Summary
BLM, the protein product of the gene mutated in Bloom syndrome, is one of five human RecQ helicases. It functions to separate double Holliday junction DNA without genetic exchange as a component of the “dissolvasome”, which also includes topoisomerase IIIα and the RMI (RecQ-mediated genome instability) subcomplex (RMI1 and RMI2). We describe the crystal structure of the RMI core complex, comprising RMI2 and the C-terminal OB domain of RMI1. The overall RMI core structure strongly resembles two-thirds of the trimerization core of the eukaryotic single-strand DNA-binding protein, Replication Protein A. Immunoprecipitation experiments with RMI2 variants confirm key interactions that stabilize the RMI core interface. Disruption of this interface leads to a dramatic increase in cellular sister chromatid exchange events similar to that seen in BLM-deficient cells. The RMI core interface is therefore crucial for BLM dissolvasome assembly and may have additional cellular roles as a docking hub for other proteins.
Introduction
Bloom syndrome (BS) is a rare autosomal recessive genetic disorder characterized by proportional dwarfism, sun sensitivity, an increased susceptibility to infections and diabetes, and a high occurrence of most types of cancer (Bachrati and Hickson, 2003; German, 1993). BS patients are often first diagnosed with cancer in their mid-twenties, which is the typical cause of death. Cells derived from persons with BS show a remarkable increase in chromosomal rearrangements, gaps, and breaks. The hallmark feature of BS cells, which is used in diagnosis, is a ten-fold elevated frequency of sister chromatid exchanges (SCEs). The gene mutated in BS, BLM, encodes one of five human RecQ DNA helicases. Two other RecQ-linked genetic diseases are caused by defects in WRN and RecQ4, causing Werner’s syndrome and Rothmund-Thomson syndrome, respectively. All three of these genetic disorders are characterized by increased genomic instability and cancer predisposition, highlighting the importance of RecQ proteins in genome maintenance (Bachrati and Hickson, 2003).
RecQ DNA helicases are ubiquitously conserved enzymes that catalyze the ATP-dependent unwinding of DNA with a preference for DNA structures resembling replication and recombination intermediates such as replication forks, Holliday junctions, D-loops, and G-quadruplexes (Ouyang et al., 2008). For the BLM protein, these activities drive multiple cellular activities. BLM is important in homologous recombination (HR)-dependent DNA repair, with roles in branch migration (Bachrati et al., 2006; Karow et al., 2000; van Brabant et al., 2000), RAD51 filament disruption (Bugreev et al., 2007), and in restarting stalled DNA replication processes (Ralf et al., 2006). For several of these reactions, BLM is thought to function as a component of a larger protein complex termed the “dissolvasome”, in which it partners with topoisomerase IIIα (Top3α) and the RMI (RecQ-mediated genome instability) subcomplex, comprised of RMI1 and RMI2 (Raynard et al., 2006; Singh et al., 2008; Wu et al., 2006; Xu et al., 2008; Yin et al., 2005).
The dissolvasome specifically acts on double Holliday junction intermediates in HR, creating non-crossover products in a process termed “dissolution” (Wu et al., 2006; Wu and Hickson, 2003). This is of particular interest both for the normal function of the dissolvasome in HR and for BS pathology: since only non-crossover products are formed, a defect in this process could explain the increase in SCEs in Bloom syndrome. Dissolution activity is proposed to occur through branch migration by BLM, which creates a hemicatenated structure that can then be resolved by Top3α (Wu and Hickson, 2003). RMI1 stimulates this process (Wu et al., 2006) and the more recently discovered RMI2 is thought to provide stability to the complex (Singh et al., 2008; Xu et al., 2008).
Sequence analysis of RMI1 and RMI2 indicates that the proteins have three oligonucleotide/oligosaccharide binding (OB) folds: two in RMI1 (OB1 and OB2) and one in RMI2 (OB3) (Figure 1A). RMI1 OB1 interacts with BLM and Top3α, whereas OB2 interacts with OB3 from RMI2 (Xu et al., 2008). Interestingly, the OB2 and OB3 domains both share sequence homology with domains found within Replication Protein A (RPA), the heterotrimeric eukaryotic single-stranded DNA binding protein (Xu et al., 2008). RPA contains six OB folds, which are also known as DNA binding domains (DBDs) because of their importance in ssDNA binding. A subset of these domains in RPA is responsible for the assembly of RPA heterotrimers, with one OB domain from each RPA subunit (RPA70 DBD-C, RPA32 DBD-D, and RPA14) contributing to a core trimerization structure. RPA trimerization is primarily driven by formation of a parallel bundle of α-helices from the OB domains (Bochkarev and Bochkareva, 2004; Bochkareva et al., 2002). RMI OB2 and OB3 domains share significant sequence similarity with two of the three trimerization domains from RPA (RPA70 DBD-C and RPA32 DBD-D, respectively), but whether the RMI complex interface was similar to RPA was not known.
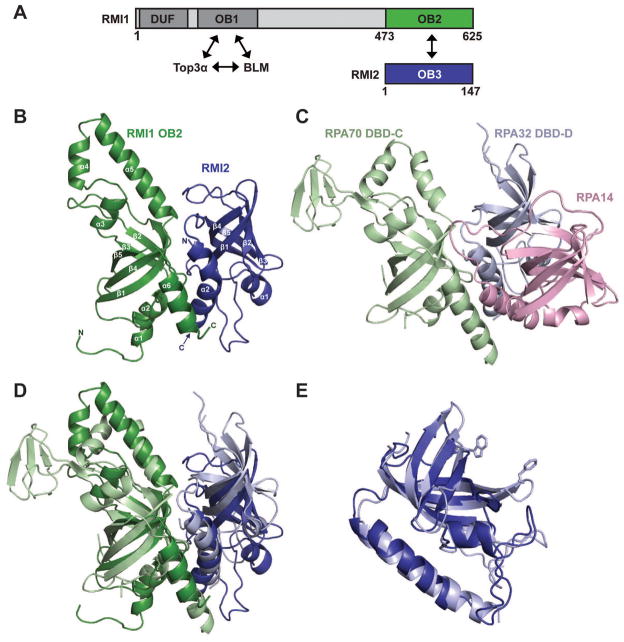
Figure 1. Structural features of the RMI core complex.
(A) Schematic diagram of the domain structures of human RMI1 and RMI2. Interacting OB domains comprising the RMI core complex are shown in green (RMI1 OB2) and blue (RMI2 OB3). Additional RMI1 domains (domain of unknown function, DUF, and an OB domain that interacts with Top3α and BLM (OB1)) are indicated. (B) Ribbon diagram of the crystal structure of the RMI core complex. RMI1 OB2 (green, residues 475-625), and RMI2 (blue, residues 17-147) are rendered using PyMol (Delano, 2002). (C) Ribbon diagram of the crystal structure of the RPA trimerization core (Bochkareva et al., 2002) shown with RPA70 in the same orientation as RMI1 OB2 in (B). RPA70 DBD-C (pale green), RPA32 DBD-D (pale blue), and RPA14 (pink) are shown. (D) Superposition of RMI1 OB2 with RPA70 DBD-C. The color scheme and orientation are presented as in (B) and (C). (E) Superposition of RMI2 with RPA32 DBD-D (Deng et al., 2007). DNA-stacking side chains are shown for RPA32 DBD-D. There are no analogous aromatic residues in RMI2. See also figure S1.
Here we describe the high-resolution X-ray crystal structure of the human RMI core complex, composed of RMI1 OB2 and RMI2. Similarly to the arrangement observed in RPA, the RMI core complex associates via OB folds that are flanked by C-terminal α-helices that form a helical bundle. This overall structure resembles two-thirds of the RPA trimerization core (RPA70 DBD-C/RPA32 DBD-D), but the specific interactions at the interface are very different. The RPA interface mostly consists of hydrophobic interactions between C-terminal helices, whereas the RMI interface is much larger and includes electrostatic interactions as well as an extensive hydrophobic interface. We have identified residues critical to the RMI interface and show that disruption of the RMI subcomplex leads to an overall increase in SCEs in vivo. These findings point to a primary role for the RMI core complex in stabilizing the BLM dissolvasome and also hint at possible functions for the core as a hub for docking additional cellular factors.
Results
Defining the RMI core complex
We initially set out to investigate the structure of full-length human RMI1 in complex with RMI2 (the RMI subcomplex). However, isolated RMI1 and RMI1 in complex with RMI2 were prone to proteolytic degradation, making either a poor crystallographic target. Limited proteolysis was used to define a stable complex that would be more appropriate for crystallographic trials. The RMI subcomplex was purified and subjected to limited proteolytic treatment with subtilisin, trypsin, or α-chymotrypsin and then analyzed by SDS-PAGE. The stability of RMI2 was remarkable, as it was almost completely resistant to all three proteases, while RMI1 was rapidly degraded by each of the proteases. Trypsin treatment produced a fragment migrating near the 16.5-kDa marker, which appeared after 5 minutes and was stable throughout the experiment (Figure S1). Mass spectrometric analysis determined that this fragment included the C-terminus of RMI1 (residues 473-625), which comprises the C-terminal RMI1 OB-fold (OB2) (Figure 1A). This protease-resistant complex consisting of RMI1 OB2 and RMI2 defined a minimal RMI core complex. The RMI core complex was co-expressed and purified for subsequent structural and biochemical experiments. The complex eluted with 1:1 stoichiometry in size-exclusion chromatographic analysis (Figure S1) and trypsin proteolysis yielded no degradation, consistent with formation of a stable minimal heterodimeric RMI core.
Crystal structure of the RMI core complex
The RMI core complex was crystallized and its structure was determined to 1.55-Å resolution using single wavelength anomalous dispersion to phase a selenomethionine-substituted variant of the complex (Table 1). The refined structure contains residues 475-625 of RMI1 and residues 17-147 of RMI2. Electron density defining the N-terminus of RMI2 was missing, which could indicate that this region of the domain is structurally dynamic. The same N-terminal segment is not well conserved among RMI2 homologs (Xu et al., 2008). The RMI core complex structure was refined with good bond geometry and crystallographic statistics (Table 1).
Table 1.
X-ray data collection and structure determination statistics
| Native | SeMet | |
|---|---|---|
| Data Collection | ||
| Wavelength, Å | 0.97856 | 0.97933 |
| Resolution Range (high resolution bin), Å | 30-1.55 (1.58-1.55) | 50-2.14 (2.18-2.14) |
| Space Group | P212121 | P212121 |
| Unit Cell (a, b, c (Å)) | 42.10, 42.22, 152.24 | 41.92, 42.03, 152.87 |
| (α, β, γ(°)) | 90, 90, 90 | 90, 90, 90 |
| Completeness, % | 97.0 (83.0) | 99.9 (99.6) |
| Total/Unique Reflections | 423,921/39,281 | 213,642/15,791 |
| Redundancy | 10.8 (5.3) | 13.5 (12.6) |
| <I/σI> | 25.7 (2.4) | 21.5 (20.2) |
| Rsym†, % | 11.5 (37.8) | 10.0 (20.8) |
| Phasing | ||
| Figure of Merit | 0.756 | |
| Refinement | ||
| Resolution, Å | 30-1.55 | |
| Rwork/Rfree, ‡ % | 19.5/23.1 | |
| Rms deviations | ||
| Bonds, Å | 0.008 | |
| Angles, ° | 1.20 | |
| Ramachandran statistics, % | ||
| Most favored | 93.8 | |
| Allowed | 5.8 | |
| Generously allowed | 0.4 | |
| Disallowed | 0.0 | |
| # atoms | ||
| protein | 2269 | |
| solvent | 259 | |
| <B factor>, Å2 | ||
| protein | 17.4 | |
| solvent | 28.9 | |
Rsym = ΣΣj|Ij − <I>|ΣIj, where Ij is the intensity measurement for reflection j and <I> is the mean intensity for multiply recorded reflections.
Rwork/Rfree = Σ||Fobs| − |Fcalc||/|Fobs|, where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections (5% of the total) were held aside throughout refinement.
Within the RMI core complex, both the RMI1 OB2 domain and RMI2 fold to form OB domains that are flanked by additional helical elements (Figure 1B). The canonical OB fold is defined by a five-stranded β-barrel structure that is often supported by an α-helix present in the region between the third and fourth strands (Murzin, 1993). The RMI1 OB2 domain has each of these elements along with two additional helices between strands 3 and 4 and a C-terminal helix. These helices are structurally similar to elements that are present in RPA, as is described below. RMI2 lacks the canonical OB-fold helix but clearly forms the core β-barrel OB-fold structure with an N-terminal helix that packs against the β-barrel RMI2 core. As is the case with the RMI1 OB2 domain, RMI2 also has an additional C-terminal helix that is similar to a helix found in RPA.
Comparison of the RMI core complex and the RPA trimerization core
Structural comparison of the RMI core complex to other OB-domain proteins revealed a striking similarity with the core trimerization domain of RPA, the major eukaryotic single-strand DNA-binding protein. The RPA trimerization core interface is formed among OB folds present in RPA70, RPA32, and RPA14, with C-terminal α-helices that flank the core OB folds. These helices form a parallel trihelical bundle that provides a large portion of the interface (Bochkareva et al., 2002) (Figure 1C). In a similar fashion, the interface between RMI1 and RMI2 is stabilized by contacts between the two C-terminal α-helices, which align in parallel. A least-squares alignment (Holm et al., 2008) of RPA70 DBD-C with the RMI1 OB2 domain correspondingly aligns RMI2 with RPA32 DBD-D, with the two C-terminal helices aligning analogously (Figure 1D). The RPA70 DBD-C and RMI1 OB2 domains align with a root mean square deviation (rmsd) of 3.2 Å for 122 common Cα atoms, which includes structurally similar helical motifs positioned in the loop between β-strands 3 and 4 (L34 loop) of both proteins. The RMI1 and RPA70 L34 loops are distinct, however, as the helices in RMI1 L34 extend further from the core OB domain than those from RPA70. A second major difference between these two domains is that the loop connecting β-strands 2 and 3 (L23 loop) in RPA70 DBD-C contains a zinc ribbon motif whereas the corresponding loop in RMI1 lacks this motif. This RPA70 zinc-binding region is thought to contribute to DNA binding in RPA (Bochkareva et al., 2002; Lao et al., 1999; Pestryakov et al., 2003).
Least-squares alignment of RMI2 with RPA32 DBD-D, which is the most similar protein to RMI2 in the Protein Data Bank, show that the two proteins align with a rmsd of 2.2 Å for 112 common Cα atoms (Figure 1E). Interestingly, RMI2 also shares significant structural similarity with S. pombe Stn1N (2.5 Å for 115 common Cα atoms) and Ten1 (2.3 Å for 102 common Cα atoms) which, along with Cdc13, form a heterotrimeric RPA-like complex called CST involved in telomere maintenance (Gao et al., 2007; Sun et al., 2009). The CST complex is conserved in humans as Stn1, Ten1, and Ctc1, and is thought to assemble through an RPA-like clustering of OB domains (Miyake et al., 2009; Surovtseva et al., 2009).
There are two key functional differences between the RMI and RPA complexes that appear to be reflected in their respective core structures. The first is that RPA binds DNA whereas previous studies indicate that RMI does not (Xu et al., 2008). Accordingly, the aromatic residues used for DNA-binding in RPA are not conserved in the structure of the RMI core complex (Figure 1E). The second major difference is that the RMI core complex exists as a heterodimer, but the RPA core is a heterotrimer (Figures 1B and 1C). Whether a third protein also docks against the RMI core complex to complete a heterotrimer is not yet known.
The RMI core complex interface is distinct from RPA
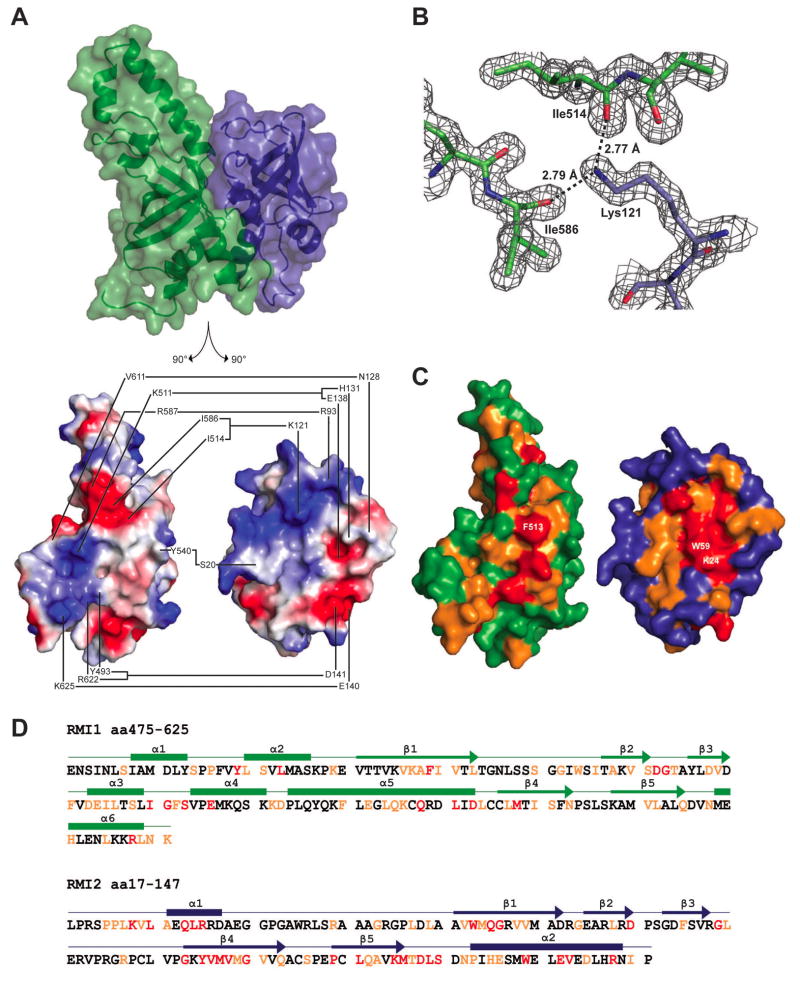
The RMI interface is large, burying 33 residues of RMI1 OB2 and 36 residues of RMI2, which together corresponds to a buried surface area of 2610 Å2 (computed using PISA (Krissinel and Henrick, 2007)). Surface electrostatic representation of the structure reveals the complexity of the interface, as hydrophobic, basic, and acidic surfaces on RMI1 and RMI2 are all involved in complex formation (Figure 2). Lys121 of RMI2 has been previously posited to play a role at the interface (Singh et al., 2008; Xu et al., 2008), and indeed it forms apparent interactions with carbonyl groups from Ile514 and Ile586 of RMI1 in the core complex structure (Figure 2B). A second important ionic interaction is formed between the side chains of RMI2 Asp141 and RMI1 Arg622 (Figure S2), both of which are in the C-terminal helices of the proteins. Other contributing residues from this helix in RMI2 are Glu140, which interacts with RMI1 Lys625, and residues His131 and Glu138, which interact with Lys511 of RMI1. Additionally, Asn128, which is adjacent to the RMI2 C-terminal α-helix interacts with the carbonyl oxygen of Val611 of RMI1. There are two other potentially important interfacing residues away from the C-terminus: the carbonyl oxygen of RMI2 Ser20 interacts with the Tyr540 side chain of RMI1, and the main chain amide of RMI2 Arg93 interacts with the RMI1 Asp587 side chain. Finally, two highly conserved RMI2 residues that have been previously mutated and shown to disrupt complex formation (Lys24 and Trp59) were located in the center of the RMI core complex interface (Figure 2C) (Singh et al., 2008). Trp59 forms a hydrophobic network in the interface, stacking between Phe513 from RMI1 and the hydrophobic portion of the Lys24 side chain from RMI2 at the center of the interface. Notably, the residues that comprise the RMI core complex interface are evolutionarily well conserved (Figure 2C and 2D), indicating that the overall arrangement of subunits with the core observed in the structure is likely to be representative of most RMI complexes.
Figure 2. RMI core complex interface.
(A) Top: structure of the RMI core complex showing semi-transparent surface and ribbon representations in the same orientation and color scheme as in 1B. Bottom: split view of the RMI core complex in which both proteins are separated and rotated 90° to show interacting surfaces. The interface between RMI1 and RMI2 is shown with connected lines indicating residues forming hydrogen bonds or ionic interactions. The interface is depicted using surface electrostatics, showing electropositive (blue) and electronegative (red) surface potential. (B) 2Fo − Fc electron density map (1.5 σ) showing interaction of RMI1 Ile514 and Ile586 with RMI2 Lys121. (C) The interface between RMI1 and RMI2 is shown in the same orientation as 2A, bottom. RMI1 and RMI2 are colored to indicate residues that are invariant (red), highly conserved (orange), or poorly conserved (green for RMI1 and blue for RMI2) among identified RMI1 and RMI2 proteins from human, bovine, mouse, zebrafish, and arabidopsis sequences. Three of the invariant residues that form a hydrophobic interaction network are labeled: RMI1 Phe513, RMI2 Lys24, and RMI2 Trp59. (D) The sequence for the RMI core complex is shown, with secondary structural features labeled above as boxes (α-helices) and arrows (β-strands). Red and orange lettering indicates invariant and highly conserved residues, respectively. See also Figure S2.
Although the overall arrangement of RMI and RPA OB domains is similar, the size and chemical composition of the interfaces are distinct. In contrast to the large RMI interface (2610 Å2), the corresponding RPA interface (between RPA70 DBD-C and RPA 32 DBD-D) is much smaller (900 Å2), with fewer amino acids from each OB fold contributing to the interaction. In addition, the RMI interface is composed of many electrostatic and hydrophobic interactions, whereas the RPA interface is dominated by hydrophobic contacts (Bochkareva et al., 2002). Furthermore, the residues that make up the RPA trimerization core are primarily from the C-terminal α-helices that extend from the RPA OB domains, whereas those composing the RMI core complex are from both the OB domains and the C-terminal helices.
Coimmunoprecipitation experiments demonstrate the importance of RMI core complex interface
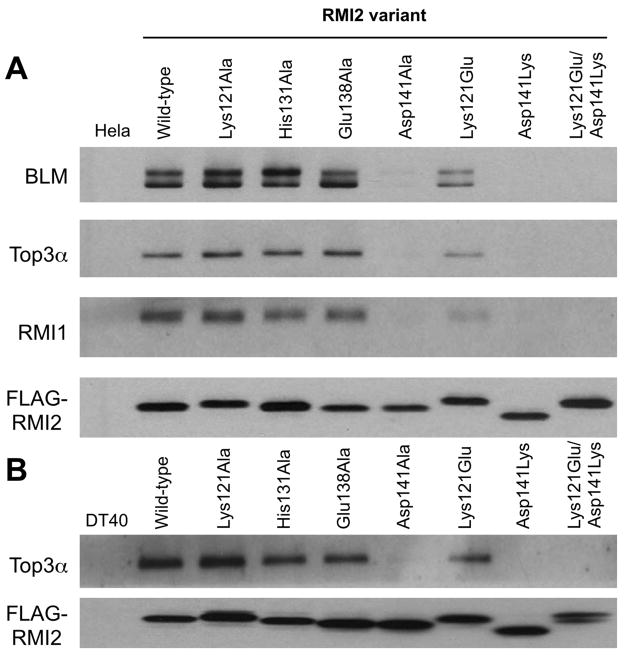
Coimmunoprecipitation experiments were used to test the importance of the identified interface of the RMI core complex in assembly of the BLM dissolvasome. Based on the structural insights described above, expression constructs were produced that encode FLAG-epitope labeled single-site variants of RMI2 that are predicted to destabilize the complex. These RMI2 variants were then transiently transfected into HeLa cells and the integrity of the dissolvasome was measured by immunoprecipitating the RMI2 variants and probing for the presence of other dissolvasome members (BLM, Top3α, and RMI1) by Western analysis.
Initial experiments tested RMI2 variants in which Lys121, His131, Glu138, or Asp141 were individually changed to alanine. In agreement with earlier studies, BLM, Top3α, and RMI1 coimmunoprecipitate with wild-type FLAG-RMI2 (Xu et al., 2008) (Figure 3A). Interestingly, when the RMI2 alanine variants were used in the assay, only the Asp141Ala variant showed a major defect in coimmunoprecipitation with BLM, Top3α and RMI1. Given the large interface in the RMI core complex structure, we reasoned that alanine-substitution variants may not be sufficient to disrupt the interface in all cases. To test this idea, charge reversal RMI2 variants Lys121Glu, Asp141Lys, and a variant that included both changes were tested in the coimmunoprecipitation assay. With these more dramatic changes, coimmunoprecipitation was either greatly diminished (Lys121Glu RMI2 variant) or entirely eliminated (Asp141Lys and double-reversal variants) (Figure 3A). These results thus support a structural model in which Lys121 and Asp141 are key residues at the interface between RMI1 and RMI2.
Figure 3. Coimmunoprecipitations of the dissolvasome complex components with RMI2 variants.
(A) Western analysis of dissolvasome complex components (BLM, Top3α, RMI1, and FLAG-RMI2) from immunoprecipitates of FLAG-tagged RMI2 transiently expressed in HeLa cells. (B) Western analysis of dissolvasome complex components (Top3α and FLAG-RMI2) from immunoprecipitates of FLAG-tagged RMI2 stably expressed in chicken DT40 cells. RMI1 and BLM were not detected in DT40 cells because the antibodies raised against human proteins fail to recognize the chicken homologs. See also Figure S3.
Disrupting the RMI core complex interface increases SCE formation in vivo
The consequences of destabilizing the RMI core complex interface on dissolvasome function in vivo were tested next. Disruption of the RMI2 gene in chicken DT40 cells leads to a ~5-fold increase in SCE formation that can be suppressed by expressing human RMI2 protein in the mutant cells (Xu et al., 2008). As was observed in HeLa cells, experiments in DT40 cells verified that immunoprecipitation of the human RMI2 protein coprecipitated a member of the dissolvasome (Top3α) whereas RMI core complex-destabilizing RMI2 variants failed to do so (Figure 3B). These controls allow the use of the DT40 system to analyze the chromosomal effects of the RMI2 interface variants.
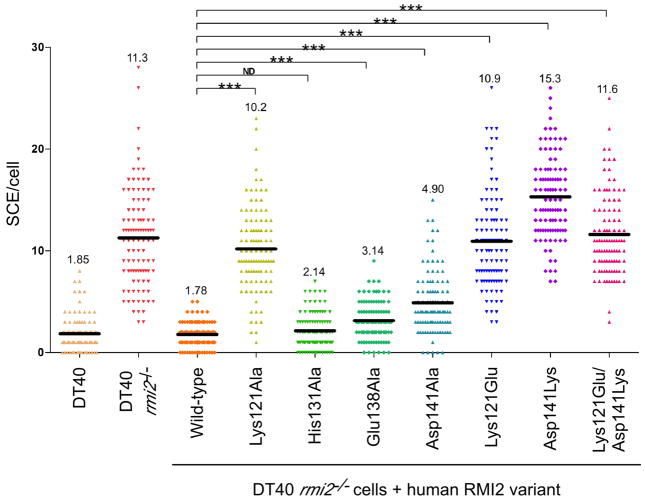
Assays were next performed to test the effects of RMI2 variants on SCE levels in vivo. As was observed previously (Xu et al., 2008), the mean number of SCE per cell increased ~6-fold in the rmi2−/− cells compared to that of the wild type DT40 cells, but this difference was suppressed by expression of the FLAG-tagged human RMI2 protein (Figure 4). Consistent with retention of the RMI subcomplex, the His131Ala RMI2 variant showed no significant increase in the mean number of SCEs/cell when compared to wild type RMI2. Similarly, the Glu138Ala RMI2 variant, which also retained complex integrity, showed only a slight increase in mean SCEs/cell (less than 2-fold). In contrast, drastic changes in the mean number of SCEs/cell were observed with the Lys121 and Asp141 RMI2 variants. The Asp141Ala and Asp141Lys variants had 2.8- and 8.6-fold increases, respectively. The Lys121Ala, Lys121Glu, and Lys121Glu/Asp141Lys mutations all had 5- to 6-fold increases in mean SCEs/cell relative to wild type RMI2-complemented experiments. These values are very similar to the SCE levels of rmi2−/− cells. With one exception (Lys121Ala), these data parallel the coimmunoprecipitation results; RMI2 variants that failed to assemble with the dissolvasome led to increases in cellular SCE events. Folding defects or aberrant RMI core complex formation by the Lys121Ala RMI2 variant were ruled out as possible explanations for its unusual behavior. The circular dichroic spectrum of the isolated Lys121Ala RMI2 variant showed that the protein was folded, and an X-ray crystal structure of the variant in complex with the RMI1 OB2 domain was nearly indistinguishable from that of the wild type RMI core complex except for the Lys-to-Ala alteration (Figure S3). While the possibility still exists that there may be differences in thermodynamic stability between the two proteins, the effects of the Lys121Ala RMI2 variant appear to indicate a role for the Lys121 residue beyond RMI complex stabilization.
Figure 4. SCE frequencies observed with different RMI2 variants.
The number of SCEs per cell in chicken DT40 cells are shown as a histogram in which each point in a column is the number of SCE events in a single cell. The mean level of SCEs/cell is displayed as a dark black line and numerically at the top of the column. Statistical significance is indicated with *** (p<0.001) and ND (p>0.05). “DT40” denotes wild type DT40 cells, “rmi2−/−” denotes DT40 cells lacking RMI2, “wild-type” denotes rmi2−/− cells complemented with human RMI2 and the remainder denote the human RMI2 variants that have been substituted. See also Figure S3.
Discussion
RMI1 and RMI2 define a subcomplex that is an integral component of the BLM dissolvasome (Singh et al., 2008; Xu et al., 2008; Yin et al., 2005). The RMI subcomplex is stabilized by binding of the C-terminal OB fold of RMI1 (OB2) to RMI2, which itself comprises an OB fold (OB3) (Figure 1A) (Singh et al., 2008; Xu et al., 2008). Here we report the 1.55 Å-resolution X-ray crystal structure of the RMI core complex, revealing a large interface that mediates RMI1/RMI2 heterodimer formation. This interface has been verified using coimmunoprecipitation experiments in HeLa and DT40 cells. Furthermore, disruption of the RMI interface was shown to be linked to a profound increase of SCEs in DT40 cells. Our data support a model in which the RMI core complex interface is critical for stabilizing the BLM dissolvasome, because RMI2 destabilizing mutations at the RMI interface lead to increased SCE levels similar to those observed in cells lacking RMI2 altogether. Additional possible roles for the RMI core complex domains in higher order protein complex assembly are also supported as described below.
Comparison of the RMI core complex and the RPA trimerization core
It has been suggested previously that the RMI OB domains would resemble those from RPA (Xu et al., 2008). Consistent with this hypothesis, the overall structures of the RMI OB domains are strikingly similar to those of the RPA OB domains, and the two complexes assemble using analogous surfaces (Figure 1). Interestingly, in spite of this overall similarity, the RMI core complex and the RPA trimerization core interfaces also display significant differences. First, while the RPA interface is primarily hydrophobic in nature, the RMI interface consists of a network of hydrogen bonds and salt bridges in addition to hydrophobic interactions. Second, while nearly all of the interactions within the RPA interface are mediated by residues in the α-helices C-terminal to the OB domains, significant contacts between the RMI core subunits are contributed both by the OB core domains and the C-terminal helices. This difference extends the buried surface area in the RMI core complex relative to that of RPA. These distinctions could contribute to the particular functions of the two complexes.
One important functional difference between RPA and RMI is that the RMI subcomplex lacks DNA binding activity. Previous work has shown that isolated human and yeast RMI1 bind DNA weakly in vitro (Chen and Brill, 2007; Mullen et al., 2005; Raynard et al., 2008; Wu et al., 2006), but that the RMI subcomplex is unable to bind DNA (Xu et al., 2008). Accordingly, the RMI core complex lacks analogous DNA-binding residues found in RPA, as the surface-exposed aromatic residues that might stack with ssDNA bases are buried in the RMI core complex interface. Exposure of these interface residues in the isolated RMI1 could explain its previously observed weak DNA binding (Raynard et al., 2008).
Perhaps the most noticeable difference between the RMI and RPA core structures is the lack of a third partner in the RMI subcomplex. RPA forms a heterotrimeric complex that, in addition to the RPA70 and RPA32, includes RPA14 (Figure 1C). In contrast, the RMI core complex lacks a third partner to complete a possible trimeric structure. Since the RMI subcomplex further associates with other proteins, it is tantalizing to speculate that another protein could dock onto the RMI core structure in a similar manner to that of RPA14 binding RPA70/RPA32. Possibilities include BLM or Top3α, which comprise the remaining dissolvasome members. However these proteins have been shown to associate with an N-terminal OB domain (OB1) fragment of RMI1 and in the absence of RMI2 (Raynard et al., 2008). A second possibility is that the RMI1 OB1 domain or some other OB-fold-containing protein might bind to the RMI core complex; such an interaction could potentially regulate dissolvasome complex formation or activation. A final, non-exclusive possibility is that other proteins could form complexes with the dissolvasome by docking onto the “open” site on the RMI core complex. These could include FANCM, for example, which has recently been shown to be able to bind to the RMI1 OB2 domain (Deans and West, 2009). Future experiments will be required to determine whether the heterodimeric RMI core complex described here is able to associate with an additional protein partner (or partners) in an RPA-like assembly.
Importance of RMI subcomplex interface in suppressing SCE levels
Based on the RMI core complex structure, RMI2 variants that have lost the ability to assemble with RMI1 and the rest of the dissolvasome were shown to lead to an increase in SCE levels in vivo. RMI2 variants Asp141Ala, Lys121Glu, Asp141Lys, and Lys121Glu/Asp141Lys either fail to assemble with the dissolvasome or do so with diminished stability (Figure 3) and lead to 2.8- to 8.6-fold increase in mean SCE events/cell compared to wild-type RMI2 (Figure 4). In contrast, RMI2 variants His131Ala and Glu138Ala retain the ability to assemble with the dissolvasome and have more modest increases in mean SCE events/cell of 1.2- and 1.8-fold, respectively. These data thus define a relationship between destabilization of the RMI subcomplex and an increase in cellular SCE events, and indicate a role for the RMI proteins in stabilization and/or activation of the dissolvasome within cells.
The Lys121Ala RMI2 variant presented an intriguing exception to the parallel between RMI subcomplex destabilization and elevated SCE levels. This variant retained a normal ability to assemble into the dissolvasome (Figure 3) but had mean SCE/cell levels that were nearly the same as the rmi2−/− control (Figure 4). Indeed, even the Lys121Glu charge reversal variant retained a weakened ability to assemble within the dissolvasome. Similar observations for RMI2 Lys121Ala variants have been noted in the past (Singh et al., 2008; Xu et al., 2008). What could account for this behavior? One possibility is that the Lys121 RMI2 variants are misfolded or that they form aberrant complexes with RMI1 that have an altered interface. However solution studies and a crystal structure of the Lys121Ala RMI2 in complex with RMI1 OB2 domain rule out this explanation, at least in vitro (Figure S3). Another possibility is that the observed difference is caused by a change in the rates of association or dissociation of the RMI core complex that cannot be detected by the coimmunoprecipitation assay. It is possible that the dissolvasome could be regulated by this key interaction through the dynamics of association instead of a constant equilibrium state. A final hypothesis is that Lys121 from RMI2 is important for functions beyond RMI complex stabilization. Notably, RMI2 Lys121 is positioned on the periphery of the RMI1/RMI2 interface and its interaction with two carbonyl groups in RMI1 appears to contort the Lys121 side chain away from the surrounding solvent (Figure S3). It is possible that in the context of the full dissolvasome Lys121 assumes an alternate conformation in which its charged group is presented on the surface of the RMI subcomplex to foster interactions with other cellular binding partners. Such a possibility could explain the dispensability of RMI2 Lys121 for dissolvasome formation but its clear importance in cellular activity. Furthermore, the Lys121 surface is on the opposite side of the RMI core complex from the putative RPA-like interaction site, which could indicate that the complex has two protein interaction sites and could possibly act as a hub for docking multiple protein partners.
Concluding Remarks
The structure of the RMI core complex and analysis of RMI protein variants has revealed the importance of RMI subcomplex integrity in genome stability. It is interesting that not only are RMI and RPA both complexes that interact via OB folds, but both complexes also mediate BLM function. RPA physically interacts with BLM and stimulates its ability to unwind DNA (Brosh et al., 2000). Furthermore, there are other OB-fold-containing complexes with roles in genome maintenance such as the CST complex and TPP-POT1, both of which act at telomeres. The CST complex has been proposed to form an RPA-like complex specifically at telomeres (Miyake et al., 2009; Surovtseva et al., 2009), and a recent S. pombe Stn1-Ten1 structure has also identified a similarity to the RPA interface (Sun et al., 2009). This and other studies point to a growing number of genome maintenance roles for OB-fold-containing proteins that extend beyond the ssDNA-binding activities for which the domain is named.
Experimental Procedures
Plasmids, Protein Overexpression, and Purification
Purification of the RMI subcomplex
A plasmid pCDFDuet-1 (Novagen) encoding RMI1 and RMI2 (Xu et al., 2008) was transformed into Rosetta 2 (DE3) E. coli transformed with pLysS (Novagen). Cells were grown at 30°C in Luria Broth supplemented with 25 μg/ml streptomycin and 50 μg/ml chloramphenicol. At an OD600 of ~0.6 cells were induced to overexpress protein with 0.3 mM isopropyl β-D-thiogalactopyranoside at ~25°C for 3 hours. Cells were harvested with centrifugation, frozen at −80°C, and resuspended at 4°C in PBS (10 mM phosphate, pH 7.4, 138 mM NaCl and 2.7 mM KCl) with 5% glycerol, 1 mM phenylmethanesulfonylfluoride (PMSF), and protease-inhibitor tablets (Roche). All subsequent purification steps were performed at 4°C. Cells were lysed by sonication and centrifuged for 30 min at 15,000 rpm. Ammonium sulfate (35% w/v) was added to the soluble lysate and the resulting pellet was resuspended in PBS with 5% glycerol, then incubated with Ni-NTA resin (Qiagen) for 45 min. The resin was poured into a column and washed twice with PBS containing 10 mM imidazole and 500 mM NaCl. The RMI subcomplex eluted in 300 mM imidazole in TBS buffer (50 mM Tris-HCl, pH 7.5, 5% glycerol, 150 mM NaCl). The eluate was diluted 1:5 in Q buffer (25 mM Tris-HCl, pH 7.5, 5% glycerol) and loaded onto a HiPrep Q FF column (Amersham). The bound protein was eluted with a linear (0.1 M to 0.5 M) NaCl gradient. The peak fractions were used for limited proteolysis.
Purification of the RMI core complex
Open-reading frames encoding RMI1 OB2 and RMI2 (or an RMI2 variant) were separately subcloned from the above plasmid into a modified pGEX4T-1 plasmid (Amersham) that expresses an amino-terminal glutathione S-transferase (GST) tagged RMI1 OB2 and amino-terminal 6x-histidine tagged RMI2. This plasmid was transformed into Rosetta 2 (DE3) E. coli cells transformed with pLysS (Novagen) and grown at 30°C in LB medium supplemented with 100 μg/ml ampicillin and 50 μg/ml chloramphenicol. At an OD600 of ~0.6 cells were induced to overexpress protein with 0.5 mM isopropyl β-D-thiogalactopyranoside at 30°C for 3 hours. Cells were harvested with centrifugation, frozen at −80°C, and resuspended at 4°C in lysis buffer (50 mM Tris-HCl pH 7.5, 20% sucrose, and 600 mM KCl) supplemented with 1 mM PMSF and protease-inhibitor tablets (Roche). All subsequent purification steps were performed at 4°C. Cells were lysed by sonication and centrifuged for 30 min at 15,000 rpm. The soluble lysate was incubated with Ni-NTA resin (Qiagen) for 30 min, then poured into a column and washed twice with wash buffer (25 mM Tris-HCl pH 7.5, 500 mM NaCl, 10 mM imidazole, 100 mM dextrose, and 10% glycerol). The complex was eluted with 300 mM imidazole in TBS buffer, then incubated with Glutathione Sepharose 4B resin (Amersham) for 40 min. The resin was then poured into a column, washed twice with GS4B buffer (25 mM Tris-HCl pH 7.5, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 10% glycerol) with 500 mM KCl, and eluted with GS4B buffer supplemented with 150 mM KCl and 20 mM reduced glutathione. The eluate was incubated with thrombin overnight to remove both affinity tags, leaving Gly-Ser-His peptides on the amino terminus of both proteins. The eluate was dialyzed against SPFF buffer (20 mM MES pH 6.0, 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol) and then loaded on a HiPrep SP FF column (Amersham). The bound protein was eluted with a linear (0.05 M to 0.50 M) NaCl gradient. Fractions containing the complex were pooled, concentrated to <2 ml, then further purified on a Sephacryl S-100 column in SPFF buffer. Selenomethionine-incorporated protein was expressed as previously described (Van Duyne et al., 1993) and purified identically to native protein. Analytical size-exclusion chromatography was performed on with a Sephacryl S-200 column in SPFF buffer.
Limited Proteolysis Assay
The purified RMI subcomplex was incubated at a 150:1 molar ratio (complex:protease) with subtilisin, trypsin, or α-chymotrypsin in 25 mM Tris-HCl pH 8, 8 mM DTT, 1 mM EDTA, 150 mM NaCl, and 10% glycerol. Proteolysis was terminated after 1, 5, 15, and 30 minutes by adding 2X sample buffer (12.5 mM Tris-HCl pH 6.8, 20% glycerol, 5% w/v SDS, 10% β-mercaptoethanol, bromophenol blue) and freezing on dry ice. The products were analyzed by 15% SDS-PAGE and mass spectrometry at the UW Biotechnology Center.
Protein Crystallization and Structure Determination
Wild type RMI core complex or a RMI core complex formed with Lys121Ala RMI2 was dialyzed against water. Selenomethionine-incorporated RMI core complex was dialyzed against 10 mM HEPES pH 7.1, 1 mM DTT. Core complex samples (5 mg/ml) were mixed with mother liquor (10 mM Hepes pH 7.1, 0.05 M NaSCN, 20% ethylene glycol, 2 mM DTT, and 13% polyethylene glycol 3350) at a 1:1 (vol) ratio, and benzamidine (1%) was added to the solution. Crystals were formed by hanging drop vapor diffusion. Crystals were transferred to a cryoprotectant solution (10mM Hepes pH 7.1, 0.05 M NaSCN, 30% ethylene glycol, 2mM DTT, 15% polyethylene glycol 3350, 1% benzamidine) and flash-frozen in liquid nitrogen. The highest quality crystals were obtained by microseeding with native crystals.
The structure of the selenomethionine-incorporated RMI core complex was determined by single-wavelength anomalous dispersion (SAD) phasing. Data were indexed and scaled using HKL2000 (Otwinowski and Minor, 1997). Selenium positions were identified using SHELX within Crank (Ness et al., 2004) and an initial structure was built using ARP/wARP (Perrakis et al., 1999). The structure was improved by rounds of manual fitting using Coot (Emsley and Cowtan, 2004) and refinement against the native data set using REFMAC5 (Winn et al., 2001). The structure of the RMI core complex that includes RMI2 Lys121Ala variant was determined by molecular replacement using Phaser (McCoy et al., 2007). The two proteins in the RMI core complex were positioned as individual search models so that any differences in packing would be revealed. Refinement was limited to positioning the RMI core complex subunits. Coordinate and structure factor files have been deposited in the Protein Data Bank (PDB ID 3MXN).
Coimmunoprecipitation and SCE assays
FLAG-tagged RMI2 variants were transiently expressed in Hela cells or stably expressed in chicken DT40 rmi2−/− cells. Nuclear extract was prepared as described previously (Guo et al., 2009) for coimmunoprecipitation. Coimmunoprecipitation experiments were performed using the anti-FLAG M2-agarose (Sigma) as described (Guo et al., 2009). Polyclonal antibodies for Western blotting against BLM, Topo 3α, RMI1 or RMI2 were described elsewhere (Meetei et al., 2003; Wu et al., 2000; Xu et al., 2008; Yin et al., 2005). The different lanes were normalized by comparison of FLAG-RMI2 levels. The SCE assay was performed as described previously (Guo et al., 2009).
Highlights.
The human RMI core complex shares a striking overall similarity with RPA
Important differences in the subunit interfaces distinguish the two complexes
Integrity of the RMI interface is critical for Bloom dissolvasome activity in vivo
The RMI core complex may act as a docking site for additional protein partners
Supplementary Material
Acknowledgments
We thank Advanced Photon Source staff (LS-CAT beamline) for assistance with data collection, and Dr. David Schlessinger, James Berger, David Fox, and members of the Keck Lab for critical reading of this manuscript. This work was funded by a grant from the NIH (GM068061, JLK) and by the Intramural Research Program of the National Institute on Aging (Z01 AG000657-08, WW). KAH was supported in part by an NIH training grant in Molecular Biosciences (GM07215).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucl Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes & Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMol Molecular Graphics System. San Carolos, CA: DeLano Scientific; 2002. [Google Scholar]
- Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl GE. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J Mol Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–21. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Guo R, Xu D, Wang W. Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods. 2009;48:72–79. doi: 10.1016/j.ymeth.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1 Stn1 Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. Ob(Oligonucleotide Oligosaccharide Binding)-Fold - Common Structural and Functional Solution for Nonhomologous Sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness SR, de Graaff RA, Abrahams JP, Pannu NS. CRANK: new methods for automated macromolecular crystal structure solution. Structure. 2004;12:1753–1761. doi: 10.1016/j.str.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: Cancer and aging through the prism of human RecQ helicases. Mech Ageing & Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Pestryakov PE, Weisshart K, Schlott B, Khodyreva SN, Kremmer E, Grosse F, Lavrik OI, Nasheuer HP. Human replication protein A. The C-terminal RPA70 and the central RPA32 domains are involved in the interactions with the 3′-end of a primer-template DNA. J Biol Chem. 2003;278:17515–17524. doi: 10.1074/jbc.M301265200. [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes & Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes & Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes & Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.