Abstract
Founder mutations, particularly 35delG in the GJB2 gene, have to a large extent contributed to the high frequency of autosomal recessive nonsyndromic hearing loss (ARNSHL). Mutations in transmembrane channel-like gene 1 (TMC1) cause ARNSHL. The p.R34X mutation is the most frequent known mutation in the TMC1 gene. To study the origin of this mutation and determine whether it arose in a common ancestor, we analyzed 21 polymorphic markers spanning the TMC1 gene in 11 unrelated individuals from Algeria, Iran, Iraq, Lebanon, Pakistan, Tunisia, and Turkey who carry this mutation. In nine individuals, we observed significant linkage disequilibrium between p.R34X and five polymorphic markers within a 220 kb interval, suggesting that p.R34X arose from a common founder. We estimated the age of this mutation to be between 1075 and 1900 years, perhaps spreading along the third Hadramaout population movements during the seventh century. A second founder effect was observed in Turkish and Lebanese individuals with markers in a 920 kb interval. Screening for the TMC1 p.R34X mutation is indicated in the genetic evaluation of persons with ARNSHL from North African and Southwest Asia.
Introduction
Hearing loss is the most common sensory disorder worldwide. In children, mutation in a single gene is a major cause, and many different responsible genes have been identified. Most frequently the disorder is nonsyndromic and of autosomal recessive inheritance. Founder effects have been reported for some of the common mutations associated with autosomal recessive nonsyndromic hearing loss (ARNSHL) (Abe et al., 2000; Dreyer et al., 2001; Borck et al., 2003; Ouyang et al., 2003; Yan et al., 2003; Rodríguez-Ballesteros et al., 2008; Joseph and Rasool, 2009). Some ancient founder mutations, such as 35delG in the GJB2 gene, have been described in many parts of the world (Gasparini et al., 2000; Van Laer et al., 2001; RamShankar et al., 2003; Belguith et al., 2005).
Twenty-nine different mutations of the transmembrane channel-like gene 1 (TMC1) gene (GenBank accession number: NM_138691.2) have been reported in 48 families with ARNSHL (Kurima et al., 2002; Kalay et al., 2005; Meyer et al., 2005; Santos et al., 2005; Kitajiri et al., 2007; Hilgert et al., 2008; Tlili et al., 2008; Sirmaci et al., 2009). The most common recessive mutation for hearing loss in the TMC1 gene is p.R34X. This nonsense mutation results from a T-to-C transition at position 100 from the first ATG (c.100C → T) and is located in exon 7. It accounts for over 30% of mutant alleles of TMC1 and occurs in populations throughout Asia and North Africa (Kurima et al., 2002; Kitajiri et al., 2007; Hilgert et al., 2008; Tlili et al., 2008; Sirmaci et al., 2009). The high frequency of the p.R34X mutation in Tunisian and Pakistani populations has been related to a common ancestral founder (Kitajiri et al., 2007; Tlili et al., 2008).
In this study, we performed haplotype analysis in unrelated hearing-impaired individuals carrying the p.R34X mutation originating from Algeria, Iran, Iraq, Lebanon, Pakistan, Tunisia, and Turkey. We analyzed polymorphic markers both within and flanking the coding sequence of the TMC1 gene and found two distinct p.R34X haplotypes in these populations. We have estimated the age of the more prevalent haplotype to be between 1075 and 1900 years.
Materials and Methods
Subjects
The study was performed on 156 unrelated individuals with ARNSHL from seven countries in North Africa and Asia. Among this cohort, 11 individuals carry the p.R34X mutation (Table 1). Blood samples were collected from each subject after written informed consent.
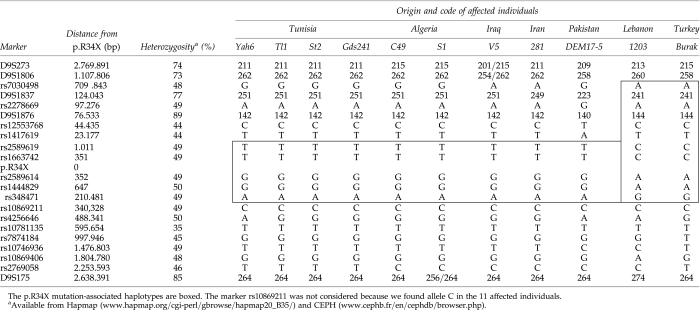
Table 1.
Haplotypes Associated with the p.R34X Alleles in 11 Unrelated Patients of Geographically Diverse Origins
Mutation analysis
For specific detection of the p.R34X mutation, primers 5′-AAGCACTTTCTGACATTACTC ATTG-3′ and 5′-TGGAACTTTTGAAAGAATATCAGA-3′ were used to amplify a 250-bp fragment including exon 7 of the TMC1 gene. After polymerase chain reaction (PCR), the samples were restriction digested with the enzyme TaqI (Jena Bioscience, Munich, Germany) followed by electrophoresis on a 3% agarose gel. Digestion cleaves the normal allele into two fragments (79 and 171 bp), whereas the mutant allele remains uncleaved (250 bp).
Genotyping
To examine haplotypes associated with the p.R34X mutation, genotyping was performed for 5 microsatellite markers (D9S273, D9S1806, D9S1837, D9S1876, and D9S175) and 16 single-nucleotide polymorphisms (SNPs) (rs7030498, rs2278669, rs12553768, rs1417619, rs2589619, rs1663742, rs2589614, rs1444829, rs348471, rs10869211, rs4256646, rs10781135, rs7874184, rs10746936, rs10869406, and rs2769058) flanking the TMC1 gene. Positions and average heterozygosity of the markers are listed in Table 1. For the microsatellite markers, heterozygosity varied from 73% to 89% (www.cephb.fr/en/cephdb/browser.php). For SNPs, it varied from 35% to 50% (www.hapmap.org/cgi-perl/gbrowse/hapmap20_B35/). PCR for the microsatellite markers was performed using fluorescently labeled forward primers. Genotyping was done using an ABI 3100 Genetic Analyzer and analysis was performed using Genotyper (version 3.5). The SNPs were genotyped by sequencing using Big Dye Terminator Sequencing V3.1 Kit (Applied Biosystems, Foster City, CA).
Estimation of the age of p.R34X mutation
Calculation of the age of the p.R34X mutation was performed using a C program developed by Genin et al. (2004). When compared with other methods using haplotype information, this approach is efficient with a very small number of affected individuals. Results are presented as the mean number of generations (with empirical 95% confidence interval) estimated over 5000 replicates.
Results
Using PCR–restriction fragment length polymorphism, the screening of the p.R34X mutation was performed in 72 Tunisian, 54 Algerian, and 22 Pakistani unrelated cases with ARNSHL. Two Algerian and one Pakistani patients carrying the p.R34X mutation were detected. In addition to these three cases, eight hearing-impaired subjects carrying the p.R34X mutation and originating from Tunisia (n = 4), Iraq (n = 1), Iran (n = 1), Lebanon (n = 1), and Turkey (n = 1) were included into this study (Scott et al., 1996; Hilgert et al., 2008; Tlili et al., 2008; Sirmaci et al., 2009).The p.R34X mutation in all family probands from these populations was confirmed by direct sequencing.
To explore whether the high frequency of the p.R34X mutation of TMC1 in North African and Asian populations is the result of a founder effect or a mutational hot spot, we searched for evidence of a shared common haplotype of p.R34X with flanking polymorphisms. We analyzed 5 microsatellite markers and 16 SNPs in all probands. In the four Tunisian individuals, an identical haplotype was identified with marker loci D9S273 to rs348471 (centromeric to telomeric) (Table 1). The two Algerians shared 13 of 14 alleles with the Tunisians, while the Iraqi, Iranian, and Pakistani individuals shared 12 of 14, 11 of 14, and 7 of 14 alleles with the Tunisians, respectively (Table 1). The common haplotype in these individuals spans the 220 kb interval between rs2589619 and rs348471. For the two families from Turkey and Lebanon, a second haplotype was observed within the 920 kb interval between rs7030498 and rs348471 (Table 1).
Using the method of Genin et al. (2004), we estimated the age of the p.R34X mutation in the first haplotype to be about 43 generations when the marker mutation rate is set at 10−3. Assuming that one generation is 25 years, this corresponds to 1075 years. When marker mutations are set at 10−6, the estimated age is 76 generations corresponding to about 1900 years.
Discussion
Mutations in TMC1 are a common cause of ARNSHL in India, Pakistan, Tunisia, and Turkey where they account for the hearing-loss phenotype in 3–6% of families (Kurima et al., 2002; Kalay et al., 2005; Santos et al., 2005; Kitajiri et al., 2007; Hilgert et al., 2008; Tlili et al., 2008; Sirmaci et al., 2009). Of the 29 reported mutations in this gene, the p.R34X mutation is the most frequent and accounts for over 30% of all TMC1 ARNSHL-causing mutations. This mutation has been reported in hearing-impaired persons originating from Iran, Iraq, Lebanon, Pakistan, Tunisia, and Turkey (Scott et al., 1996; Kurima et al., 2002; Kitajiri et al., 2007; Hilgert et al., 2008; Tlili et al., 2008; Sirmaci et al., 2009), and we have also identified this mutation in Algeria. Its detection in normal control samples of African-American and northern European origin raises the probability that p.R34X is a prevalent contributor to the genetic load of hearing loss in multiple world populations (Kitajiri et al., 2007).
High carrier rates for recessive mutations are associated with high rates of consanguinity and endogamy (Ben Arab et al., 2004), thereby conserving the haplotype flanking those mutations. Consistent with this founder effect, the p.R34X mutation has been reported in 10 Pakistani families where it segregates on a common haplotype (Kitajiri et al., 2007). A common haplotype has also been observed in four Tunisian families segregating the p.R34X mutation (Tlili et al., 2008).
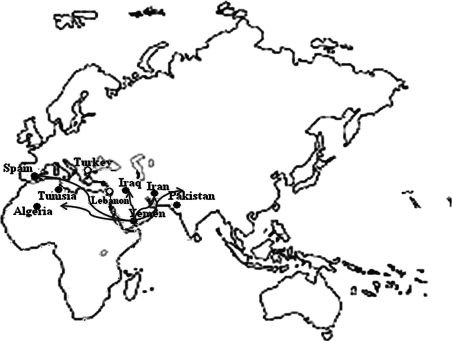
In this study, we analyzed 21 polymorphic markers surrounding the p.R34X mutation in 11 unrelated individuals from Algeria, Iran, Iraq, Lebanon, Pakistan, Tunisia, and Turkey and identified two disease-associated haplotypes. The hypothesis that the mutation arose a very long time ago in a single individual was not considered. In fact, we detected two different haplotypes at 350 bp from the p.R34X mutation. The first haplotype, a 220 kb interval flanked by markers rs2589619 and rs348471, was detected in nine of 11 individuals from Algeria, Iran, Iraq, Pakistan, and Tunisia. It is the first time that a common haplotype was described in individuals from these different countries. The Algerian, Iraqi, Iranian, and Pakistani haplotypes shared 13 of 14, 11 of 14, 10 of 14, and 6 of 14 alleles, respectively, with the Tunisian founder haplotype. This result shows that there is correlation between genetic diversity and geographic distance from Tunisia. The small chromosomal interval of the first haplotype is consistent with an ancient origin of a single founder mutation. The age of the mutation in this haplotype was estimated to be between 1075 and 1900 years. Possibly, the mutation was spread throughout these countries along the third Hadramaout population movement in the seventh century (Fig. 1). Hadramaout, a historical region of the South Arabian Peninsula, was the focal point for the origins and development of the Islamic faith in the seventh century (Hussain, 2005). The two families originating from Turkey and Lebanon segregate a second haplotype of 920 kb defined by markers rs7030498 and rs348471. The mutation may have spread in the Ottoman Empire that lasted from 1299 to 1922 when many countries including Lebanon and Turkey were part of this empire.
FIG. 1.
Distribution of hearing-impaired patients carrying the p.R34X mutation. Dots indicate the origin of patients presenting the p.R34X mutation. Arrows correspond to the main Hadramaout population movement routes during the seventh century.
In conclusion, our study shows that the p.R34X mutation in TMC1 in North African and Asian individuals arose from at least two different founders. Screening for this mutation should be included in the evaluation of North African and Asiatic persons segregating ARNSHL. Its detection would facilitate genetic counseling in these populations.
Acknowledgments
This work was supported by the Ministère de l'Enseignement Supérieur, la Recherche Scientifique et la Technologie, Tunisia, and the European Commission FP6 Integrated Project EUROHEAR, LSHG-CT-2004-512063. R.J.H.S. is the Sterba Hearing Research Professor, University of Iowa College of Medicine. This project was supported by a grant from the National Institutes of Health–National Institute on Deafness and Other Communication Disorders (RO1 DCOO2842, to R.J.H.S.).
Disclosure Statement
No competing financial interests exist.
References
- Abe S. Usami S. Shinkawa H, et al. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet. 2000;37:41–43. doi: 10.1136/jmg.37.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguith H. Hajji S. Salem N, et al. Analysis of GJB2 mutation: evidence for a Mediterranean ancestor for the 35delG mutation. Clin Genet. 2005;68:188–189. doi: 10.1111/j.1399-0004.2005.00474.x. [DOI] [PubMed] [Google Scholar]
- Ben Arab S. Masmoudi S. Beltaief N, et al. Consanguinity and endogamy in Northern Tunisia and its impact on non-syndromic deafness. Genet Epidemiol. 2004;27:74–79. doi: 10.1002/gepi.10321. [DOI] [PubMed] [Google Scholar]
- Borck G. Roth C. Martiné U, et al. Mutations in the PDS gene in German families with Pendred's syndrome: V138F is a founder mutation. J Clin Endocrinol Metab. 2003;88:2916–2921. doi: 10.1210/jc.2002-021334. [DOI] [PubMed] [Google Scholar]
- Dreyer B. Tranebjaerg L. Brox V, et al. A common ancestral origin of the frequent and widespread 2299delG USH2A mutation. Am J Hum Genet. 2001;69:228–234. doi: 10.1086/321269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini P. Rabionet R. Barbujani G, et al. High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet. 2000;8:19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- Genin E. Tullio-Pelet A. Begeot F, et al. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–449. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N. Alasti F. Dieltjens N, et al. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74:223–232. doi: 10.1111/j.1399-0004.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain R. The effect of religious, cultural and social identity on population genetic structure among Muslims in Pakistan. Ann Hum Biol. 2005;32:145–153. doi: 10.1080/03014460500075167. [DOI] [PubMed] [Google Scholar]
- Joseph AY. Rasool TJ. High frequency of connexin26 (GJB2) mutations associated with nonsyndromic hearing loss in the population of Kerala, India. Int J Pediatr Otorhinolaryngol. 2009;73:437–443. doi: 10.1016/j.ijporl.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Kalay E. Karaguzel A. Caylan R, et al. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26:591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- Kitajiri S. McNamara R. Makishima T, et al. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007;72:546–550. doi: 10.1111/j.1399-0004.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- Kurima K. Peters LM. Yang Y, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Meyer CG. Gasmelseed NM. Mergani A, et al. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25:100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- Ouyang XM. Hejtmancik JF. Jacobson SG, et al. USH1C: a rare cause of USH1 in a non-Acadian population and a founder effect of the Acadian allele. Clin Genet. 2003;63:150–153. doi: 10.1046/j.0009-9163.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- RamShankar M. Girirajan S. Dagan O, et al. Contribution of connexin26 (GJB2) mutations and founder effect to non-syndromic hearing loss in India. J Med Genet. 2003;40:e68. doi: 10.1136/jmg.40.5.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ballesteros M. Reynoso R. Olarte M, et al. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008;29:823–831. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- Santos RL. Wajid M. Khan MN, et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat. 2005;26:396. doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA. Carmi R. Elbedour K, et al. An autosomal recessive nonsyndromic-hearing-loss locus identified by DNA pooling using two inbred Bedouin kindreds. Am J Hum Genet. 1996;59:385–391. [PMC free article] [PubMed] [Google Scholar]
- Sirmaci A. Duman D. Oztürkmen-Akay H, et al. Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. Int J Pediatr Otorhinolaryngol. 2009;73:699–705. doi: 10.1016/j.ijporl.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Tlili A. Rebeh IB. Aifa-Hmani M, et al. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol. 2008;13:213–218. doi: 10.1159/000115430. [DOI] [PubMed] [Google Scholar]
- Van Laer L. Coucke P. Mueller RF, et al. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet. 2001;38:515–518. doi: 10.1136/jmg.38.8.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D. Park HJ. Ouyang XM, et al. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum Genet. 2003;114:44–50. doi: 10.1007/s00439-003-1018-1. [DOI] [PubMed] [Google Scholar]