Abstract
Although transgenic animal production through somatic cell nuclear transfer (SCNT) has been successful, the process is still inefficient. One major limitation is the use of somatic donor cells that have a finite life span. Identification and isolation of a cell type capable of rapid proliferation while possessing immortal or prolonged life span in culture and is capable of being genetically modified would be very valuable for utilization in the production of genetically modified pigs. Here we report the birth of live piglets after cloning by using porcine skin-derived stem cells (SSC) as a donor cell type. In the present study, cell cycle analysis indicates that the porcine SSC proliferate rapidly in vitro. The porcine SSC are capable of producing live offspring and can be genetically modified with positive selection. Utilization of porcine SSC may prove to be an excellent cell type for genetic modification followed by nuclear transfer for the production of transgenic pigs.
Introduction
The early work on nuclear transfer (NT) focused on using early embryos as a source of donor nuclei and produced cloned embryos and offspring (Prather et al., 1989), but by using this strategy expansion of the genotype would require serial NTs to result in a significant number of clones. Additionally, the ability to genetically modify the donor nuclei was very limited. Subsequently fetal-derived cells, such as fetal-derived fibroblast cells, were cultured and used to clone animals, and several research groups succeeded in cloning piglets from wild-type and genetically modified somatic cells (Lai et al., 2002b; Park et al., 2001). Currently somatic cell nuclear transfer (SCNT) in pigs relies primarily on the utilization of fetal-derived fibroblast cells, and the resultant clones tend to exhibit a significant level of phenotypic instability. This instability may be due to epigenetic reprogramming and/or genomic damage in the donor cells (Cho et al., 2007). The advantage of using fetal-derived fibroblast cells for making genetically modified cloned animals is that precise genetic modifications can be accomplished and confirmed in the cultured cells prior to generating the animal. This is very important, because true embryonic stem cells have not yet been isolated from any domestic animal (Prather, 2007; Prather et al., 2008). Expansion of NT techniques for domestic animals is rapidly progressing through the utilization of SCNT to create genetically modified cloned animals. In addition to the compromised phenotypic instability, production of transgenic clones through SCNT using fibroblast donor nuclei is inefficient, because the restricted life span of somatic donor cells in culture can be limiting when the genetic modification requires selection. In contrast, stem cells would proliferate rapidly and not undergo senescence at a high rate, so the selection process can be extended. Because there is no report of an embryonic stem (ES) cell line derived in the pig that contributes to the germline, we decided to investigate the utility of porcine skin-derived stem cells (SSC). The SSC express the neural progenitor marker, nestin, as well as a gene that is critical for pluripotency, Oct4. In addition, these SSC proliferated actively in vitro and retained a normal karyotype after long-term culture (Dyce et al., 2004). Thus, SSC provide a potential source of donor cells amenable to genetic modification and subsequent cloning by NT. This study investigated the porcine SSC potential as donor nuclei for SCNT through the characterization of the porcine SSC cell cycle, ultrastructure, the potential of gene transfection, and the development of cloned embryos.
Materials and Methods
Preparation of porcine skin-derived stem cells
The porcine SSC were isolated as previously described (Dyce et al., 2004). Both male and female fetuses were removed from the uterus of 35- to 50-day pregnant Yorkshire sows, and fetal skin from the back was carefully dissected from other tissue and cut into pieces and digested with 0.2% trypsin for 40 min at 37°C, followed by 0.1% DNase for 1 min at room temperature. Tissue pieces were washed with Hank's Buffered Salt Solution (HBSS), and with Dulbecco's Modified Eagle's Medium (DMEM)/F12 (1:1, containing antibiotics; Gibco, Grand Island, NY), followed by mechanical dissociation via vortexing and pipetting. Following dissociation, the porcine SSC were strained through a 45-μm strainer, and cultured in SSC medium [DMEM/F12 containing B-27 (Gibco), 20 ng/mL EGF (Sigma, St. Louis, MO), and 40 ng/mL bFGF (Sigma)] at 37.5°C, 5% CO2 in a water-saturated atmosphere until small floating spheres formed. Spheres were transferred to a new culture dish containing fresh SSC medium and were cultured for 10–14 days. Purified populations of floating spheres were obtained after 3–4 days. The porcine SSC were collected by centrifugation of the culture medium at 500 × g for 6 min to remove spheres from suspension. The resulting cell pellet was resuspended in cryopreservation medium (10% dimethyl sulfoxide (DMSO; Sigma) in DMEM), and stored in liquid nitrogen as passage 2. Porcine SSC at passage 2 were thawed and cultured continually, and frozen again in liquid nitrogen from passage 2 to passage 8. These cells maintained morphology (see documentation below) consistent with previous descriptions of skin-derived stem cells (Dyce et al., 2004).
Growth and cell cycle analysis of porcine skin-derived stem cells
Freshly isolated SCC were cultured in vitro from passage 2 to passage 15 at an initial seeding density of 0.67 × 106 cells/mL. To determine the amount of time for cell population doubling the cells were passaged every 4 days by gentle mechanical dissociation and resuspension in fresh SSC medium, and we took two 100 μL at each passage and then counted the cell number by using a hemocytometer, the population doubling time was calculated. Porcine SSC were collected at passages 2, 5, 10, and 15, and fixed with ice-cold 70% ethanol overnight at −20°C. The samples were then centrifuged at 1500 rpm for 10 min, stained with PI Stain Master Mix, and the cell cycle data was collected by using a FAC-Scalibur Flow Cytometer and analyzed by using ModFit LT (Ormerod, 1992).
Morphology of porcine skin-derived stem cell
Live cell imaging
The images of live individual cells (at day 0) and spheres (at day 5) produced during culture were taken by using a Nikon TMS-F conventional light microscope (Nikon Inc., Japan) and edited by using Adobe Photoshop.
Epifluorescence microscopy
Individual spheres were transferred into a nine-well Pyrex plate and fixed in 2% formaldehyde as previously described (Sutovsky, 2004). Spheres were permeabilized for 40 min in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (Sigma) (PBST) and incubated with 5 μg/mL DAPI (Invitrogen-Molecular Probes, Carlsbad, CA) diluted in PBST. After a wash in PBST, spheres were mounted on microscopy slides in VectaShield antifade medium (Vector Lab, Burlingame, CA), and examined by using a Nikon Eclipse 800 epifluorescence-equipped light microscope. Images were acquired by a Cool-Snap HQ CCD camera (Roper Scientific, Tucson, AZ) operated by MetaMorph software (Universal Imaging Corp., Downington, PA) using appropriate epifluorescence filters, and edited by using Adobe Photoshop.
Electron microscopy
Spheres were fixed for 90 min in a mixture of 0.6% glutaraldehyde and 2% paraformaldehyde in cacodylate buffer, postfixed in 1% osmium tetroxide, dehydrated, and embedded in PolyBed 812. Ultrathin sections were cut on a Leica Ultracut UCT ultramicrotome, stained by uranyl acetate, and lead citrate, and photographed in a Jeol 1200 EX electron microscope. Photographs were scanned by Umax Magic Scan flat-bed scanner (Umax Technologies Inc., Fermont, CA) and edited by using Adobe Photoshop.
Gene transfection in porcine skin-derived stem cells
Identification of Genticin concentration for marker selection
To identify a minimum but sufficient concentration of genticin (G418, Gibco, Grand Island, NY) to select for stably transfected porcine SSC, two spheres of similar morphology were transferred into media containing 0, 25, 50, 75, 100, 125, or 150 μg/mL G418. For 15 days, media was changed every 3–4 days and cell viability and morphology was assessed and recorded. To assess the cell viability, the cells were stained with 0.4% trypan blue solution (Sigma, Lot # T8154) for 5 min, and counted the dead cell (blue) number and total cell number in the hemocytometer, and then calculated the cell viability (the percentage of the alive cells in total cells).
Production of green fluorescent protein-transfected skin stem cells
Porcine SSC were digested to produce a single cell population from spheres, washed with Opti-MEM (Invitrogen, Carlsbad, CA), and resuspended in transfection media (75% cytosalts (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4; pH 7.6, 5 mM MgCl2) and 25% Opti-MEM) at a concentration of 1 × 106 cells/mL. A combination of both supercoiled and linearized GFP vector containing a SV40 promoter driven neomycin selectable marker (Whitworth et al., 2009) was utilized to enable the evaluate transfection efficiency at 24 h (Ross et al., unpublished) and to promote the production of transfectants through negative selection by using 50 μg/mL G418. Supercoiled (12.5 μg/mL, to monitor transient transfection efficiency) and linearized GFP (12.5 μg/mL, to produce stable transfection SSC) vector DNA was added to 200 μL of cell suspension in transfection media and electroporated at 200 and 300 volts for 1 ms in a 2-mm cuvette. Because linearized DNA is degraded after a few days in culture if not integrated (Tsulaia et al., 2003), this process should provide an estimate of integration. Following electroporation, the cells were incubated at room temperature for 3–5 min. Transfected cells were diluted in 1300 μL SSC culture media, centrifuged at 500 × g for 5 min, and resuspended in 500 μL SSC culture medium, and split into two separate wells of a 48-well culture plate. Twenty-four hours later, cells from one well for each voltage were harvested, digested as described, and resuspended in 100-μL SSC culture media. Ten microliters were loaded onto a hemocytometer and cell concentration was determined, and GFP-expressing cells were identified and quantified. Cell survival is expressed as the percentage of cells relative to untransfected control cells while percent transfected was determined by the percentage of GFP expressing cells in the whole population of cells that survived. After 24 h cells from the other 48-well dish were cultured in the presence of G418 (200 μg/mL) to establish stably integrated SSC.
In vitro development of cloned embryos with porcine skin-derived stem cells
Collection of oocytes and in vitro maturation (IVM)
Prepubertal gilt ovaries were collected at a local abattoir. Cumulus–oocytes complexes (COCs) were aspirated from 3- to 6-mm diameter antral follicles by using a 10-mL disposable syringe with an 18-gauge needle. COCs with an evenly distributed cytoplasm and at least three compact layers of cumulus cells were selected and washed three times in TL HEPES supplemented with 0.1% (w/v) polyvinyl alcohol (PVA). Fifty to 70 oocytes were transferred into 500 mL of maturation medium (TCM-199; Gibco) that had been covered with mineral oil in a four-well multidish (Nunc, Roskilde, Denmark) per well. The TCM-199 was supplemented with PVA (0.1%), D-glucose (3.05 mM), sodium pyruvate (0.91 mM), cysteine (0.57 mM), luteinizing hormone (0.5 mg/mL), follicle stimulating hormone (0.5 mg/mL), epidermal growth factor (10 ng/mL), penicillin (75 mg/mL), and streptomycin (50 mg/mL). Oocytes were cultured for 38–44 h at 38.5°C under 5% CO2 in air, the maturation oocytes were used partly for IVF and SCNT at same time.
In vitro fertilization (IVF)
To compare development of SSC SCNTs to IVF-derived embryos cumulus-free metaphase II oocytes were derived after vortexing and were washed three times in IVF medium (Hao et al., 2007). Approximately 30–35 oocytes were transferred into 50-μL droplets of IVF medium covered with tissue culture grade mineral oil. One frozen semen pellet was thawed at 39°C in 10 mL sperm washing medium. After washing twice by centrifugation (1900 rpm, 4 min), cryopreserved ejaculated spermatozoa were resuspended with IVF medium to a concentration of 2 × 106 cells/mL. Fifty microliters of the sperm sample was added to the fertilization droplets containing the oocytes, giving a final sperm concentration of 1 × 106 cells/mL. Oocytes were coincubated with sperm for 6 h. At 6 h postinsemination, oocytes were washed three times and cultured in 500 μL of embryo culture medium in four-well Nunclon dishes (Nunc) at 38.5°C in 5% CO2 in air.
Nuclear transfer embryo development in vitro utilizing SSC as donor cells
To obtain donor cells for NT, porcine SSC at passage 2 were thawed and cultured 1 day before NT and then pipetted vigorously in PBS–EDTA to isolate individual cells. Enucleation and injection were carried out in TCM-199 supplemented with HEPES, 0.3% BSA, and 7.5 mg/mL of cytochalasin B at 38°C (Lai et al., 2002a). Denuded metaphase II oocytes were enucleated by aspirating the first polar body and neighboring cytoplasm in the micromanipulation medium with a glass injection pipette. Healthy cells were selected as donor cells and introduced into the perivitelline space of the enucleated recipient oocytes. The injected oocytes were placed between 0.2-mm diameter platinum electrodes for fusion. The medium used for fusion was low calcium medium (0.3 M mannitol, supplemented with 0.1 mM CaCl2, 0.1 mM MgCl2, and 0.5 mM HEPES). Two DC pulses (1 sec interval) of 1.2 kV/cm for 30 msec were applied, by using a BTX-Cell Manipulator 200 (BTX). The reconstructed embryos were activated with 200 μM thimerosal for 10 min, and then washed three times with TL-HEPES supplemented with 0.1% PVA. Embryos were transferred into 8 mM dithiothreitol (DTT) for 30 min. After washing three times they were cultured in PZM3 (108.0 mM NaCl, 10 mM KCl, 0.35 mM KH2PO4, 0.40 mM MgSO4 · 7H2O, 25.07 mM NaHCO3, 0.2 mM Na-pyruvate, 2.0 mM Ca · (lactate)2 · 5H2O, 1.0 mM L-Glutamine, 5.0 mM Hypotaurine, 20 mL/LBME amino acid soluation, 10 mL/L MEM (nonessential amino acids), with 0.3% BSA (Im et al., 2004).
Production of cloned pigs with SSC
Embryo transfer
Potential surrogate gilts were checked for estrus. Cloned embryos (105–113) that were cultured for 15–18 h and then were surgically transferred to the oviduct of four naturally cycling gilts on day 0 or day 1 of standing estrus. Surrogates were monitored daily for signs of estrus as well as weekly for pregnancy with a transabdominal ultrasound.
Microsatellite analysis
Parentage was analyzed in cloned piglets to confirm genetic identity of the piglets with the SSC used for NT by microsatellite analysis. Microsatellite marker primers were synthesized commercially with the 5′ end of the forward primer of each set labeled with one of the following fluorescent dyes: 6FAM™, VIC®, NED™, PET™ (Applied Biosystems, Foster City, CA). Each PCR reaction contained 30 ng of genomic DNA, 5 μL 10× GeneAmp Taq Buffer with 50 mM MgCl2 (Applied Biosystems), 0.2 mM of each dNTP (Roche, Indianapolis, IN), 1.25 U of FastStart Taq (Roche), 0.2–0.8 μM of each primer and was brought up to a total reaction volume of 50 μL with sterile deionized water. PCR amplification was performed in a GeneAmp PCR System (Applied Biosystems) with the following conditions: 1 cycle of 95°C, 4 min; 35 cycles of 94°C, 15 sec; 55°C, 2 min; 72°C, 2 min and 1 cycle of 72°C, 7 min. After PCR amplification, 0.5 μL of the PCR reaction was mixed with 0.5 μL GeneScan 500 LIZ Size S Standard (Applied Biosystems) and 9 μL deionized formamide (Applied Biosystems). These samples were transferred to 96-well plates, centrifuged for 2 min at 1000 rpm in an Allegra 6R Centrifuge (Beckman Coulter, Fullerton, CA), denatured at 95°C, 5 min in a thermal cycler, and quick chilled on ice until loaded. Analysis of the samples was performed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) using POP4 polymer (Applied Biosystems). Data was visualized and allele sizes were scored by using GeneMapper version 4.0 software (Applied Biosystems). All genotyping analysis was performed by the RADIL Genetic Testing Service Laboratory (Columbia, MO).
Statistical analysis
All dependent variables of population time and the percentage of the cell cycle phases were analyzed for normality by using the Shapiro-Wilk test (SAS Institute, Cary, NC). The cleavage rate and blastocyst rate were first arcsine transformed. Data for the function variables were analyzed by using the general linear model and Duncan multiple range tests of SAS. Data are expressed as least-squares (LS) mean ± SEM.
Results
Proliferation and cell cycle analysis of the porcine skin-derived stem cell
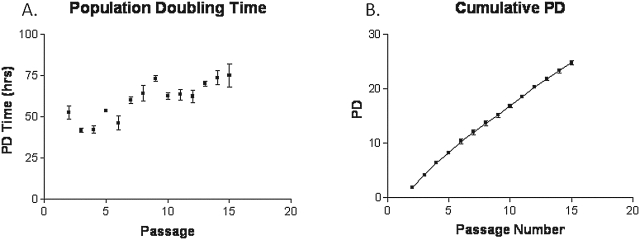
The population doubling (PD) time of the porcine SSC was 55–140 h, and the PD time was prolonged as the passage number increased (Fig. 1). The distribution of cells at G1/G0, S-phase, and G2/M phase of the cell cycle was similar between the early (P2) and later passages (P5; 87.5% vs. 87.8% at G1/G0; 9.2% vs. 10.2% at S; and 3.2% vs. 1.9% at G2/M phase; Table 1). At each passage three independent replications were tested. Each stem cell line originated from one porcine fetus.
FIG. 1.
(A) The population doubling time of the porcine SSC at different passages. (B) Growth curve of porcine SSCs. At each passage three independent replications were tested. Each stem cell line originated from one porcine fetus.
Table 1.
Cell Cycle Analysis of Porcine Skin-Derived Stem Cells
| Passage | % G1 | % S | % G2/M |
|---|---|---|---|
| 2 | 87.5 ± 0.4 | 9.2 ± 0.6a | 3.2 ± 0.2a |
| 5 | 86.1 ± 0.3 | 7.2 ± 0.3b | 6.7 ± 0.4b |
| 10 | 84.7 ± 2.6 | 7.9 ± 0.3ab | 7.4 ± 2.9ab |
| 15 | 87.8 ± 0.6 | 10.2 ± 1.1ab | 1.9 ± 0.5a |
Data are expressed as mean ± SEM from three experiments.
Within a column are different (p < 0.05).
Morphology of porcine skin-derived stem cell
Morphology of the porcine SSC spheres and isolated cells is shown in Figure 2 by light microscopy. They have a characteristic spherical shape. In Figure 3, sections and whole-mounted spheres are shown. Flat cells are on the periphery and cuboidal cells in the interior of the spheres, in some areas separated by a distinct cavity (Fig. 3A and B). The nuclei can be visualized in intact spheres with DAPI staining (Fig. 3C). Figure 3D shows the corresponding DIC image. Electron microscopy revealed two distinct cell types within the spheres, sometimes separated by a cavity (Fig. 3E). Toward the sphere periphery, mostly elongated cells with invaginated nuclear envelopes and prominent nucleoli were observed (Fig. 3F), often with extensive plasma membrane blebbing on their apical surface (Fig. 3G). These cells displayed few, but large elongated mitochondria with transversal cristae, as well as large cisternae of rough endoplasmic reticulum (Fig. 3H). The cells in the center of the spheres were predominantly round shaped, with a large round nucleus (Fig. 3I).
FIG. 2.
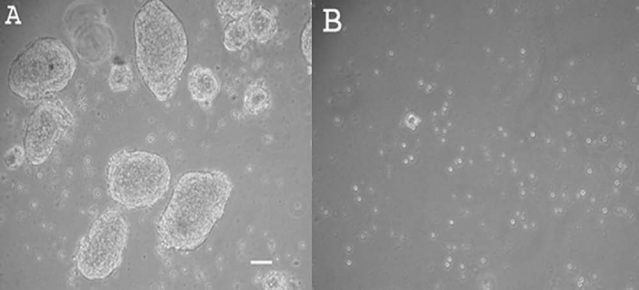
(A) Floating spheroids in suspension culture (original magnification ×100; The scale bar represents 25 μm). (B) Floating individual cells (original magnification ×40).
FIG. 3.
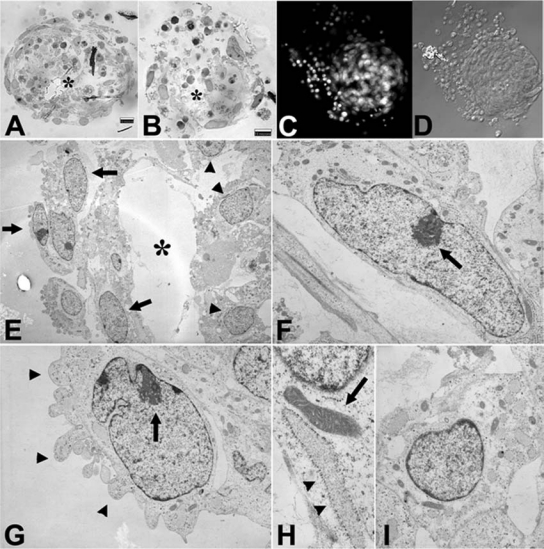
Morphology and ultrastructure of the skin stem cell spheres. (A,B) Histological cross-sections of the PolyBed-embedded spheres. Flat cells are on the periphery and cuboidal cells in the interior of the spheres, in some areas separated by a distinct cavity (asterisk). (C,D) Whole-mounted sphere labeled with DNA stain DAPI (C) and a corresponding DIC image (D). (E–I) Transmission electron micrographs showing distinct types of cell morphology in the spheres. (E) Low magnification of a sphere section. Note mostly flat cells on the periphery (arrows) and cuboidal/round cells in the center of the sphere (arrowheads), separated from the peripheral cells by a distinct cavity (asterisk). (F) An elongated peripheral cell with invaginated nuclear envelope and a prominent nucleolus (arrow). (G) Peripheral cell with extensive blebbing of apical plasma membrane (arrowheads), and large nucleolus (arrow) anchored at the inner face of the nuclear envelope. (H) Detail of an elongated mitochondrion (arrow) and large ER cistern (arrowheads) in a flat peripheral cells. (I) Cuboidal cell with a round-shaped nucleus.
Porcine SSC are capable of being transiently and stably transfected
The effect of G418 on the sphere morphology selection at different levels of G418 was tested. At day 1 of G418 selection, a few cells on the surface of the sphere were dead and separating from the rest of the sphere in the 150 and 200 μg/mL G418 groups. The number of degenerating cells increased over time, with most all cells appearing to be dead by 15 days of selection in 150 μg/mL G418 groups. Few cells in the 50 μg/mL group appeared to be dying by day 13 of G418 selection, but there were no dead cells at 25 μg/mL G418. After maintaining positive selection for 15 days, the morphology of the cells was normal in the 25 μg/mL G418 group (Fig. 4). Therefore, while 50 μg/mL G418 appears to be an appropriate dose of G418 for selection of the transfected SSC 200 μg/mL may also work.
FIG. 4.
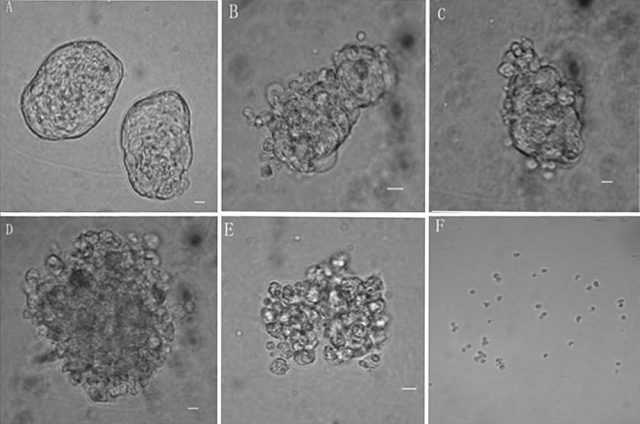
Selection of the SSC with different doses of G418. (A) An SSC sphere in 0 μg/mL G418; after 1 day of G418 selection, few cells on the surface of the sphere appear to be degenerating and separating from the sphere in 150 and 200 μg/mL G418 groups (C,D). Cells appeared to be dying at 15 days selection in 150 and 200 μg/mL G418 groups (E,F). A few cells in the 50 μg/mL group were dying at 13 days of G418 selection (B).
A single square-wave pulse of 200 or 300 volts was capable of producing transiently transfected cells as determined by GFP expression 24 h posttransfection (Table 2). Within 24 h following electroporation, mechanically dissociated cells expressed GFP as determined visually following excitation with ultraviolet light (Fig. 5). Addition of 200 μg/mL of G418 24 h later did not impede the formation of spheres 5 days posttransfection, and appeared to increase the number of cells expressing GFP. Maintaining positive selection with 200 μg/mL G418 for 21 days posttransfection selected for a population of predominately GFP expressing cells; however, cell morphology seemed to be somewhat affected compared to earlier in the selection process (Fig. 5).
Table 2.
Transfection Efficiency and Survival of Porcine SSCs (24 h Posttransfection)
| Voltagea | Total cellsb(× 104) | Green cellsc(× 104) | % Green cellsd(% transfection) | % Survivale |
|---|---|---|---|---|
| 0 | 38.5 | 0 | 0 | 100 |
| 200 | 23.5 | 4 | 17.02 | 61.0 |
| 300 | 12 | 1 | 8.33 | 31.2 |
Single 1-ms pulse was administered in a 2-mm cuvette.
Number of total cells 24 h following transfection.
The total number of cells visually determined to express GFP.
The number of green cells divided by the total cells within a voltage group.
Survival was determined by dividing the number of total cells in the electroporated group by the number of total cells in the untransfected control.
FIG. 5.
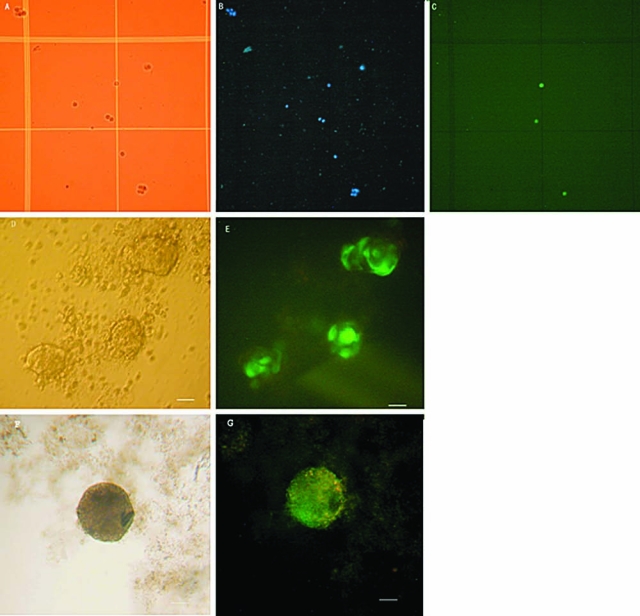
Transfection of porcine SSCs. (A,B,C) Transfected single cells at 24 h after tranfection with GFP (green: cells expressing GFP; blue: dead cell nuclei were stained with Trypan Blue; yellow: the cells under normal light, original magnification ×40.) (D,E) The skin-derived stem cell sphere expressing GFP at 5 days after G418 selection (original magnification ×100; the scale bar represents 25 μm). (F,G) SSCs expressing GFP on day 21 after G418 selection (original magnification ×400; the scale bar represents 25 μm).
Development in vitro of cloned embryos and production of cloned pigs
Although there was no difference in the percentage cleaved at 48 h between the IVF embryos and the cloned embryos (61.2 ± 3.1, 64.9 ± 8.2, respectively, N = 392), the cell number in the blastocyst stage embryos derived from cloning with the SSC was significantly higher than those of the blastocysts derived from IVF (28.5 ± 1.9, 16.8 ± 4.0, respectively, p < 0.05), although there was no significant difference in blastocyst formation rates between these groups (21–25%).
A total of 443 SSC cloned embryos were transferred to four surrogate gilts on day 1 of the estrous cycle. Three of the animals became pregnant; one did not deliver any offspring, one delivered two healthy male piglets via C-section, and the third one delivered one healthy female piglet via C-section (Table 3, Supplementary Figs. S1 and S2; see online supplementary material at www.liebertonline.com).
Table 3.
Embryo Transfer Data and Outcomes
| Donor cell ID | %Fusion | %Cleavage prior to ET | Surrogate ID | # Embryos transferred | Pregnancy | Date of delivery | Size of litter | Birth weight (Kg) |
|---|---|---|---|---|---|---|---|---|
| Male | 66.27 (112/169) | 10.48 (11/105) | O62 | 105 | + | 8/15/07 | 2 males | 1.12; 1.13 |
| Female | 70.22 (125/178) | 5.35 (6/112) | O97 | 112 | + | 9/8/97 | 1 female | 1.50 |
| Female | 77.55 (114/147) | 8.85 (10/113) | O83 | 113 | + | |||
| Female | 66.07 (111/168) | 7.97 (9/113) | O78 | 113 | − |
Microsatellite analysis
Microsatellite analysis of the male donor cells showed that 17 different markers were identical between the donor cells and the two male piglets. In addition, of these 17 markers, 9 were different from the surrogate. Microsatellite analysis of the female donor cells and the female piglet showed similarity for 8 markers, and of those, 5 were different from her surrogate (Supplementary Table S1).
Pathological examination of the cloned piglets
There were no gross abnormalities in any of the cloned piglets. Details of the examination are described in the Supplementary Table S1.
Discussion
Birth of cloned piglets after SCNT with the skin-derived stem cell nuclei in this study demonstrated that the porcine SSC are clonable. Porcine SSC may be suitable donor cells for the generation of genetically modified pigs by NT because the SSC proliferated rapidly in vitro, retained a normal karyotype after long-term culture, and it was possible to stably transfect these cells. Fetus-derived fibroblast cells can be used for genetic modification followed by cloning, but unfortunately, they generally undergo senescence after about 30 population doublings. Thus, a stem cell line that can proliferate rapidly, is amenable to genetic modification and is clonable, may prove to be very useful for the production of transgenic, knockout, and knock-in pigs for agriculture and medicine.
It has been suggested that stem cell-type cells (e.g., undifferentiated cells or partially differentiated cells) would require less nuclear remodeling and reprogramming by the oocyte compared to somatic cells, and thus may result in a higher percentage of development after NT as well as animals that have fewer phenotypic abnormalities. This concept has been confirmed by using mouse ES cells where multiple groups found that up to 20% of transferred blastocyst stage embryos derived from ES cells survive to term, compared with less than 3% of cumulus cell-derived blastocysts, and that ES cells supported term development with an efficiency 3–10 times higher than differentiated somatic cells (Eggan et al., 2001; Rideout et al., 2000; Wakayama et al., 1999). Magnani et al. (2008) suggested that the transcriptional abundance of four of six Sucrose NonFementing 2 (SNF2)-type ATPases was higher in the SSC compared to the fibroblast cells. SNF2-type ATP-dependent chromatin factors contribute to epigenetic reprogramming, and the relative amount of these factors in the donor cells may affect developmental potential of the reconstructed embryos. Several studies on cloned embryos using various types of partially differentiated donor cells indicated an improvement in the preimplantation development of the cloned embryos compared to when somatic cells were used, although there has yet been a comparison of the cloning efficiency using ES and somatic cells as donors in livestock species. Tomii et al. (2005) reported 20.8% blastocyst rate when preadipocytes were used as donors compared to 12.3% with fetal-derived fibroblast cells as the donor. A similar improvement was reported from 1.2% with fetal-derived fibroblast cells to 4.1% with porcine mesenchymal stem cells (Bosch et al., 2006), and 21 to 25% with SSC (Zhu et al., 2004) and 11.6 to 23.5% with olfactory bulb progenitor cells (Magnani et al., 2008). Unfortunately, few of these studies produced offspring to determine if the cells were clonable. In this study we report 21% blastocyst rate, and 75% of the surrogates establishing a pregnancy with 50% of them producing live offspring. Although the overall efficiency is not high (three offspring from 443 embryos transferred), the results show that the SSC can be cloned.
In conclusion, we report that pigs can by cloned by using SSC as donor cells. Porcine SSC may be a superior resource to provide donor nuclei for genetic modification by NT because they may require lesser nuclear reprogramming by the oocyte to result in normal embryo development, and they do not undergo senescence as rapidly as somatic cells. Porcine SSC are easy to isolate and available in large number (400–1000 spheres/cm2 of skin). Unlike ES cells, the porcine SSC proliferate well without a feeder layer. During long-term (5 months) in vitro culture, the porcine SSC maintain a normal karyotype, and are primarily in the G0/G1 stage of the cell cycle. It is also possible to genetically modify the SSC. Reconstructed embryos with SSC showed good pre- and postimplantation development.
Supplementary Material
Acknowledgments
We thank Dr. Clay Isom, Dr. Eric Walters, Kristin Whithworth, and Dr. Elizabeth Bryda for assistant in Microsatellite analysis, Dr. Scott Korte for newborn piglet recovery and examination of their pathology, Lonnie Dowell for managing the surrogates, and everyone in the Prather laboratory for help with the surgery, feeding, and caring for the pigs. Finally we thank the staff of Electron Microscopy Core, University of Missouri, for TEM sample processing. This work was supported in part by a grant from the NIH R01-RR13438-10 and intramural funding from the Food for the 21st Century program of the University of Missouri.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Bosch P. Pratt S.L. Stice S.L. Isolation, characterization, gene modification, and nuclear reprogramming of porcine mesenchymal stem cells. Biol. Reprod. 2006;74:46–57. doi: 10.1095/biolreprod.105.045138. [DOI] [PubMed] [Google Scholar]
- Cho S.K. Kim J.H. Park J.Y, et al. Serial cloning of pigs by somatic cell nuclear transfer: restoration of phenotypic normality during serial cloning. Dev. Dynam. 2007;236:3369–3382. doi: 10.1002/dvdy.21308. [DOI] [PubMed] [Google Scholar]
- Dyce P.W. Zhu H. Craig J, et al. Stem cells with multi-lineage potential derived from porcine skin. Biochem. Biophys. Res. Commun. 2004;316:651–658. doi: 10.1016/j.bbrc.2004.02.093. [DOI] [PubMed] [Google Scholar]
- Eggan K. Akutsu H. Loring J, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y. murphy C. Spate L.D, et al. Osteopontin improves in vitro development of porcine embryos and decreases apoptosis. Mol. Reprod. Dev. 2007;75:291–298. doi: 10.1002/mrd.20794. [DOI] [PubMed] [Google Scholar]
- Im G.-S. Lai L. Liu Z, et al. In vitro development of preimplantation porcine nuclear transfer embryos cultured in different media and gas atmospheres. Theriogenology. 2004;61:1125–1135. doi: 10.1016/j.theriogenology.2003.06.006. [DOI] [PubMed] [Google Scholar]
- Lai L.X. Kolber-Simonds D. Park K.W, et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002a;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- Lai L.X. Park K.W. Cheong H.T, et al. Transgenic pig expressing the enhanced green fluorescent protein produced by nuclear transfer using colchicine-treated fibroblasts as donor cells. Mol. Reprod. Dev. 2002b;62:300–306. doi: 10.1002/mrd.10146. [DOI] [PubMed] [Google Scholar]
- Magnani L. Lee K. Fodor W.L, et al. Developmental capacity of porcine nuclear transfer embryos correlate with levels of chromatin-remodeling transcripts in donor cells. Mol. Reprod. Dev. 2008;75:766–776. doi: 10.1002/mrd.20818. [DOI] [PubMed] [Google Scholar]
- Ormerod M.G. Flow cytometry: a practical approach. 2nd. New York: Oxford University Press; 1992. [Google Scholar]
- Park K.W. Cheong H.T. Lai L.X, et al. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Anim. Biotechnol. 2001;12:173–181. doi: 10.1081/abio-100108344. [DOI] [PubMed] [Google Scholar]
- Prather R.S. Targeted genetic modification: xenotrans-plantation and beyond. Cloning Stem Cells. 2007;9:17–19. doi: 10.1089/clo.2006.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather R.S. Sims M.M. First N.L. Nuclear transplantation in early pig embryos. Biol. Reprod. 1989;41:414–418. doi: 10.1095/biolreprod41.3.414. [DOI] [PubMed] [Google Scholar]
- Prather R.S. Shen M. Dai Y. Genetically modified pigs for medicine and agriculture. Biotechnol. Genet. Eng. Rev. 2008;25:245–266. doi: 10.7313/upo9781904761679.011. [DOI] [PubMed] [Google Scholar]
- Rideout W.M. Wakayama T. Wutz A, et al. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet. 2000;24:109–110. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- Sutovsky P. Visualization of sperm accessory structures in the mammalian spermatids, spermatozoa and zygotes. In: Schattens H., editor. Methods in Molecular Biology, vol 253: Germ Cell Protocols: Sperm and Oocyte Analysis. Humana Press; Totowa, NJ: 2004. pp. 59–77. [DOI] [PubMed] [Google Scholar]
- Tomii R. Kurome M. Ochida T, et al. Production of cloned pigs by nuclear transfer of preadipocytes established from adult mature adipocytes. Cloning Stem Cells. 2005;7:279–288. doi: 10.1089/clo.2005.7.279. [DOI] [PubMed] [Google Scholar]
- Tsulaia T.V. Prokopishyn N.L. Yao A, et al. Glass needle-mediated microinjection of macromolecules and trans-genes inot primary human mesenchymal stem cells. J. Biomed. Sci. 2003;10:328–336. doi: 10.1007/BF02256452. [DOI] [PubMed] [Google Scholar]
- Wakayama T. Rodriguez I. Perry A.C.F, et al. Mice cloned from embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:14984–14989. doi: 10.1073/pnas.96.26.14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth K. Li R. Spate L.D, et al. A comparison of three fusion/activation methods used to create enhanced green fluorescent protein pigs by somatic cell nuclear transfer. [208];Mol Reprod. Dev. 2009 [Google Scholar]
- Zhu H. Craig J.A. Dyce P.W, et al. Embryos derived from porcine skin-derived stem cells exhibit enhanced pre-implantation development. Biol. Reprod. 2004;71:1890–1897. doi: 10.1095/biolreprod.104.032227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.