with 4 μg/ml rabbit anti-pHyde antibody in 10% NGS in PBS for 1 h at room temperature, followed by with 5μg/ml FITC-labeled goat anti-rabbit IgG for 1 h at room temperature. After washing and fixing, the coverslips were mounted onto the slides and photographed under a fluorescence microscopy.
Northern blot
A premade Northern blot that contains 1 μg of poly A+ RNA per lane from various normal human tissues was purchased from Clontech (Palo Alto, CA). The hpHyde cDNA probe was labeled by α-32P-dCTP using random primer method (Prime-It II Kit, Stratagene, La Jolla, CA). The blot was hybridized with the probe in Rapid-hyb buffer (Amersham Life Science) according to the manufacturer’s protocol. After washing, the blot was exposed to a Kodak X-ray film under intensifying screen at −80°C for autoradiography. ß-actin cDNA probe was labeled as described above and used as an internal control for RNA integrity and normalization of RNA loading.
In vitro growth inhibition assay
DU145 cells were incubated with or without chemotherapeutic agent (0.1 μM cisplatin or 0.5 nM docetaxel, both from Sigma, St Louis, MO) for 24 hours. Cells were then infected with AdRSVpHyde at a multiplicity of infection (moi) of 100. After viral infection, cells were continued to be incubated at 37°C with or without chemotherapeutic agent. The viability of cells were determined by either counting cell number or MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay at day 4 following viral infection. Untreated cells and cells treated with control virus AdRSVlacZ were used as controls.
In vivo growth inhibition assay
DU145 cells (1.4×107 cells in 0.2 ml of PBS) were injected subcutaneously into the flanks of male nude mice (6-8 week old, Harlan Sprague Dawley, Indianapolis, IN). Mice were divided into four groups when the tumors reached an average volume of 100 mm3: Group 1-untreated control, mice were untreated, no virus and no cisplatin. Group 2-AdRSVpHyde only, mice were injected with one single dose of 5×109 plaque forming units (pfu) AdRSVpHyde directly into the tumors. Group 3-cisplatin only, mice were injected intraperitoneally cisplatin (1.5 mg/kg of body weight) for four consecutive days. Group 4-AdRSVpHyde and cisplatin, mice were injected with one single dose of 5×109 pfu AdRSVpHyde directly into the tumors, and were injected intraperitoneally cisplatin (1.5 mg/kg of body weight) for four consecutive days starting on the same day of viral injection. Tumor volume was measured every three days until the animals were euthanized. All the animals were sacrificed at day 27 after viral injection when some of them showed tumor burdens greater than 15% of total body weight.
TUNEL staining of tumor sections
The details of this staining technique was as previously described.4 Briefly, xenograft DU145 tumors were harvested at necropsy (27 days after viral injection), fixed, and paraffin-embedded. The tumor sections were mounted on Superfrost Plus glass slides (Fisher, Scientific, Pittsburgh, PA), deparaffinized, rehydrated, washed, and subjected to TUNEL staining using the In Situ Cell Death Detection Kit (Boehringer Mannheim, Indianapolis, IN) according to the manufacturer’s instruction. Apoptotic cells in tumor sections were visualized by fluorescence microscopy. To show the tissue morphology on the section, the light microscopic photos of the section were also taken.
DNA extraction and gel electrophoretic analysis of DNA fragmentation
The exact same cell numbers (1×105) were plated on each well of 6-well plate. Cells were either untreated or transduced by control virus or AdRSVpHyde at moi of 200. The supernatants were collected from each well 48 h post viral transduction. Soluble DNA was extracted as described below and all soluble DNA extracted from one well was loaded on one lane of the agarose gel for comparison of the apoptotic extent. Briefly, the suspended cells in medium were collected 48 h post transduction by centrifugation. The pellet was resuspended in Tris-EDTA buffer (pH 8.0). The cells were lysed in 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100 on ice for 15 min. The lysate was centrifuged at 12,000xg for 15 min to separate soluble (fragmented) DNA from pelleted (intact genomic) DNA. Soluble DNA was treated with RNase A (50 μg/ml) at 37°C for 1 h, followed by treatment with proteinase K (100 μg/ml) in 0.5% SDS, at 50°C for 2 h. The residual material was extracted with phenol/chloroform, precipitated in ethanol, and electrophoresed on a 2% agarose gel. In some cases, the inhibitor of caspase-3 protease (200 μM DEVD, succinyl-Asp-Glu-Val-Asp-aldehyde; or 100 μM VAD, benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone, both from Biomol, Plymouth Meeting, PA) were added 24 h prior to viral transduction and maintained throughout the experiment.
Results
Cloning of human pHyde cDNA gene and its homology to rat pHyde
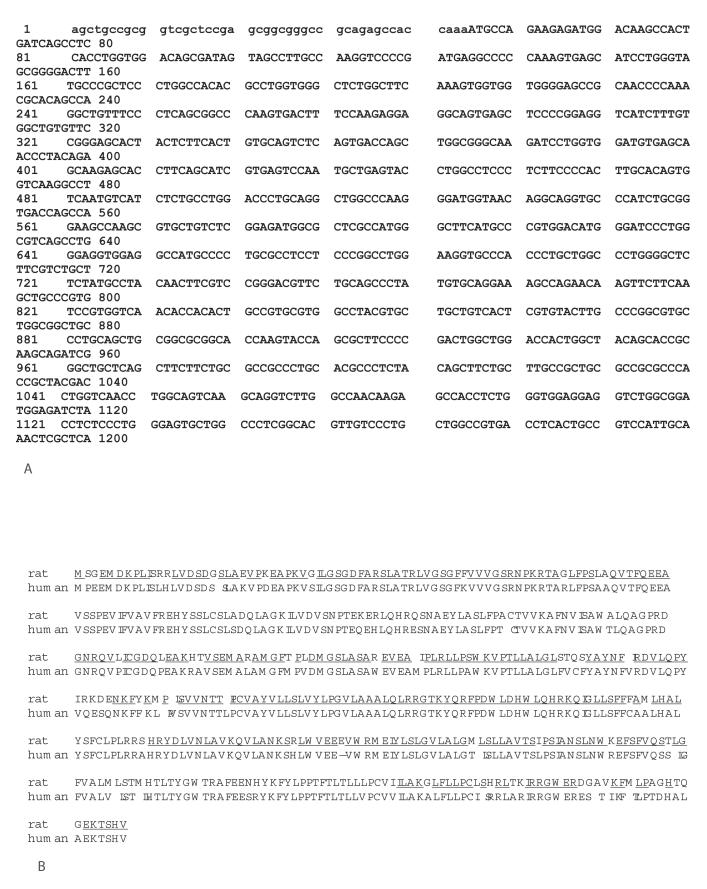
Recently we have isolated a full-length cDNA gene of human homologue (hpHyde) of rat pHyde gene. The hpHyde cDNA gene comprises of 1992 nucleotides (GenBank accession number AY082673) with an open reading frame of 1464 nucleotides (Fig. 1A) coding for a polypeptide of 487 amino acid residues. Interestingly, the length of nucleotides between stop codon and poly A signal in hpHyde gene is much shorter (374 nucleotides) than that in rat pHyde gene (1215 nucleotides, GenBank accession number AF335281); however, they have almost the same length of nucleotides for coding region and result in only one amino acid residue short in hpHyde protein compared to rat pHyde protein (Fig. 1B). The rat pHyde cDNA gene comprises of 2731 nucleotides with an open reading frame of 1467 nucleotides coding for a polypeptide of 488 amino acid residues.4 The hpHyde cDNA gene comprises of 1884 nucleotides with an open reading frame of 1464 nucleotides (Fig. 1A) coding for a polypeptide of 487 amino acid residues (Fig. 1B). The homology between human and rat pHyde amino acid sequences is 86.2% (Fig. 1B) and the homology between two species cDNA coding region sequence is 84.7%.
Figure 1.
A. Sequence of human pHyde cDNA The hpHyde cDNA gene (GenBank accession number AY082673) comprises of 1884 nucleotides with an open reading frame of 1464 nucleotides (uppercase) coding for a polypeptide of 487 amino acid residues.
B. Sequence of human pHyde protein Homology between rat and human pHyde amino acid sequences. The homologous sequence regions are underlined. The rat pHyde protein comprises of 488 amino acid residues and human pHyde protein comprises of 487 amino acid residues. The homology of pHyde protein sequences between the two species is 86.2%.
Chromosomal mapping of hpHyde gene
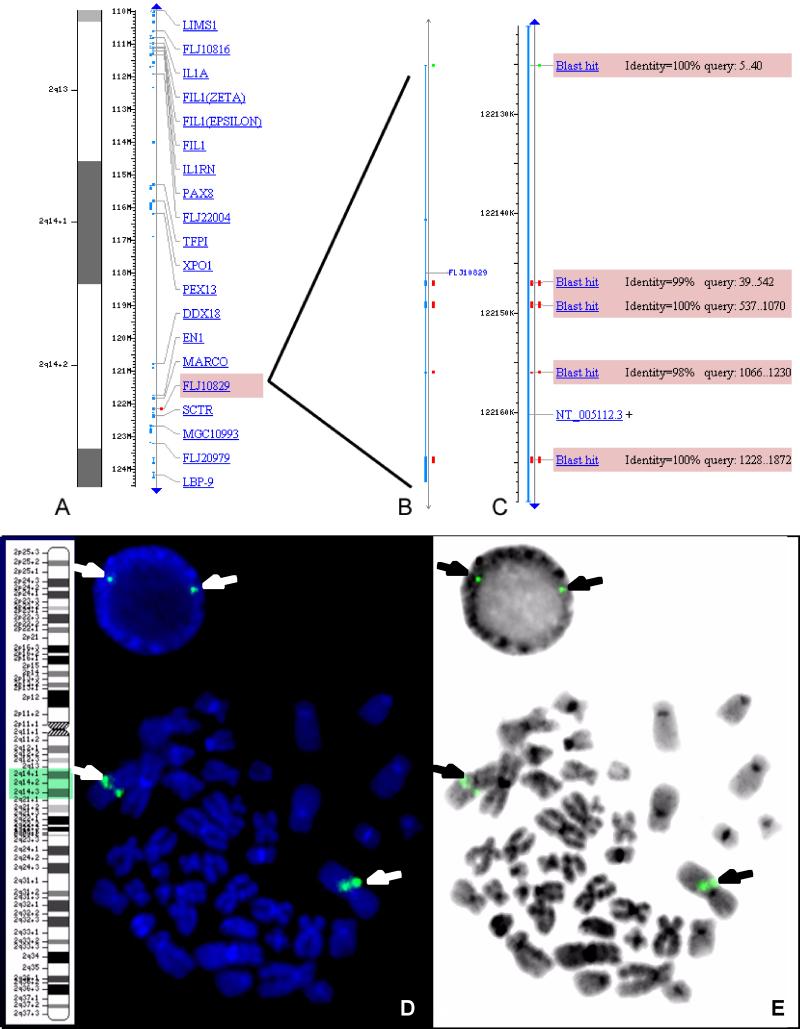
Database search using BLAST matched hpHyde cDNA sequence to a predicted gene FLJ10829 at human chromosome 2q14.2, at approximately 122.1 MB from the 2pter (Fig. 2A). The predicted gene FLJ10829 was annotated by Human Genome Project to this location. The BLAST result suggests that hpHyde genomic gene contains four exons (Blast hits, Fig. 2B and 2C) and three introns, and the BLAST positions are along the genomic backbone contig NT_005112.3 (Fig. 2C).
Figure 2. Localization of hpHyde gene in human chromosomes.
(Fig. A, B and C) Blast database search indicates that hpHyde cDNA match a predicted gene “FLJ10829” at contig NT_005112.3 of chromosome 2q14 (Fig. A, B, C). By using hpHyde cDNA as probe to screen RPCI-11 human BAC genomic libraries (Roswell Park Cancer Institute, Buffalo, NY), a matched clone, BAC RPCI-11-17N4, which contain larger part of NT_005112 contig, was identified. (Fig. D and E) FISH confirmation of localization of hpHyde gene on chromosome 2q14. The hpHyde genomic sequence was amplified by PCR from BAC RPCI-11-17N4 clone using primer specific to hpHyde cDNA sequences. The resulted PCR product was labeled with biotin and in situ hybridized with normal human metaphase. The FISH signals were detected with fluorescein isothiocyanate (FITC). Shown are a normal metaphase chromosome spread and an interphase nucleus. Fig. D is a DAPI-stained image. Fig. E is an inverted DAPI image to make it easier to delineate the chromosomes. The expected two signals (arrows) for the hpHyde gene were observed: 2 signals at 2q14 were seen on each chromosome 2 (2 per chromosome since 1 per sister chromatid).
To confirm the above in silico prediction, fluorescence in situ hybridization (FISH) experiment was performed to locate the hpHyde gene on a normal human metaphase chromosome spread (Fig. 2D and 2E). By using hpHyde cDNA as probe to screen RPCI-11 human genomic BAC libraries, a matched clone, BAC RPCI-11-17N4, which contains larger part of NT_005112 contig, was identified. Consequently, the hpHyde genomic sequence was amplified by PCR from BAC RPCI-11-17N4 clone using primers specific to hpHyde cDNA sequences. The resulted PCR product was labeled with biotin and in situ hybridized with normal human metaphase. Shown at Fig. 2D and 2E are a normal metaphase spread and an interphase nucleus. Fig. 2D is a DAPI-stained image. Fig. 2E is an inverted DAPI image to make it easier to delineate the chromosomes. The potential gene (signal) was assigned to chromosomal band by superimposing the FISH signals and the DAPI banding pattern. The expected signals (arrows) for the hpHyde gene were observed at metaphase corresponding to 2q14 on each chromosome 2 (2 per chromosome since 1 per sister chromatid because it is at metaphase) (Fig. 2D and 2E). Therefore, both database search and FISH results consistently indicate and confirm that hpHyde genomic gene localizes at chromosome 2q14.
Protein structure analysis of hpHyde protein predicted that hpHyde may be a plasma membrane protein with calcium channel-like function
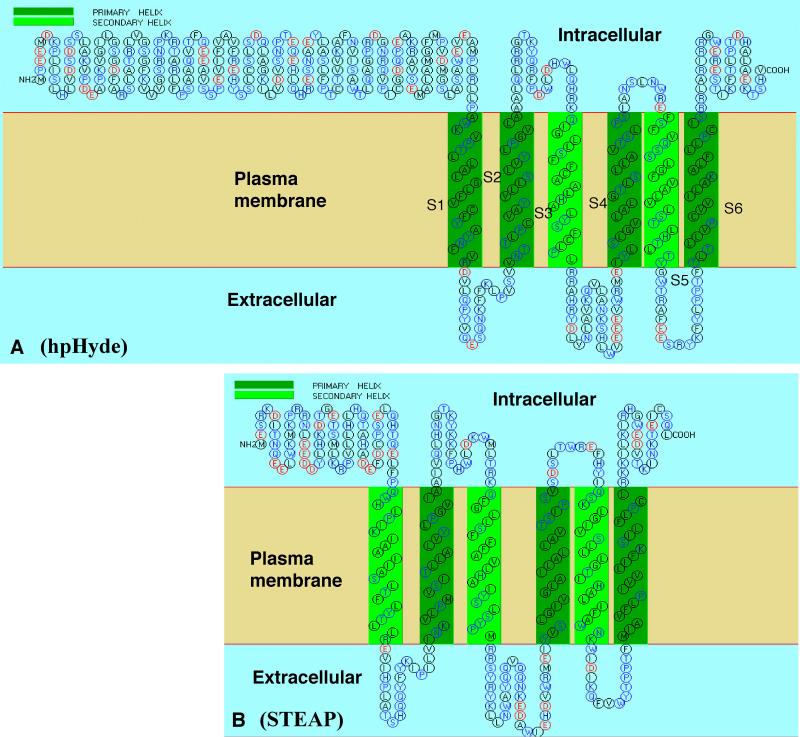
hpHyde protein sequence was analyzed by multiple database for functional domains/motifs, sorting signal and structural features. The protein structure analysis showed that hpHyde protein is a membrane protein which has six transmembrane helices, as indicated by regions from S1 to S6, respectively (Fig. 3A), a classic and typical feature of Ca2+ channel proteins and a mimicry to Ca2+ channel α1 subunit, which contains 4 repeated motifs of six-transmembrane-helix domain,11,12 the main structure which forms the calcium-conducting pore.12,13 More noticeably, the secondary structure of hpHyde, with six transmembrane segments, three extracellular loops and two intracellular loops, and both intracellular N- and C-terminus, shares a strikingly similarity to those of recently identified cell-membrane proteins, STEAP (six transmembrane epithelial antigen of the prostate)14 and CaT-L (epithelial calcium channel-like protein), a calcium channel protein.15 Both STEAP and CaT-L were found in human prostate epithelial cell membrane and are suggested to be involved in prostate cancer progression.14,15 In particular, database search using BLAST program indicates that hpHyde has a high sequence homology (46% contiguous identity) among amino acid 208-473 (the region that contains all six transmembrane helices) to STEAP, with an almost identical secondary structure in terms of transmembrane helix domains and C-terminal except that STEAP has a shorter N-terminal (compare Fig. 3A with Fig. 3B). However, hpHyde gene localizes at chromosome 2 (see above) while both STEAP and CaT-L genes were found to localize on chromosome 7.14,15 In addition, the combined results from amino acid sequence analyses showed that hpHyde protein does not have a N-terminal signal peptide, nuclear localization signal, mitochondria preseq cleavage site, endoplasmic reticulum retention motif in the C-terminus, transport motif from cell surface to Golgi, peroxisomal targeting signal, no vacuolar targeting motif, no RNA-binding and actin-binding motifs, ribosome protein motif, and coil-coil regions. Moreover, our immunofluorescence staining for AdRSVpHyde-transduced DU145 cells with primary antibody raised against rat pHyde4 demonstrated a cell-surface staining (not shown). These data, taken together, suggest that hpHyde is most likely to be a plasma membrane protein with calcium channel-like function.
Figure 3. Predicted secondary structutre of hpHyde protein.
Amino acid sequence and predicted secondary structure of (A) hpHyde protein and (B) STEAP, a known plasma-membrane protein by SOSUI transmembrane protein prediction site: (http://sosui.proteomebio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_ submit.html). The sequence analysis showed that hpHyde protein is a membrane protein which have six transmembrane helices, as indicated by regions from S1 to S6, respectively (A). Notice that STEAP shares an almost identical protein secondary structure to hpHyde, only with a shorter intracellular N-terminal. Notice that “Intracellular” is in the upper region and “Extracellular” is in the bottom region, so both N- and C- terminus are intracellular.
hpHyde expressed differentially in various normal tissues
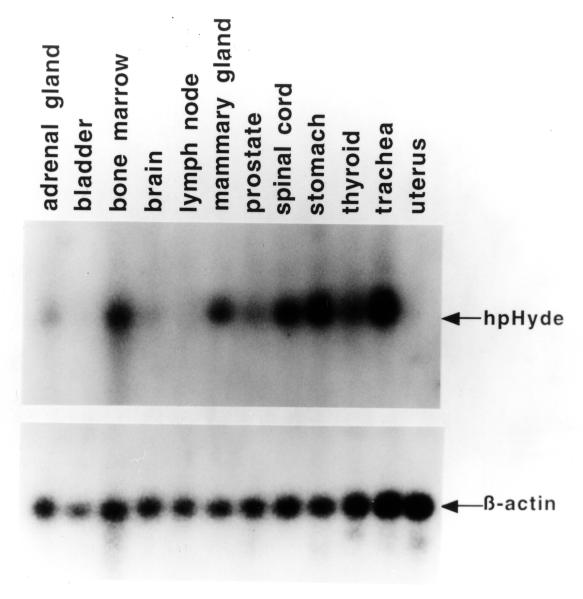
By using hpHyde cDNA probe, we analyzed endogenous expression of hpHyde at the mRNA level by Northern blot analysis in various normal human tissues (Fig. 4). The differential expression pattern in various normal tissues implies hpHyde may play roles in differentiation and development. The Northern blot (Fig. 4) showed that some normal tissues, such as trachea, stomach, thyroid, and bone marrow, have high endogenous mRNA expression of hpHyde. Whether there is a relationship between high expression of endogenous hpHyde (a proapoptotic gene) and tissues with high turnover cell population (such as bone marrow and stomach) needs to be further established.
Figure 4. Expression of hpHyde mRNA in various human tissues.
A pre-made Northern blot that contained 1 μg of poly A+ RNA per lane from 12 different human tissues was hybridized with a 32P-labeled hpHyde cDNA. The same blot was then rehybridized with ß-actin cDNA to assess RNA integrity and gel loading.
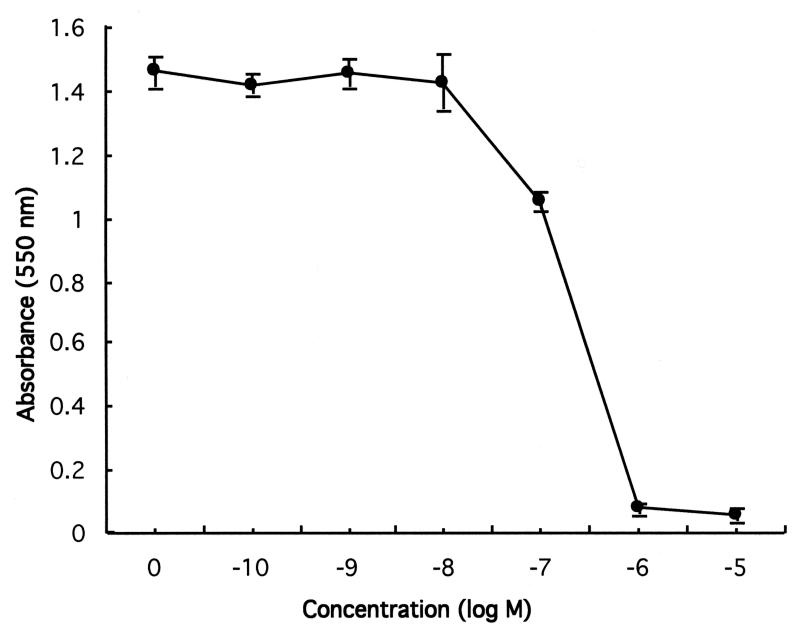
Determination of effect of cisplatin on human prostate cancer DU145 cell growth at various concentrations
Cisplatin is one of the most effective chemotherapeutic agents and plays a major role in the treatment of a variety of human solid tumors.16,17 To use the optional concentration of cisplatin to treat human prostate cancer DU145 cells, the IC50 (the concentration of drug that gives 50% growth inhibition) for cisplatin of DU145 cells was determined. Evaluation of a series of concentrations of cisplatin at 0, 10−10, 10−9, 10−8, 10−7, 10−6, and 10−5 M revealed that the IC50 for cisplatin was around 10−7 M (0.1 μM) (Fig. 5).
Figure 5. Effect of cisplatin on human prostate cancer DU145 cells.
DU145 cells were treated with a series of concentrations of cisplatin. The cells were incubated with cisplatin for three days, then MTT was added to cells followed by reading at 550 nm. The data represent the results of two independent experiments each performed in duplicate.
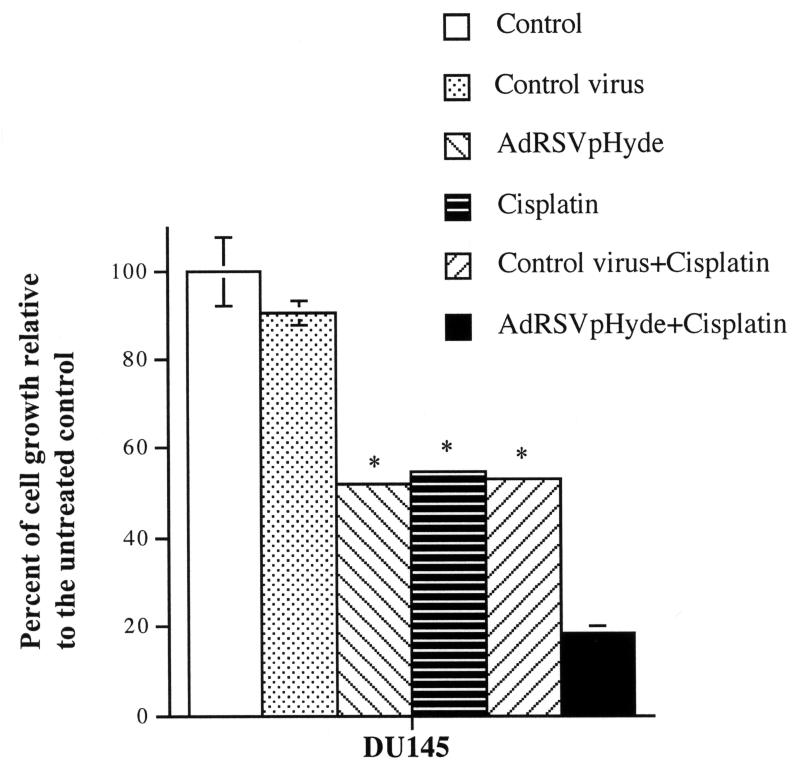
AdRSVHyde and cisplatin have synergistic antiproliferative effects on prostate cancer cell growth in vitro
We previously reported that AdRSVpHyde inhibited DU145 cell growth in vitro.4 To determine whether pHyde, a potent inducer of apoptosis, and cisplatin, a DNA damaging agent, together had synergistic inhibition of prostate cancer cell growth, DU145 cells were untreated, or treated with control virus AdRSVlacZ (moi=100) or AdRSVpHyde (moi=100) alone, cisplatin (0.1 μM) alone, or AdRSVpHyde (moi=100) plus cisplatin (0.1 μM) in vitro. Cell growth was evaluated by counting the number of cells at day 4 post viral transduction. As shown in Fig. 6, AdRSVpHyde suppressed the growth of DU145 cells (48.2% inhibition) compared to the untreated control cells, whereas AdRSVlacZ treated cells had no significant inhibition. Cisplatin alone also suppressed cell growth with 45.2% inhibition compared to untreated control. Treatment of both AdRSVpHyde and cisplatin resulted in even higher growth inhibition (81.7%) than either AdRSVpHyde or cisplatin alone.
Figure 6. Inhibitory effects of pHyde and cisplatin on growth of prostate cancer cell line DU145.
DU145 cells were treated with or without cisplatin (0.1 μM) for 24 hours. Cells were then transduced with or without adenoviral vectors (control virus AdRSVlacZ or AdRSVpHyde) at moi=100. Cell numbers were counted at day 4 after viral transduction. The data represent the results of two independent experiments each performed in duplicate. *Error bars are too small to show in this scale.
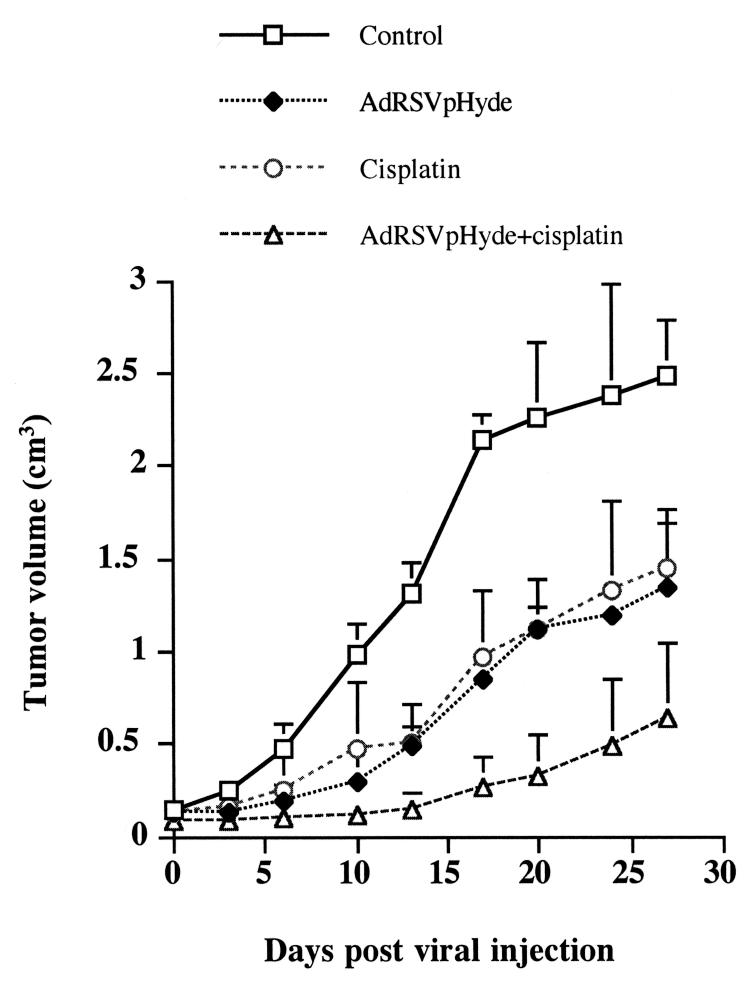
AdRSVpHyde and cisplatin additively suppressed human prostate tumor growth
To evaluate the potential synergistic inhibition of AdRSVpHyde and cisplatin treatment on prostate cancer cell growth in vivo, DU145 human prostate tumors were established in nude mice by injecting 1.4×107 DU145 cells subcutaneously into the flanks of nude mice. When mice developed tumors averaging 100 mm3 volume, the mice were divided into four groups: untreated control tumors (n=6), cisplatin alone treated (n=6), AdRSVpHyde alone treated (n=6), and AdRSVpHyde plus cisplatin treated (n=6). As shown in Fig. 7, tumors treated by cisplatin alone or AdRSVpHyde alone grew at a similar rate, but grew slower than untreated control tumors. Moreover, AdRSVpHyde plus cisplatin treated tumors resulted in smaller tumors among all the groups. By day 27 following viral injection, the nude mice bearing tumors treated by AdRSVpHyde plus cisplatin had a significant reduction in tumor volume compared to that of untreated control tumors, with a 74.3% growth suppression. Whereas tumors treated by AdRSVpHyde alone and cisplatin alone had 46.0% and 41.9% growth suppression, respectively, in tumor volume as compared to that of the untreated group (Fig. 7). Our previous study has showed that control virus reproducibly did not cause significant tumor reduction,4 therefore, the effects of AdRSVpHyde or AdRSVpHyde plus cisplatin were truly from pHyde or pHyde plus cisplatin respectively. These results showed that AdRSVpHyde or cisplatin alone is able to suppress prostate cancer, but the combination of the two has a synergistic inhibitory effect on prostate cancer growth.
Figure 7. Inhibitory effects of AdRSVpHyde and cisplatin on DU145 prostate tumor growth in vivo.
DU145 cells (1.4×107 cells) were injected subcutaneously into the flanks of nude mice. When tumors reached an average volume of 100 mm3, the mice were divided into four groups: (1) untreated control; (2) AdRSVpHyde alone treated; (3) cisplatin alone treated; and (4) AdRSVpHyde and cisplatin treated. For mouse group (2) and (4), a single dose of 5×109 pfu AdRSVpHyde was injected directly into the tumor at day 0. For mouse group (3) and (4), a consecutive 4-day intraperitoneal injection of cisplatin (1.5 mg/kg of body weight) was administrated starting at day 0. The tumor sizes were periodically measured up to day 27 days post viral injection when all the mice were sacrificed. Each point represents the average volume and standard deviation from six mice (n=6).
Cisplatin increased pHyde-mediated apoptosis in prostate cancer
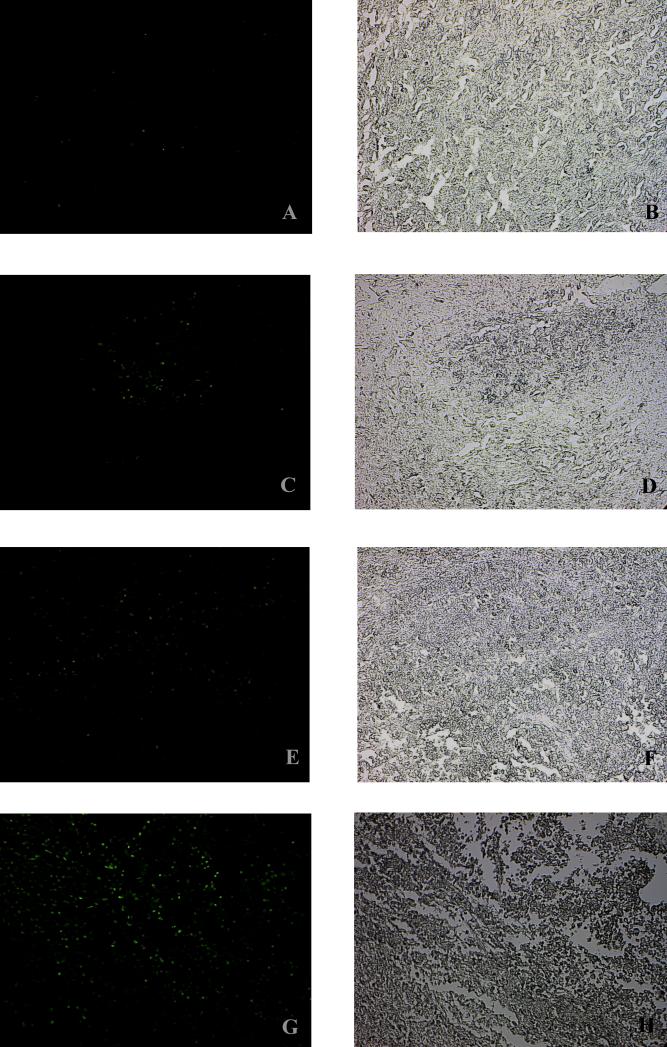
Our previous study has demonstrated that pHyde can directly induce apoptosis in human prostate cancer cells including DU145 after AdRSVpHyde transduction.4 To determine whether cisplatin, a DNA damaging agent, was able to increase pHyde-induced apoptosis in the prostate tumors, xenograft tumors growing in nude mice were harvested at day 27 after viral injection and tumor sections were analyzed by TUNEL staining assay. As shown in Fig. 8, there were some fluorescence-stained cells in AdRSVpHyde alone treated cells (Fig. 8C) but not in untreated control (Fig. 8A), indicating that AdRSVpHyde indeed induced DU145 cells to apoptosis. There was minimal staining of apoptotic cells in DU145 xenograft tumors treated by cisplatin alone (Fig. 8E), however, a significantly greater number of apoptotic cells was observed in tumors treated with AdRSVpHyde plus cisplatin (Fig. 8G). Morphologically, neither AdRSVpHyde alone (Fig. 8D) nor cisplatin alone (Fig. 8F) caused a significant histological change at day 27 after treatment, however, the combination of AdRSVpHyde and cisplatin treatment resulted in necrosis (Fig. 8H). These results demonstrate that pHyde-mediated induction of apoptosis in prostate cancer cells was enhanced by cisplatin.
Figure 8. TUNEL assay of DU145 prostate tumors.
DU145 xenograft tumors as described in Fig. 7 legend were harvested in 27 days post viral injection. Tumor sections of untreated control (A, B), AdRSVpHyde treated alone (C, D), cisplatin treated alone (E, F) and AdRSVpHyde plus cisplatin treated (G, H) were fixed and proceeded for TUNEL staining assay (A, C, E and G). Tissue histology is illustrated by the light microscopic images of the paired tumor sections (B, D, F and H). The original magnification of all images: x20.
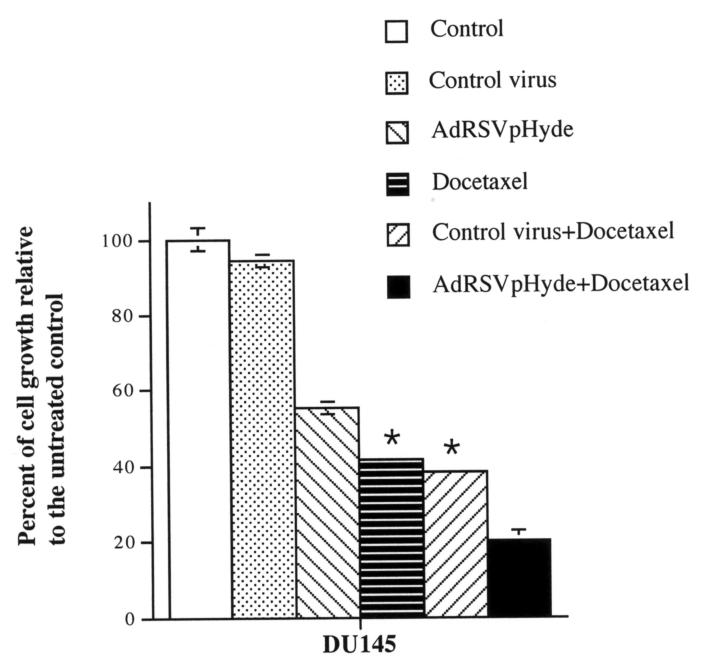
Inhibitory effect of AdRSVpHyde and docetaxel on prostate cancer DU145 cell growth
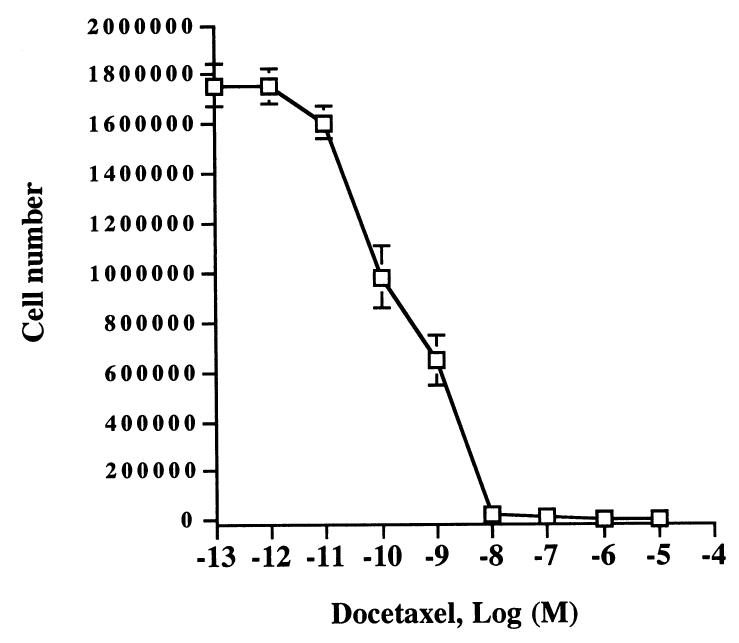
Like cisplatin which is commonly used as a treatment strategy for prostate cancer42-48, docetaxel is another chemotherapeutic drug equally common-used for treatment of prostate cancer.41,42 To investigate whether pHyde and docetaxel have a similar additively inhibitory effect on prostate cancer, we used AdRSVpHyde coupled with docetaxel to examine the anticancer effects in DU145 cells. First, we determined the IC50 concentration of docetaxel for DU145 cells is around 0.5 nM (or 5×10−10 M, which is consistent with that observed by other group36) by evaluation of a series of dilution of docetaxel on DU145 cells (Fig. 9). Then we treated DU145 cells either control virus or AdRSVpHyde alone, docetaxel (0.5 nM) alone, or AdRSVpHyde (moi=100) plus docetaxel (0.5 nM) in vitro. As shown in Fig. 10, AdRSVpHyde suppressed the growth of DU145 cells (44.6% inhibition) compared to untreated control cells, whereas control virus treated cells had no significant inhibition. Docetaxel alone also suppressed cell growth with 58.4% inhibition compared to untreated control. Treatment of both AdRSVpHyde and docetaxel resulted in even higher growth inhibition (79.7%) than either AdRSVpHyde or docetaxel alone. These data demonstrated that docetaxel can equally serve as an effective chemotherapeutic agent to additively suppress prostate cancer cell growth when in combination with AdRSVpHyde. Due to the fact that docetaxel has a much higher cytotoxicity (IC50 around 0.5 nM) than cisplatin (IC50=0.1 μM) to prostate cancer cells in terms of magnitude of concentration, docetaxel may have even more potential to serve as a chemotherapeutic agent coupled with AdRSVpHyde for prostate cancer treatment.
Figure 9. Effect of docetaxel on DU145 cells.
DU145 cells were treated with a series of concentrations of docetaxel. Cell numbers were counted four days after incubation with docetaxel. The data represent the results of two independent experiments each performed in duplicate.
Figure 10. Inhibitory effects of pHyde and docetaxel on growth of prostate cancer cell line DU145.
DU145 cells were treated with or without docetaxel (0.5 nM) for 24 hours. Cells were then transduced with or without adenoviral vectors (control virus or AdRSVpHyde) at moi=100. Cell numbers were counted at day 4 after viral transduction. The data represent the results of two independent experiments each performed in duplicate. *Error bars are too small to show in this scale.
pHyde mediated a caspase-3 dependent apoptosis in prostate cancer cells
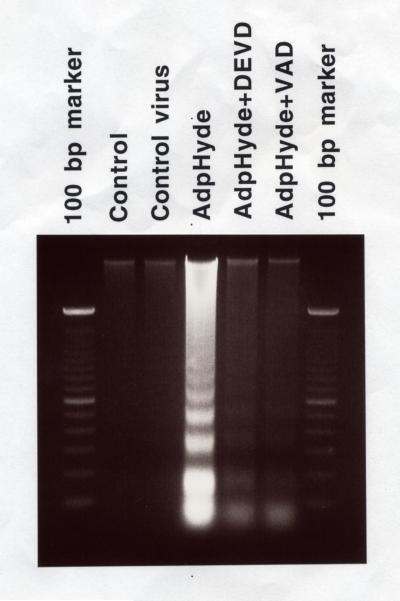
Our previous study revealed that pHyde increased enzymatic activity of caspase-3 in DU145 cells, and this elevated caspase-3 activity was blocked by DEVD and VAD the inhibitors specific to caspase-3 (not shown). To further determine whether this caspase-3 stimulation/activation was responsible for pHyde-mediated apoptosis, DU145 cells were preincubated with either DEVD or VAD for 24 h prior to AdRSVpHyde transduction. DNA was extracted from suspended cells 48 h after viral transduction and subjected to DNA fragmentation assay. DNA extracted from AdRSVpHyde (AdpHyde) transduced cells exhibited an expected laddering pattern following electrophoresis in agarose gel (lane 4, Fig. 11), a hallmark of cells undergoing apoptosis. In contrast, untreated control or control virus transduced cells did not have a laddering pattern (lane 2 and 3, Fig. 11). AdpHyde transduced cells which were pre-incubated with caspase-3 inhibitors, DEVD or VAD, however, showed reduction in DNA laddering (lane 5 and 6, Fig. 11), indicating that pHyde-mediated apoptosis was indeed blocked by caspase-3 inhibitors. Thus, the stimulation of caspase-3 activity was critical for the apoptotic induction by pHyde. Please notice that AdpHyde-only lane (lane 4, Fig. 11) showed more DNA content than other lanes, this is not due to an erroneous overloading. Because all samples are derived from the same starting cell numbers before viral transduction, and all DNA extracted from supernatants were loaded into each corresponding lane. AdRSVpHyde had the most floating (apoptotic) cells in the supernatant, so it consequently has more DNA content. These results suggest that pHyde may induce a caspase-3 dependent apoptosis.
Figure 11. Caspase-3 inhibitors block pHyde-mediated apoptosis.
The exact same cell numbers (1×105) were plated on each well of 6-well plate. Cells were either untreated or transduced by control virus or AdRSVpHyde at moi of 200, supernatants were collected from each well 48 h post transduction. Soluble DNA was extracted from cell suspensions and all extracted DNA was electrophoresed on a 2% agarose gel. In the cases of lane 5 and 6, cells were pretreated with caspase-3 inhibitor (DEVD or VAD) 24 h prior to viral transduction and maintained in 200 μM DEVD (lane 5) or 100 μM VAD (lane 6) throughout.
Discussion and Conclusions
In summary, this study showed that the genomic gene hpHyde, a human homologue of the novel apoptosis-inducing gene pHyde, was localized at human chromosome 2q14. hpHyde expressed differentially in various normal tissues and organs, implying that hpHyde may play roles in normal development and differentiation. hpHyde appears to be a cell-surface protein, however, whether it indeed has calcium channel-like function as predicted by protein structure analysis needs to be further investigated. Furthermore, an additively inhibitory effect of adenoviral vector expressing pHyde and cisplatin on prostate cancer growth was observed both in vitro and in vivo.
A delicate balance between cell proliferation and programmed cell death (apoptosis) results in the maintenance of the normal prostate epithelium. The progression of prostate cancer may be partly due to an imbalance of this dynamic equilibrium in which programmed cell death pathway is impaired. The introduction of genes to either induce apoptosis18-20 or suppress cellular anti-apoptotic pathway21 may be an effective approach to treat prostate cancer. Our previous study showed that adenoviral vector expressing rat pHyde inhibited human prostate cancer cell growth both in vitro and in vivo, and pHyde directly induced apoptosis in human prostate cancer cells.4 Similar experiments showed that pHyde also has growth inhibitory effect on cancer cells of nonprostate origin, implying that pHyde may have a more global tumor suppressor effect (our unpublished results). Therefore, pHyde may be a tumor suppressor gene that induces apoptosis across different cell types and may be a candidate gene for cancer gene therapy.
Current treatments for prostate cancer include surgery, radiation therapy, hormone therapy (androgen deprivation), chemotherapy and gene therapy. It is most likely that gene therapy will have the best potential for prostate cancer treatment when in conjunction with other therapeutic modality, such as chemotherapy. Adenoviral-mediated gene transfer of an apoptosis-inducing gene p53 (AdCMVp53) had greater efficacy when combined with chemotherapeutic agents for suppression of xenograft DU145 prostate tumors.22 AdCMVp53 combined with cisplatin, doxorubicin, 5-fluorouracil, methotrexate, or etoposide inhibited cell proliferation in vitro more effectively than either AdCMVp53 alone or chemotherapy alone in various cancer cells including DU145 prostate cancer cells.22 By using human tumor xenograft models in SCID mice and intratumoral or intraperitoneal injection of AdCMVp53 with and without chemotherapeutic drugs, Gurnani et al. found a greater in vivo anticancer efficacy by combination of AdCMVp53 and chemotherapeutic agents in four human tumor including DU145 xenograft prostate tumor.22 The mechanism behind the additive/synergistic effect includes increased chemosensitivity and radiosensitivity by p53 in cancers.23,30 Likewise, other tumor suppressor gene or apoptosis-inducing gene also showed additive/synergistic effects when in combination with chemotherapeutic agent cisplatin. Adenoviral-mediated p16 expression and cisplatin had a synergistic inhibitory effect on growth of human PPC-1 prostate xenograft tumor.24
In this study, to better evaluate the potential therapeutic effects of pHyde gene transfer for prostate cancer treatment, it was combined with a DNA damaging chemotherapeutic agent, cisplatin. Cisplatin is considered has anticancer activity against a broad spectrum of malignancy16,17,25 including prostate cancer.26,27 Cisplatin induced both p53-dependent28 and p53-independent29 growth arrest and cell death in cancer cells. The benefits of combination of apoptosis-inducing gene therapy and DNA damaging agent (chemotherapy) are apoptosis-inducing gene usually increases chemosensitivity of cancer cells, and tumor cells resistant to chemotherapeutic agents may be induced to apoptosis by apoptosis-inducing gene via a different mechanism. On the other hand, the presence of DNA damaging agent usually increases the susceptibility of cancer cells to cell death by apoptosis-inducing gene. In an effort to determine the efficacy of a combination of AdCMVp53 and cisplatin therapy, human lung cancer H358 cells were implanted into nude mice and the xenograft tumors were treated with either intratumoral injection of AdCMVp53, intraperitoneal of cisplatin or both. The combination treatment resulted in significantly smaller tumor sizes than those in any of the other treatment groups.30 Consistently, our study showed that AdRSVpHyde and cisplatin have an additive inhibitory effect on suppression of prostate cancer cell growth both in vitro and in vivo. While our study focused on the use of cisplatin, which is commonly used as a treatment strategy for prostate cancer (either alone or combined with other agents or another therapeutic modality),42-48 we also investigated anti-prostate cancer effects of another chemotherapeutic drug, docetaxel, which is equally common-used for treatment of prostate cancer.41,42 We found that AdRSVpHyde and docetaxel have an additive inhibitory effect on growth of prostate cancer cells as well.
At the time of experiment, we have used virus expressing rat pHyde that was already made and available in our laboratory for the in vitro and in vivo work to analyze pHyde’s anti-prostate cancer effects. Because of the highly conserved sequences of pHyde gene between human and rat (with a homology of pHyde protein sequences between the two species at 86%, Fig. 1), we consider the biological function of human pHyde and rat pHyde would be similar, if not identical.
Consistent with our finding, Porkka et al37 previously identified hpHyde gene is located in the chromosome 2q14 and is highly homologous at the protein level with STEAP, a six spanned-transmembrane protein. While they did not provide any functional or biological analysis of the gene as we did (in vitro and in vivo pHyde-mediated growth inhibition and apoptotic induction in prostate cancer cells), they did provide unique mutational analysis and screening for pHyde expression in prostate cancer cell lines, xenograft tumors and clinical prostate tumors. In contrast, we only screened for hpHyde expression pattern for the normal human tissues. Therefore, the complementary results from these two reports should provide better and more complete information of pHyde gene and help elucidation of the mechanism of pHyde-mediated action.
Our protein structure analysis showed that hpHyde protein is a membrane protein which has six transmembrane helices, a classic and typical feature of Ca2+ channel proteins. The same secondary structure of hpHyde also shares a striking similarity to those of two recently reported novel cell-membrane proteins STEAP14 and CaT-L, a Ca2+ channel protein,15 all having six spanned-transmembrane-helix motifs, three extracellular loops and two intracellular loops, and both intracellular N- and C-termini (Fig. 3A). In particular, hpHyde has a 46% contiguous sequence homology among amino acid 208-473, the region that contains all six transmembrane helices, to STEAP, with an almost identical secondary structure in terms of transmembrane helix domains and C-terminal except that STEAP has a shorter N-terminal (Fig. 3). Both STEAP and CaT-L were found in human prostate epithelial cell membrane and are suggested to be involved in prostate cancer progression.14,15 Based on this information, hpHyde is most likely to be a plasma membrane protein with Ca2+ channel function. While our chromosomal mapping showed that hpHyde gene localizes on a different chromosome from that of STEAP and CaT-L genes, they may present a family of Ca2+ channel-like protein involved in prostate cancer carcinogenesis and progression.
Ca2+ channels are the signal transducers that convert signals in the cell membrane into an increase in the intracellular second messenger Ca2+ and thereby activate many crucial intracellular processes including contraction, secretion, neurotransmission, regulation of enzyme activities and gene expression, cell differentiation and apoptosis.32 The role of the calcium ion (Ca2+) as intracellular regulator of many physiological processes is now well established. The effects of a variety of hormones and growth factors mediated by transient increases in the level of cytosolic Ca2+. The perturbation of intracellular Ca2+ homeostasis may be a common step in the development of cytotoxicity.33 While roles of Ca2+ and other ion channels have been extensively studied for their functions in normal cells, it is not well known of the roles played by ion channels in cancer. However, accumulating evidences have emerged that Ca2+ channels play a central role in the regulation of apoptosis, which is directly associated with normal and malignant prostate cell proliferation.34 Recently, a mouse gene, Trp12, which has a similar six-transmembrane secondary structure, was identified as a Ca2+ permeable channel protein. Trp12 transfected cells reveal a significantly elevated cytosolic Ca2+ concentration compared to nontransfected cells.31 The human homologue of mouse Trp12 localizes on chromosome 12.31 Like hpHyde, another ion channel has been reported recently that acts as a proapoptotic gene and causes tumor suppression: a pair of closely related Ca2+-activated chloride channels, mCLCA1 and mCLCA2, with differential regulation in normal and transformed mouse cells, were shown to be able to reduce colony formation and tumorigenecity on mice as compared to their vector transfected control.35 Based on our results that rat pHyde expression induces a caspase-3 dependent apoptosis (Fig. 8 and Fig. 11) and our protein structure analysis that hpHyde may be a Ca2+ channel transmembrane protein (Fig. 3), it is very exciting that we may find a novel gene product, which has both identified biological functions (growth inhibition and apoptotic induction) and physiological functions (Ca2+ channel transmembrane protein), that may provide mechanism of action of a novel apoptotic signal transduction pathway. Taken together, our hypothesis is that hpHyde is a plasma membrane Ca2+ channel protein. Overexpression of hpHyde may lead to an influx of extracellular Ca2+ into cells, the increased cytosolic Ca2+ concentration leads to apoptosis in prostate cancer cells via a caspase-3 dependent apoptotic pathway. We are currently in the process of investigating whether hpHyde is indeed a Ca2+ channel protein, exploring the mechanism of hpHyde-mediated action, and identifying the missing links between pHyde expression and apoptosis.
Prostate cancer has become the most frequently diagnosed malignancy and the second leading cause of cancer deaths in American men today. Identification and characterization of novel genes that may successfully inhibit or destroy prostate cancer cells may have potential utility in clinical therapeutic treatment. Several tumor suppressor genes including PTEN,32 DOC-2,38 E-cadherin,39 and C-CAM40 have been shown to suppress prostate cancer growth. As such, the finding and characterization of novel gene that inhibits cancer growth and induces apoptosis such as hpHyde may have a therapeutic potential. In summary, this study has not only demonstrated that pHyde, a novel tumor suppressor gene that induces growth inhibition and apoptosis in prostate cancer cells, but also, that the combination of AdRSVpHyde and cisplatin may be more effective against advanced prostate cancer than either agent alone. These studies support the use of this combined therapy in men who have locally advanced prostate cancer.
Acknowledgements
This research was supported by National Institutes of Health grants (DK65962 and CA107162) (YL). We gratefully thank Dr. Jeremy Squire of Princess Margaret Hospital and Ontario Cancer Institute (Toronto, Canada) for assistance in database search and FISH analysis.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer Statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Carter BH, Piantadosi S, Isaacs JT. Clinical evidence for and implications of the multistep development of prostate cancer. J Urol. 1990;143:742–6. doi: 10.1016/s0022-5347(17)40078-4. [DOI] [PubMed] [Google Scholar]
- 3.Sandberg AA. Chromosome abnormalities and related events in prostate cancer. Hum Pathology. 1992;23:368–80. doi: 10.1016/0046-8177(92)90083-f. [DOI] [PubMed] [Google Scholar]
- 4.Steiner MS, Zhang X, Wang Y, Lu Y. Growth inhibition of prostate cancer by adenovirus expressing a novel tumor suppressor gene pHyde. Cancer Res. 2000;60:4419–25. [PubMed] [Google Scholar]
- 5.Lu Y, Lotan R. Transcriptional regulation by butyrate of mouse galectin-1 gene in embryonal carcinoma cells. Biochim Biophy Acta. 1999;1444:85–91. doi: 10.1016/s0167-4781(98)00257-7. [DOI] [PubMed] [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Akeson EC, Davisson MT. Chromosome preparation from solid adult tissues or tumors. In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG, Smith DR, editors. Current protocols in human genetics. John Wiley & Sons, Inc.; New York: 2000. pp. 4.10.8–4.10.9. [Google Scholar]
- 8.Lu Y, Carraher J, Zhang Y, Armstrong J, Lerner J, Roger W, Steiner MS. Delivery of adenoviral vectors to the prostate for gene therapy. Cancer Gene Ther. 1999;6:64–72. doi: 10.1038/sj.cgt.7700011. [DOI] [PubMed] [Google Scholar]
- 9.Graham FL, Prevec L. Manipulation of adenovirus vectors. In: Murray EJ, editor. Methods in molecular biology, gene transfer and expression protocols. The Human Press Inc; New Jersey: 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y, Lotan D, Lotan R. Differential regulation of constitutive and retinoic acid-induced galectin-1 gene transcription in murine embryonal carcinoma and myoblastic cells. Biochim Biophys Acta. 2000;1491:13–9. doi: 10.1016/s0167-4781(00)00055-5. [DOI] [PubMed] [Google Scholar]
- 11.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev. 2000;16:521–55. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz A. Molecular studies of the calcium antagonist binding site on calcium channels. Am J Cardiol. 1994;73:12B–14B. doi: 10.1016/0002-9149(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 13.Catterall W, Epstein PN. Ion channels. Diabetologia. 1992;35(Suppl 2):23–33. doi: 10.1007/BF00586276. [DOI] [PubMed] [Google Scholar]
- 14.Hubert RS, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, Jakobovits A, Saffran DC, Afar DE. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA. 1999;96:14523–8. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wissenbach U, Niemeyer BA, Fixemer T, Schneidewind A, Trost C, Cavalie A, Reus K, Meese E, Bonkhoff H, Flockerzi V. Expression of CaT-like, a novel calcium-selective channel, correlates with the malignancy of prostate cancer. J Biol Chem. 2001;276:19461–8. doi: 10.1074/jbc.M009895200. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg B. Fundamental studies with cisplatin. Cancer. 1985;55:2303–l6. doi: 10.1002/1097-0142(19850515)55:10<2303::aid-cncr2820551002>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Lee CK, Park KK, Lim SS, Park JH, Chung WY. Effects of the licorice extract against tumor growth and cisplatin-induced toxicity in a mouse xenograft model of colon cancer. Biol Pharm Bull. 2007;30:2191–5. doi: 10.1248/bpb.30.2191. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Cirielli C, Capogrossi MC, Passaniti A. Adenovirus-mediated wild-type p53 expression induces apoptosis and suppresses tumorigenesis of prostatic tumor cells. Cancer Res. 1995;55:4210–3. [PubMed] [Google Scholar]
- 19.Hedlund TE, Meech SJ, Srikanth S, Kraft AS, Miller GJ, Schaack JB, Duke RC. Adenovirus-mediated expression of Fas ligand induces apoptosis of human prostate cancer cells. Cell Death Differ. 1999;6:175–82. doi: 10.1038/sj.cdd.4400477. [DOI] [PubMed] [Google Scholar]
- 20.Hyer ML, Voelkel-Johnson C, Rubinchik S, Dong J-Y, Norris JS. Intracellular Fas ligand expression causes Fas-mediated apoptosis in human prostate cancer cells resistant to monoclonal antibody-induced apoptosis. Mol Ther. 2000;2:348–58. doi: 10.1006/mthe.2000.0139. [DOI] [PubMed] [Google Scholar]
- 21.Dorai T, Perlman H, Walsh K, Shabsigh A, Goluboff ET, Olsson CA, Buttyan R. A recombinant defective adenoviral agent expressing anti-bcl-2 ribozyme promotes apoptosis of bcl-2-expressing human prostate cancer cells. Int J Cancer. 1999;82:846–52. doi: 10.1002/(sici)1097-0215(19990909)82:6<846::aid-ijc13>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 22.Gurnani M, Lipari P, Dell J, Shi B, Nielsen LL. Adenovirus-mediated p53 gene therapy has greater efficacy when combined with chemotherapy against human head and neck, ovarian, prostate, and breast cancer. Cancer Chemother Pharmacol. 1999;44:143–51. doi: 10.1007/s002800050959. [DOI] [PubMed] [Google Scholar]
- 23.Hamada M, Fujiwara T, Hizuta A, Gochi A, Naomoto Y, Takakura N, Takahashi K, Roth JA, Tanaka N, Orita K. The p53 gene is a potent determinant of chemosensitivity and radiosensitivity in gastric and colorectal cancers. J Cancer Res Clin Oncol. 1996;122:360–65. doi: 10.1007/BF01220804. [DOI] [PubMed] [Google Scholar]
- 24.Adams C, Lu Y, Steiner MS, Allay JA. Adenovirus-mediated p16 expression and cisplatin synergize to inhibit human prostate cancer xenograft growth. Mol Ther. 2000;1:163. [Google Scholar]
- 25.Rosenberg B, VanCamp L, Trosko JE, Mansour VH. Platinum compounds; A new class of potent antitumor agents. Nature. 1969;222:385–86. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 26.Chao D, von Schlippe M, Harland SJ. A phase II study of continuous infusion 5-fluorouracil (5-FU) with epirubicin and cisplatin in metastatic, hormone-resistant prostate cancer: an active new regimen. Eur J Cancer. 1997;33:1230–3. doi: 10.1016/s0959-8049(97)00097-x. [DOI] [PubMed] [Google Scholar]
- 27.Geldof AA, Slotman BJ. Radiosensitizing effect of cisplatin in prostate cancer cell lines. Cancer Lett. 1996;101:233–9. doi: 10.1016/0304-3835(96)04140-7. [DOI] [PubMed] [Google Scholar]
- 28.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–18. [PubMed] [Google Scholar]
- 29.Wang X, Liu Y, Chow LS, Wong SC, Tsao SW, Kwong DL, Wang J, Sham JS, Nicholls JM. Cisplatin-induced p53-independent growth arrest and cell death in cancer cells. Int J Oncol. 1999;15:1097–102. doi: 10.3892/ijo.15.6.1097. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara T, Grimm EA, Mukhopadhyay T, Zhang WW, Owen-Schaub LB, Roth JA. Induction of chemosensitivity in human lung cancer cells in vivo by adenovirus-mediated transfer of the wild-type p53 gene. Cancer Res. 1994;54:2287–2291. [PubMed] [Google Scholar]
- 31.Wissenbach U, Bodding M, Freichel M, Flockerzi V. Trp12, a novel Trp related protein from kidney. FEBS Lett. 2000;485:127–34. doi: 10.1016/s0014-5793(00)02212-2. [DOI] [PubMed] [Google Scholar]
- 32.Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA. 1998;95:5246–50. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orrenius S, Nicotera P. The calcium ion and cell death. J Neural Transm. 1994;(Suppl 43):1–11. [PubMed] [Google Scholar]
- 34.Tapia-Vieyra JV, Mas-Oliva J. Apoptosis and cell death channels in prostate cancer. Arch Med Res. 2001;32:175–85. doi: 10.1016/s0188-4409(01)00274-0. [DOI] [PubMed] [Google Scholar]
- 35.Elble RC, Pauli BU. Tumor suppression by a pro-apoptotic calcium-activated chloride channel in mammary epithelium. J Biol Chem. 2001;276:40510–7. doi: 10.1074/jbc.M104821200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Stanfield J, Frenkel E, Kabbani W, Hsieh J-T. Enhanced therapeutic effect on androgen-independent prostate cancer by depsipeptide (FK228), a histone deacetylase inhibitor, in combination with docetaxel. [DOI] [PubMed]
- 37.Porkka KP, Nupponen NN, Tammela TLJ, Vesselia RL, Vishkorpi T. Human pHyde is not a classical tumor suppressor gene in prostate cancer. Int J Cancer. 2003;106:729–35. doi: 10.1002/ijc.11278. [DOI] [PubMed] [Google Scholar]
- 38.Tseng CP, Ely BD, Li Y, Pong RC, Hsieh JT. Regulation of rat DOC-2 gene during castration-induced rat ventral prostate degeneration and its growth inhibitory function in human prostatic carcinoma cells. Endocrinology. 1998;139:3542–53. doi: 10.1210/endo.139.8.6159. [DOI] [PubMed] [Google Scholar]
- 39.Luo J, Lubaroff DM, Hendrix MJ. Suppression of prostate cancer invasive potential and matrix metalloproteinase activity by E-cadherin transfection. Cancer Res. 1999;59:3552–6. [PubMed] [Google Scholar]
- 40.Kleinerman DI, Zhang WW, Lin SH, Nguyen TV, von Eschenbach AC, Hsieh JT. Application of a tumor suppressor (C-CAM1)-expressing recombinant adenovirus in androgen-independent human prostate cancer therapy: a preclinical study. Cancer Res. 1995;55:2831–6. [PubMed] [Google Scholar]
- 41.Hayes-Lattin BM, Kovach PA, Henner WD, Beer TM. Successful treatment of metastatic hormone-refractory prostate cancer with malignant pericardial tamponade using docetaxel. Urology. 2002;59:137–8. doi: 10.1016/s0090-4295(01)01481-9. [DOI] [PubMed] [Google Scholar]
- 42.Culine S, El Demery M, Lamy PJ, Iborra F, Avancès C, Pinguet F. Docetaxel and cisplatin in patients with metastatic androgen independent prostate cancer and circulating neuroendocrine markers. J Urol. 2007;178:844–8. doi: 10.1016/j.juro.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 43.Sakura M, Tsukamoto T, Yonese J, Ishikawa Y, Aoki N, Fukui I. Successful therapy of a malignant phyllodes tumor of the prostate after postoperative local failure. Urology. 2006;67:845.e11–3. doi: 10.1016/j.urology.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Weber B, Serafin A, Michie J, Van Rensburg C, Swarts JC, Bohm L. Cytotoxicity and cell death pathways invoked by two new rhodium-ferrocene complexes in benign and malignant prostatic cell lines. Anticancer Res. 2004;24:763–70. [PubMed] [Google Scholar]
- 45.Urakami S, Shiina H, Sumura M, Honda S, Wake K, Hiraoka T, Inoue S, Ishikawa N, Igawa M. Long-term control or possible cure? Treatment of stage D2 prostate cancer under chemotherapy using cisplatin and estramustine phosphate followed by maximal androgen blockade. Int Urol Nephrol. doi: 10.1007/s11255-007-9301-z. Epub 2007 Dec 19. [DOI] [PubMed] [Google Scholar]
- 46.Moriyama-Gonda N, Shiina H, Terashima M, Satoh K, Igawa M. Rationale and clinical implication of combined chemotherapy with cisplatin and oestrogen in prostate cancer: primary evidence based on methylation analysis of oestrogen receptor-alpha. BJU Int. 2008;101:485–91. doi: 10.1111/j.1464-410X.2007.07256.x. [DOI] [PubMed] [Google Scholar]
- 47.Sessa C, Minoia C, Ronchi A, Zucchetti M, Bauer J, Borner M, de Jong J, Pagani O, Renard J, Weil C, D’Incalci M. Phase I clinical and pharmacokinetic study of the oral platinum analogue JM216 given daily for 14 days. Ann Oncol. 1998;9:1315–22. doi: 10.1023/a:1008441416790. [DOI] [PubMed] [Google Scholar]
- 48.Culine S, El Demery M, Lamy PJ, Iborra F, Avancès C, Pinguet F. Docetaxel and cisplatin in patients with metastatic androgen independent prostate cancer and circulating neuroendocrine markers. J Urol. 2007;178:844–8. doi: 10.1016/j.juro.2007.05.044. [DOI] [PubMed] [Google Scholar]