Abstract
Survival of Pseudomonas aeruginosa in cystic fibrosis (CF) chronic infections is based on a genetic adaptation process consisting of mutations in specific genes, which can produce advantageous phenotypic switches and ensure its persistence in the lung. Among these, mutations inactivating the regulators MucA (alginate biosynthesis), LasR (quorum sensing) and MexZ (multidrug-efflux pump MexXY) are the most frequently observed, with those inactivating the DNA mismatch repair system (MRS) being also highly prevalent in P. aeruginosa CF isolates, leading to hypermutator phenotypes that could contribute to this adaptive mutagenesis by virtue of an increased mutation rate. Here, we characterized the mutations found in the mucA, lasR, mexZ and MRS genes in P. aeruginosa isolates obtained from Argentinean CF patients, and analyzed the potential association of mucA, lasR and mexZ mutagenesis with MRS-deficiency and antibiotic resistance. Thus, 38 isolates from 26 chronically infected CF patients were characterized for their phenotypic traits, PFGE genotypic patterns, mutations in the mucA, lasR, mexZ, mutS and mutL gene coding sequences and antibiotic resistance profiles. The most frequently mutated gene was mexZ (79%), followed by mucA (63%) and lasR (39%) as well as a high prevalence (42%) of hypermutators being observed due to loss-of-function mutations in mutL (60%) followed by mutS (40%). Interestingly, mutational spectra were particular to each gene, suggesting that several mechanisms are responsible for mutations during chronic infection. However, no link could be established between hypermutability and mutagenesis in mucA, lasR and mexZ, indicating that MRS-deficiency was not involved in the acquisition of these mutations. Finally, although inactivation of mucA, lasR and mexZ has been previously shown to confer resistance/tolerance to antibiotics, only mutations in MRS genes could be related to an antibiotic resistance increase. These results help to unravel the mutational dynamics that lead to the adaptation of P. aeruginosa to the CF lung.
Introduction
The opportunistic pathogen Pseudomonas aeruginosa is the leading cause of morbidity and mortality in CF patients, producing chronic pulmonary infections that can persist over decades [1], [2]. After a period of recurrent infections, CF patients become infected permanently by a single lineage of P. aeruginosa, generally acquired from environmental reservoirs [3]. During the establishment of the infection, this single P. aeruginosa lineage diversifies into phenotypes specifically adapted to the hostile environment of the CF lung, thus allowing its long-term persistence [4]. These phenotypes possess traits that differ from environmental isolates, but which are commonly observed among CF patients, suggesting a conserved evolutionary pattern in the adaptive process of P. aeruginosa to the CF lung [5], [6]. Well-known examples of this diversification process include conversion to the mucoid phenotype [1], [7], loss of the O antigen [8], inactivation of quorum sensing functions [6], [9], [10], loss of motility [11], [12], auxotrophies [13], resistance to antibiotics [14], and a rise in the mutation rate leading to a hypermutator phenotype.
The acquisition of these P. aeruginosa CF adaptive phenotypes is mainly based on the occurrence (and further selection) of loss-of-function mutations in specific genes. Among these, the most commonly mutated is mucA, which encodes for a negative regulator of the biosynthesis of the exopolysaccharide alginate and whose inactivation is the basis of the conversion to the mucoid phenotype [7]. Another gene, commonly altered even at the early stages of P. aeruginosa CF infections, is lasR, which is the main quorum sensing regulator controlling the expression of several virulence traits [6], [9], [10]. Moreover, genes which regulate the expression of multidrug-efflux pumps are frequently mutated, such as mexZ, a negative regulator of the MexXY-OprM multidrug-efflux pump, whose inactivation increases the resistance to aminoglycoside and other drugs [6], [15].
Another highly prevalent phenotype observed in P. aeruginosa isolates from CF patients is the hypermutator phenotype [16]–[19], with hypermutator strains displaying high mutation rates, mainly due to alterations in the genes of the DNA mismatch repair system (MRS) [20]. This phenotype has been reported in CF patients from both Europe and North America [16], [19], [21]–[24], suggesting that this could be a worldwide phenomenon. Moreover, this high prevalence of hypermutators seems to be essentially associated to chronic infections, since it has only very occasionally been observed in either acute infections or environmental samples (≤1%) [19], [24], indicating that it could be linked with the particular genetic adaptive process taking place during the establishment and persistence of P. aeruginosa infections. In fact, the link between the acquisition of antimicrobial resistance and hypermutators has been well-established by in vitro and in vivo approaches [16], [17], [19], [25]–[28]. However, antibiotic resistance is not only a phenomenon restricted to chronic infections, but is also a frequent outcome of acute infections [29]. Working with a P. aeruginosa MRS-deficient strain in vitro, we previously established an association between hypermutability and both mucoid conversion [30] and lasR inactivation [31], [32], both hallmarks of P. aeruginosa infections in the CF airways. Similarly, Waine et al. [23] observed an association between the hypermutator and the mucoid phenotypes by examining P. aeruginosa isolates from adult CF patients with chronic lung infections. Nevertheless, the role of hypermutability in P. aeruginosa phenotypic diversification in the natural scenario of a human infection has only very recently been investigated by Mena et al. [22], who recently showed that this genetic adaptation is catalyzed by hypermutators. However, the hypermutability was not linked to any particular adaptive trait (including genes involved in the antimicrobial resistance and lasR, the latter of which showed a higher number of mutations). Hence, these results show that the involvement of hypermutability in the acquisition of the mutations which drive the adaptive process of P. aeruginosa into the CF airways is complex. Also, exactly how P. aeruginosa acquires those mutations which enable it to persist into the CF lung environment is still an open question. These issues need to be resolved in order to shed light on the mutational mechanisms involved in P. aeruginosa adaptation.
In the present work, we carried out an exhaustive survey and characterization of mutations in mucA (mucoid phenotype), lasR (quorum sensing deficient phenotype), mexZ (multi-drug resistant phenotype) and the MRS genes (hypermutator phenotype) in a collection of P. aeruginosa isolates obtained from Argentinean CF patients. mexZ turned out to be the most frequently mutated gene followed by mucA and lasR, but mutations in all these three genes were highly prevalent in our collection. Also, the occurrence of hypermutators among Argentinean CF patients was very high, with hypermutability being a consequence of mutations affecting the MRS genes (mostly the mutL gene, but also mutS). However, the mutational spectra of each gene were clearly different, suggesting that more than one mechanism could be involved in the mutational process of each gene. Our results show a lack of association between hypermutability and the frequency of adaptive mutations occurring in mucA, lasR and mexZ, and, although hypermutability was associated with an increased resistance to most of the antibiotics tested on P. aeruginosa isolates from the Argentinean CF population, this association was not observed for mucoid variants, quorum sensing deficiency or the mexZ related multi-drug resistant phenotype.
This is the first detailed characterization of P. aeruginosa CF isolates in Latin America which is focused on mucoidy, lasR-deficiency, hypermutability and antibiotic resistance, critical issues regarding CF chronic infections, and provides new insights into the mutational mechanisms required for bacterial persistence.
Results
Phenotypic and genotypic characterization of the P. aeruginosa isolates
Thirty-eight P. aeruginosa isolates were collected from the sputum of twenty-six CF patients during the course of chronic pulmonary infection (see Material and Methods). The samples were obtained from child/teenage patients (18 isolates, 12 patients) as well as from adult patients (20 isolates, 14 patients) obtained at two Hospitals located in different Argentinean cities. In order to avoid duplications, two isolates from the same patient were considered different if they fulfilled at least one of the following conditions: 1) they were isolated with a separation period of at least one year; 2) they were obtained on the same date, but showed different colony morphotypes (see Material and Methods).
We characterized all P. aeruginosa isolates based on their mucoidy, iridescence at white light exposure, pigmentation, and colony size. As shown in Table 1, this first phenotypic characterization displayed the typical colony variant diversification described for pulmonary chronic infection isolates, which differed from the monomorphic variants isolated from P. aeruginosa acute infections [5], [6].
Table 1. Genotypic and morphotypic characterization of the 38 P. aeruginosa CF isolates.
| Patient | Isolate | Genotypea | Phenotype | Source | Year of isolation | ||||
| Mucoidity | SCV | Pigmentation | Iridescence | Mutation Frequencyb | |||||
| 1 | a | ND | + | - | - | - | (1.3±0.7)×10−5 | HNC | 2004 |
| b | A1 | + | - | - | - | (1.6±0.7)×10−5 | HNC | 2007 | |
| c | A1 | + | + | - | - | (2.7±1.9)×10−9 | HNC | 2007 | |
| 2 | a | A1 | - | + | - | + | (1.0±0.4)×10−5 | HNC | 2007 |
| b | A2 | + | - | - | - | (1.7±1.4)×10−9 | HNC | 2007 | |
| 3 | a | B1 | + | - | - | - | (1.0±0.7)×10−5 | HNC | 2004 |
| b | B1 | - | + | - | - | (1.6±0.3)×10−6 | HNC | 2007 | |
| c | B1 | + | - | + (green) | - | (2.8±1.6)×10−7 | HNC | 2008 | |
| 4 | a | C1 | + | - | - | - | (2.8±1.4)×10−8 | HNC | 2007 |
| b | C1 | - | - | - | - | (9.0±6.3)×10−9 | HNC | 2007 | |
| 5 | a | D1 | + | - | - | - | (4.5±2.6)×10−6 | HNC | 2007 |
| 6 | a | E1 | - | - | - | - | (6.4±4.5)×10−9 | HNC | 2007 |
| 7 | a | Q1 | - | - | + (green) | - | (2.9±0.8)×10−8 | HNC | 2007 |
| 8 | a | R1 | + | - | - | - | (2.2±0.5)×10−8 | HNC | 2007 |
| 9 | a | F1 | + | - | - | - | (3.5±0.3)×10−6 | HNC | 2004 |
| 10 | a | L1 | + | + | - | - | (6.5±3.8)×10−6 | HNC | 2008 |
| 11 | a | N1 | + | - | - | - | (1.0±0.5)×10−9 | HNC | 2008 |
| 12 | a | O1 | + | - | - | - | (3.9±2.1)×10−9 | HCN | 2008 |
| 13 | a | G1 | - | - | - | + | (4.1±0.3)×10−9 | HABA | 2006 |
| 14 | a | H1 | - | - | + (green) | - | (5.0±3.2)×10−9 | HABA | 2006 |
| 15 | a | S1 | - | + | + (red) | - | (1.1±0.6)×10−7 c | HABA | 2006 |
| 16 | a | I1 | + | - | + (green) | - | (6.7±1.8)×10−9 | HABA | 2006 |
| 17 | a | J1 | - | - | + (green) | - | (2.6±1.5)×10−9 | HABA | 2006 |
| b | J1 | + | - | - | - | (6.0±2.8)×10−6 | HABA | 2006 | |
| 18 | a | K1 | + | + | - | - | (5.5±2.1)×10−9 | HABA | 2006 |
| b | K1 | - | + | - | - | (2.9±2.3)×10−8 | HABA | 2006 | |
| c | K1 | + | - | - | - | (1.2±1.1)×10−9 | HABA | 2008 | |
| d | K1 | - | + | - | + | (1.3±1.2)×10−6 | HABA | 2008 | |
| 19 | a | M1 | + | - | - | - | (2.6±0.4)×10−6 | HABA | 2006 |
| 20 | a | P1 | + | - | + (green) | - | (9.0±4.0)×10−10 | HABA | 2008 |
| b | P1 | - | - | + (green) | - | (2.2±1.6)×10−6 | HABA | 2008 | |
| 21 | a | T1 | + | + | - | - | (1.5±0.3)×10−6 | HABA | 2008 |
| b | T1 | - | + | - | + | (1.1±0.9)×10−6 | HABA | 2008 | |
| 22 | a | U1 | + | - | - | - | (3.7±2.2)×10−6 | HABA | 2008 |
| 23 | a | V1 | - | - | + (green) | - | (1.9±1.3)×10−9 | HABA | 2008 |
| 24 | a | W1 | - | - | - | + | (4.0±2.0)×10−10 | HABA | 2008 |
| 25 | a | X1 | - | - | - | + | (1.1±0.2)×10−7 c | HABA | 2008 |
| 26 | a | Y1 | + | - | - | - | (2.3±1.7)×10−9 | HABA | 2008 |
All isolates were genotyped by PFGE using the SpeI enzyme.
Mutation frequency was measured as the occurrence of spontaneous resistance to rifampin 100 µg/ml.
Isolates 15a and 25a with mutation frequencies nearly under the breakpoint (≥2×10−7) were discarded to be mutS or mutL deficient strains by gene sequencing (see Table 2).
SCV, Small Colony Variant; ND, not determined; HNC, Hospital de Niños de Córdoba; HABA, Hospital Alemán de Buenos Aires.
Those isolates with mutation frequencies ≥2×10−7 were defined as hypermutators and indicated by bold type.
Mutation frequencies for PAO1 reference strain and MPAOMS and MPAOML hypermutator strains were (2.4±1.3)×10−8, (2.8±1.6)×10−6 and (1.3±0.6)×10−6 respectively.
In order to explore the extent of clonality and the epidemiology of the P. aeruginosa collection, we carried out a PFGE analysis of the 38 CF isolates. Each patient carried a single unique genotype that persisted over time, even for those isolates that were obtained from the same patient over several years (patients 3 and 18). Moreover, the strains from different patients were epidemiologically unrelated (Table 1), indicating that the infection in each patient was established independently by bacterial lineages from different environmental reservoirs.
Exceptions to this distribution were the isolates from patients 1 and 2, which demonstrated a closely related PFGE pattern (Table 1). This exceptional case may indicate a very low prevalence (5%) of transmission among patients in the Argentinean population.
P. aeruginosa hypermutator strains in the Argentinean CF population
In order to establish the prevalence of hypermutator strains in the Argentinean P. aeruginosa CF isolates, we determined the frequency of mutation to rifampin resistance for the 38 isolates. Although this assay only allows the detection of base substitutions in the rpoB gene [33], it is widely used as an efficient method to detect MRS deficient strains [19], [34].
Thus, the results were compared with those obtained for the PAO1 reference strain (2.4×10−8±1.3×10−8) as well as with those from the MPAOMS (2.8×10−6±1.6×10−6) and MPAOML (1.3×10−6±0.6×10−6) hypermutator mutant strains. In this sense, clinical isolates were considered hypermutators when their mutation frequencies were ≥2×10−7, with this being an arbitrary value which has been previously established to be reached/surpassed mostly by MRS-defficient strains [19]. As observed in Table 1, 16 hypermutators were identified among the 38 P. aeruginosa isolates (42%), with these hypermutable P. aeruginosa strains being obtained from 12 patients (46%). It is important to mention that the prevalence of P. aeruginosa hypermutator strains was very high in the adult population as well as in the child/teenaged population.
Molecular characterization of the hypermutator strains
It has been described that the stable hypermutators found among P. aeruginosa CF isolates are commonly deficient in the genes belonging to the MRS, particularly mutS [18], [20]. Thus, we explored the genetic bases for the hypermutability of the 16 P. aeruginosa hypermutator isolates by carrying out gene complementation assays as well as sequencing of the mutS and mutL genes. As shown in Table 2, 15 hypermutator isolates (94%) were confirmed to be defective in the MRS, of which 6 isolates showed a loss of function in the mutS gene, and 9 in the mutL. This observation differs from previous studies in which mutS was shown to be the main target for the acquisition of a stable hypermutable state [18], [20]. For the remaining hypermutator isolate (3c), although genetic complementation was not achieved due to this isolate showing a natural resistance to gentamicin (not shown), a region of its mutS gene could not be amplified by PCR, suggesting that this strain could also be another mutS-deficient isolate. Mainly frameshift mutations (62.5%, see below) were responsible for the hypermutator phenotype in both the mutS and mutL genes, including deletions and insertions (Table 2). Furthermore, in three hypermutator isolates, missense mutations accounted for the inactivation of the MRS genes: isolate 5a had a single point mutation leading to a change in amino acid 493 from tyrosine to proline (T493P), located in the clamp domain IV of the MutS protein; isolate 10a contained a non-synonymous mutation in mutS, G618S, one of the ADP-contacting residues of ATPase domain V [35] that is highly conserved in all ABC ATPases inhibited by vanadate [36]; and the remaining isolate 2a had a single point mutation in mutL, which produced a change in amino acid 625 from leucine to glutamine (L625Q). This amino acid is one of the last 10 C-terminal residues that abolishe MutL homodimerization upon deletion, a phenomenon that has a key function in communicating mismatch recognition by MutS to downstream repair processes. To our knowledge, these three missense mutations have not been previously reported.
Table 2. Mutations in the mucA, lasR and mexZ genes of the 38 P. aeruginosa CF isolates.
| Isolate | mucA | lasR | mexZ | MRS genes |
| 1a | C→T at 349 (TAA at 349) | A→G at 587 (E196G) | C→G at 151 (H51D) | mutS, 11 bp del at 1555 (TAA at 1854); A→G at 1276 (T426A) |
| 1b | C→T at 349 (TAA at 349) | A→G at 587 (E196G) | C→G at 151 (H51D) | mutS, 11 bp del at 1555 (TAA at 1854); A→G at 1276 (T426A) |
| 1c | 1 bp del at 426 (TGA at 440) | NF | 1 bp del at 261 (TAA at 332) | ND |
| 2a | NF | 1 bp del at 308 (TGA at 341) | 1 bp del at 242 (TAA at 332) | mutL, T→A at 1874 (L625Q) |
| 2b | 1 bp del at 426 (TGA at 440) | NF | 1 bp del at 261 (TAA at 332) | ND |
| 3a | 1 bp del at 426 (TGA at 440) | NF | NF | mutL, 2 bp ins at 619 (TGA at 784) |
| 3b | 1 bp del at 426 (TGA at 440) | NF | NF | mutL, 11 bp del at 619 (TGA at 782) |
| 3c | NF | NF | NF | mutS, NA |
| 4a | 1 bp del at 426 (TGA at 440) | NF | 10 bp del at 595 | ND |
| 4b | 1 bp del at 426 (TGA at 440) | NF | 10 bp del at 595 | ND |
| 5a | 1 bp del at 426 (TGA at 440) | NF | NF | mutS, A→C at 1477 (T493P) |
| 6a | NF | NF | 2 bp del at 60 | ND |
| 7a | 1 bp del at 426 (TGA at 440) | A→G at 626 (N209S) | T→C at 110 (L37P) | ND |
| 8a | C→T at 352 (TAG at 352) | NF | 10 bp del at 184 | ND |
| 9a | C→T at 349 (TAA at 349) | 2 bp del at 602 (TAG at 617) | 14 bp del at 340 | mutL, 17 bp del at 1240 andG→T at 1632 (TGA at 1632) |
| 10a | C→T at 340 (TAG at 340) | NF | 11 bp del at 328 | mutS, G→A at 1852 (G618S) |
| 11a | 1 bp del at 426 (TGA at 440) | NF | 1 bp deletion at 609 | ND |
| 12a | NF | NF | NF | ND |
| 13a | NF | NA | A→G at 633 (TAG 211 W) | ND |
| 14a | NF | NF | A→G at 34 (T12A); G→A at 44 (G15D) | ND |
| 15a | NF | NF | NA | NF |
| 16a | 1 bp del at 201 (TGA at 284) | NF | 11 bp del at 365 | ND |
| 17a | NF | NF | 38 bp del at 417 | ND |
| 17b | NF | NF | 13 bp del at 340 | mutS, 2 bp ins at 1611 (TGA at 1687); T→C at 986 (L329P) |
| 18a | 1 bp del at 339 (TGA at 386) | NF | 4 bp del at 595 | ND |
| 18b | 1 bp del at 339 (TGA at 386) | NF | 4 bp del at 595 | ND |
| 18c | 1 bp del at 430 (TGA at 440) | NF | 15 bp del at 60 (ΔRVFLE at 22) | ND |
| 18d | G→T at 421 (TAG at 421) | C→T at 617 (A206V) | NF | mutS, 1 bp del at 1171 (TGA at 1250) |
| 19a | 1 bp del at 426 (TGA at 440) | NF | 368 bp del at 248 | mutL, 5 bp ins at 627 (TGA at 782) |
| 20a | C→T at 367 (TAG at 367) | T→G at 645 (I215M) | C→T at 187 (C63R) | ND |
| 20b | NF | T→G at 645 (I215M) | C→T at 187 (C63R); C→T at 283 (TAG at 283) | mutL, 96 bp dup at 75 (TGA at 378) |
| 21a | NF | C→T at 221 (P74L) | 21 bp del at 631 | mutL, 4 bp del at 1475 |
| 21b | NF | C→T at 221 (P74L) | 21 bp del at 631 | mutL, 4 bp del at 1475 |
| 22a | 1 bp del at 358 (TGA at 386) | C→T at 280 (TAG at 280) | 12 bp del at 425 (ΔRAVE at 143; R142Q) | mutL, C→T at 1474 (TGA at 1474) |
| 23a | G→C at 304 (G102R) | A→G at 575 (K192R) | T→G at 413 and C→A at 414 (L150R); 7 bp del at 451 | ND |
| 24a | NF | C→A at 317 (TGA at 315) | C→A at 494 (TAG at 493) | ND |
| 25a | NF | A→T at 697 (N233Y) | NF | NF |
| 26a | 1 bp del at 201 (TGA at 284) | NF | 12 bp del at 383 (ΔPLEK at 128) | ND |
NF, no mutation found; ND, not determined; NA, not PCR amplified; del, deletion; ins, insertion; dup, duplication.
Finally, it is interesting to highlight the results obtained for patients 1 and 3. From patient 1, we isolated a previous hypermutator strain in 2004 (1a) and a subsequent hypermutator strain in 2007 (1b) (Table 1). Both these isolates contained the same 11 bp deletion in MutS and an identical single point mutation (T426A), suggesting that the same hypermutator lineage was maintained over three years of chronic infection. On the other hand, the hypermutator isolates from patient 3 obtained in 2004 (3a) and 2007 (3b) harbored two different frameshifts, produced by 11 bp and 2 bp deletions inactivating mutL, respectively. In addition, a third hypermutator isolate (3c) was obtained from the same patient in 2008, which possessed an unaltered mutL sequence but which failed to amplify a region of the mutS gene. This result indicates that during four years of chronic infection in patient 3, three different hypermutator lineages emerged, suggesting a strong selection for hypermutability in the CF lung.
Screening of mutations in the mucA, lasR and mexZ genes
The coding sequences of the mucA, lasR and mexZ genes were analyzed in our collection of isolates to score for mutations that could be involved in the phenotypic switches to mucoidy, deficiency in the quorum sensing and antimicrobial (aminoglycoside) resistance respectively, three phenotypes considered to be characteristics of P.aeruginosa chronic CF infections. Thus, only nonsynonymous mutations which are expected to cause partial or complete loss of function in these genes were considered in the analysis (see Material and Methods).
As shown in Table 2, mexZ was the most frequently mutated gene in our collection of isolates, with 30 out of 38 isolates harboring mutations in this gene (79%). This was followed by mucA, which showed mutations in 24 of the 38 isolates (63%), and finally lasR, which was mutated in 15 isolates (39%). Taken together, these results and the data from the MRS gene mutational inactivation (42%) confirm that the occurrence of mutations in all the analyzed genes was highly prevalent among Argentinean CF patients.
Mutational spectra are remarkably different among mucA, lasR, mexZ and MRS genes
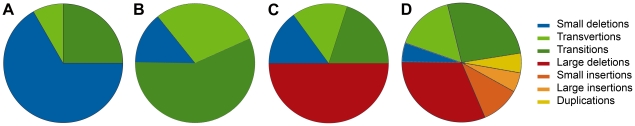
Next we examined the mutational spectrum produced in each gene. Interestingly, these mutational spectra presented large differences among mucA, lasR, mexZ, and mutS/mutL, which were particular to each gene (Figure 1). In mucA, small deletions prevailed (64.7%) with the rest of the mutations consisting of different kinds of transitions (25%) and transversions (8.3%) (Figure 1A). Notably, both frameshift mutations and substitutions generated premature stop codons in 92% of the cases (Table 2), indicating the particular way in which mucA was mutated. In lasR, missense substitutions prevailed (86%), with a 2∶1 ratio for transitions (57%) over transversions (29%) (Figure 1B), which mainly produced changes in the amino acid sequences (Table 2). In contrast with mucA, only 14% of mutations in lasR were small deletions (Figure 1B). mexZ, on the other hand, showed mostly deletions (65%), of which there was a high prevalence of large deletions (>4 bp) (50%) respect to small deletions (1–4 bp) (15%), followed by transitions (20%) and transversions (15%) (Figure 1C). However, in mexZ, almost 15% of mutations generated premature stop codons (Table 2). As mentioned above, mostly frameshift mutations (62.5%) were responsible for mutS and mutL inactivation (Table 2), including mainly large (31.5%) but also small deletions (5.3%), small insertions (10.5%) large insertions (5.3%) and duplications (5.3%), thus showing a wider mutational spectrum (Figure 1D).
Figure 1. Mutational spectra of mucA, lasR, mexZ and MRS genes observed in P. aeruginosa isolates obtained from Argentinean CF patients.
Pie charts indicate the observed percentage for each kind of mutation respect to the total number of mutations occurring in (A) mucA, (B) lasR, (C) mexZ and (D) MRS genes. The analyses on MRS genes include mutations observed in mutS and mutL.
By comparing the mutational spectra obtained in the Argentinean isolates with those reported in other studies [6], [22], [37]–[39], we observed that our results coincided with those previously described for mucA, lasR, mexZ and MRS, indicating that each gene has a particular way of being mutated. Taken together, these observations suggest that different mechanisms of mutagenesis are responsible for the loss-of-function mutations in the analyzed genes.
Mutations in the mucA, lasR and mexZ genes are not linked to hypermutability in the Argentinean CF population
Theoretical studies suggest that, under stressful conditions, selective pressure favors hypermutator strains over nonmutator strains [40]–[42]. In the particular case of P. aeruginosa chronic lung infection, it has been proposed that the proliferation of the hypermutators in the population infecting an individual patient may be sustained by their association (hitchhike) with mutations adaptive to growth in the lung [19], [20]. Related to this, in two previous recent works, we showed in vitro that the MRS loss-of-function was a key determinant in targeting mucA for mucoid conversion and lasR for quorum sensing deficiency [30]–[32].
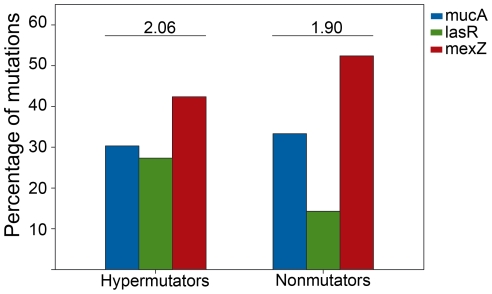
In order to investigate if an association could be established between hypermutability and the mutations observed in the mucA, lasR and mexZ genes in the Argentinean CF isolate collection, we analyzed the proportion of mutations that occurred in each gene in the hypermutator and nonmutator isolates. As shown in Figure 2, we observed that in the hypermutator isolates, the proportion of mutations that occurred in the mexZ and mucA genes was not significantly different to that observed in the nonmutator isolates (P = 0.17 and P = 1.00 respectively). In the case of lasR, we saw that among the hypermutators, the number of mutations per isolate was higher (0.56) than nonmutators (0.27), although this difference was not statistically significant (P = 0.09), indicating that, as in the case of mexZ and mucA, it was not possible to establish an association between the occurrence of mutations in lasR and the hypermutator phenotype. Furthermore, as shown in Figure 2, the number of mutations per hypermutator isolate (2.06) was not higher than that observed per nonmutator isolate (1.90).
Figure 2. Distribution of the number of mutations among mucA, lasR and mexZ genes in hypermutator and nonmutator isolates.
The number of mutations occurring in mucA (blue bars), lasR (green bars) and mexZ (red bars) is expressed as a percentage respect to the total number of mutations found in hypermutator and nonmutator isolates. Above the bars, the total number of mutations per isolate for the hypermutator and nonmutator subpopulations is indicated.
We now investigated if hypermutability could be associated with any particular kind of mutations in mucA, lasR and mexZ genes. It was observed that although the hypermutator isolates showed a tendency towards substitutions (53%), which were either transversions or transitions, with nonmutator isolates displaying a higher number of deletions (62%) (mainly small deletions between 1 to 4 bp), there was not a statistically significant difference between hypermutators and nonmutators with respect to the kind of mutations occurring in the analyzed genes. Furthermore, whatever the type of mutations, their distribution among any particular analyzed gene in hypermutators and nonmutators isolates did not show any statistically significant differences (Figure 2). Therefore, it was not possible to establish any association between hypermutability and the nature of the mutations that occurred in mucA, lasR and mexZ during the process of chronic lung infection, suggesting that in the population analyzed in this work, hypermutation was not linked (hitchhiked) to any of the specific adaptive traits studied.
Hypermutability is associated to antimicrobial resistance mainly through the presence of resistant mutant subpopulations (RMS)
As mentioned above, several reports have established a link between antimicrobial resistance and the occurrence of hypermutators. In fact, it has been observed that hypermutator P. aeruginosa strains isolated from CF patients not only possess a substantially higher antibiotic resistance than nonmutator isolates [19], but also increase the rate of acquisition of new antibiotic resistance [25]. Thus, in order to investigate the association between hypermutability and antibiotic resistance in the Argentinean P. aeruginosa CF isolates, we tested the 38 isolates by measuring the inhibition zone diameters and scoring for RMS within the inhibition zones of five different antimicrobial agents (ceftazidime, ciprofloxacin, imipenem, meropenem and tobramycin). As shown in Table 3, the proportion of hypermutator strains of P. aeruginosa with detectable levels of ciprofloxacin and ceftazidime resistance was significantly higher than the proportion observed in non-mutator strains (P = 0.02 and P = 0.02 respectively), indicating that in the Argentinean CF population, hypermutability correlated with ciprofloxacin and ceftazidime resistance. In fact, 62.5% of the hypermutators were ciprofloxacin resistant compared to a proportion of 22.7% found among nonmutators. Although a smaller proportion of hypermutators (25%) were ceftazidime resistant, all nonmutator strains were susceptible to this antibiotic (Table 3). The prevalence of antibiotic resistant strains for hypermutator and nonmutator isolates for imipenem, meropenem, and tobramycin was biased towards the hypermutators, although this was not statistically significant (P = 0.06, P = 1.00 and P = 0.61 respectively). Nevertheless, it is important to point out that only one isolate (isolate 19a) that was a hypermutator showed resistance to all the antibiotics tested (Table 3).
Table 3. Antibiotic disk diffusion test and quantification of the resistant mutant subpopulations of the 38 P. aeruginosa CF isolates.
| Isolate | MRS genesa | Ceftazidime | Ciprofloxacin | Imipenem | Meropenem | Tobramycin | |||||
| DD | RMS | DD | RMS | DD | RMS | DD | RMS | DD | RMS | ||
| 1a | - | S | +++ | S | ++ | S | +++ | S | +++ | S | ++ |
| 1b | - | S | +++ | I | ++ | S | +++ | S | +++ | R | * |
| 1c | + | S | ++ | S | - | S | - | S | ++ | S | - |
| 2a | - | R | * | R | * | S | +++ | S | +++ | S | +++ |
| 2b | + | S | + | S | - | S | - | S | + | S | - |
| 3a | - | I | ++ | I | - | S | +++ | S | +++ | I | +++ |
| 3b | - | R | * | I | ++ | I | +++ | S | +++ | I | +++ |
| 3c | - | S | +++ | S | +++ | S | +++ | S | +++ | S | ++ |
| 4a | + | S | - | S | - | S | - | S | - | S | - |
| 4b | + | S | - | S | - | S | - | S | - | S | - |
| 5a | - | S | - | R | * | S | ++ | S | +++ | S | + |
| 6a | + | S | - | S | - | S | - | S | - | S | - |
| 7a | + | S | - | I | - | S | - | S | - | S | - |
| 8a | + | S | - | R | * | S | +++ | S | +++ | S | - |
| 9a | - | S | ++ | S | +++ | S | +++ | S | +++ | S | +++ |
| 10a | - | S | ++ | S | ++ | S | +++ | S | - | S | +++ |
| 11a | + | S | - | S | - | S | - | S | - | R | * |
| 12a | + | S | - | S | - | S | - | S | - | S | - |
| 13a | + | S | - | R | * | S | - | S | - | R | * |
| 14a | + | S | - | S | - | S | - | S | - | S | - |
| 15a | + | S | - | S | - | S | - | S | - | S | - |
| 16a | + | S | - | S | - | S | - | S | - | S | - |
| 17a | + | S | - | I | - | R | * | I | - | S | - |
| 17b | - | S | - | S | - | I | +++ | S | ++ | S | + |
| 18a | + | S | - | S | - | S | - | S | - | S | - |
| 18b | + | S | - | R | * | S | - | S | - | S | - |
| 18c | + | S | - | S | - | S | - | S | - | S | - |
| 18d | - | S | +++ | R | * | R | * | S | +++ | R | ++ |
| 19a | - | R | * | R | * | R | * | R | * | R | * |
| 20a | + | S | - | S | - | S | - | S | - | S | - |
| 20b | - | S | +++ | R | * | R | * | S | +++ | S | +++ |
| 21a | - | S | +++ | R | * | S | ++ | S | +++ | S | +++ |
| 21b | - | S | +++ | R | * | S | +++ | S | +++ | S | +++ |
| 22a | - | S | - | S | - | S | ++ | S | +++ | S | +++ |
| 23a | + | S | - | S | - | S | - | S | ++ | S | - |
| 24a | + | S | - | S | - | S | - | S | - | S | - |
| 25a | + | S | - | S | - | S | - | S | - | S | - |
| 26a | + | S | - | S | - | S | - | S | - | S | - |
MRS genes was considered positive (+) when the sequences of mutS and mutL genes were wild type and negative (−) when were mutated.
DD, diffusion diameter zone; RMS, resistant mutant subpopulation: +<10 mutants; ++ 10 to 100 mutants; +++ >100 mutants.
Zone diameter interpretative criteria was according to CLSI: S, susceptible; I, intermediate resistance; R, resistant.
*lack of diffusion zone that prevents the resistant mutant subpopulation to be quantified.
By quantifying the emergence of the resistant mutant subpopulation (RMS) [43], we observed that most of the hypermutator isolates showed RMS within the inhibition zones of the five antibiotics tested. In contrast, RMS was rarely detected when the nonmutator strains were tested for the same antibiotics. In fact, every hypermutator isolate of this study showed RMS for at least three different antibiotics, with the proportion of hypermutator P. aeruginosa strains that showed RMS being significantly higher than the non-mutator strains (P<0.001) (Table 3). These observations are in agreement with a previous study, in which the disk diffusion test was proposed as a reliable method for detecting hypermutator strains from clinical isolates [43].
Finally, it is important to remark that in the Argentinean population, the disk diffusion tests showed similar results for the child/teenage and the adult groups of CF patients.
Antimicrobial resistance is not linked to mucoidy, lasR inactivation or mexZ related multi-drug resistant phenotype
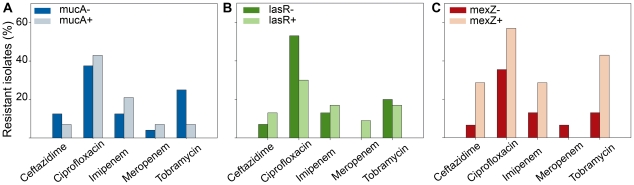
Previous studies have reported an enhanced resistance to antibiotics for mucoid strains of P. aeruginosa isolated from sputa of CF patients [44]. Also, it has been proposed that mexZ inactivation is involved in the development of stable aminoglycoside resistance among CF strains of P. aeruginosa [15]. An increased tolerance to β-lactam has been attributed to inactivation of lasR, which was not sufficient to alter the MIC, but resulted in a greater production of spontaneous resistant colonies [9]. Moreover, it has been recently observed that lasR loss-of-function mutations confered resistance to ciprofloxacin and tobramycin when oxygen was limited and there was increased nitrate utilization [45]. Related to this, we investigated if the inactivation of mucA, lasR or mexZ could be associated with an altered antibiotic resistance in our collection. As shown in Figure 3, antibiotic resistance to β-lactams ceftazidime, imipenem and meropenem, as well as flouroquinolone ciprofloxacin and aminoglycoside tobramycin was not associated with the mutagenic inactivation of mucA, lasR or mexZ. Moreover, scoring for RMS within the inhibition zones of the five antimicrobial agents tested showed that the presence of resistant mutants was not a phenomenon related to the inactivation of any of these three regulators, not even when the lasR mutants were tested for β-lactam antibiotics (not shown).
Figure 3. Association between antibiotic resistance and inactivation of mucA, lasR and mexZ.
(A) Differences in antibiotic resistance in the P. aeruginosa isolates harboring mutations in mucA (mucA-) respect to those without mutations in mucA (mucA+). Data is expressed as the percentage of resistant isolates to ceftazidime, ciprofloxacin, imipenem, meropenem and tobramycin in mucA- and mucA+ isolates. (B) The same analysis was carried out in lasR- and lasR+ isolates and (C) mexZ- and mexZ+ isolates. In the three genes, the observed differences were not significant (Fisher's exact test) for any of the tested antibiotics.
Discussion
P. aeruginosa establishes chronically in the CF airways by a diversification process that allows fine tuning-adaptation and long-term survival within the host. Since this process is mainly mutagenic [6], a better understanding of the means by which P. aeruginosa acquires these mutations may help to identify the functions required for bacterial persistence.
In this study, we carried out a survey and a molecular characterization of the mutations that occurred in the genes which had been previously described as important determinants for the adaptation of P. aeruginosa to the CF lung, due to their being the most mutated among CF patients [6]. The mucA, lasR and mexZ genes as well those implicated in the inactivation of MRS, mutS and mutL, were analyzed in a collection of P. aeruginosa isolates obtained from Argentinean CF patients, with this constituting the first study of this nature in Latin America. According to our results, mutations in the first three genes were highly prevalent in the Argentinean population, with mexZ being the most frequently mutated followed by mucA and lasR (Table 2). In addition, a high prevalence of hypermutator strains was observed in our collection (Table 1), in agreement with what has been previously reported in other geographical regions [16]–[19]. All the hypermutator strains were the result of a deficiency in MRS, and despite most reports have described mutations in the mutS gene as the main cause of hypermutability, followed by mutations in mutL and uvrD [18], [20], in our collection, mutations in mutL were actually the most frequent cause of MRS deficiency, with the mutS gene alterations playing a secondary role (Table 2). It is worth mentioning that, based on the defined mutation frequency breakpoint (≥2×10−7), our analysis was focused on strong MRS deficient hypermutators, not considering weak mutators (usually with alterations in the mutT, mutY and mutM genes of the GO system) which can also be found among the CF P. aeruginosa isolates [16], [19], [46].
It is proposed that the high prevalence of stable hypermutator strains of P. aeruginosa in the airways of CF patients is a consequence of their second-order selection with other adaptive mutations, since a higher mutation rate would increase their probability to be acquired [42]. Related to this, an association between hypermutability and mutations that confer antibiotic resistance has been established by several reports [21], [25], [26], [43], [47]. Here, we also observed that MRS-deficiency was linked to resistance to several antibiotics (Table 3), thus confirming that in our collection, hypermutability was a key factor in the generation of mutation-mediated resistance. However, antibiotic resistance could not be associated to the mucoid phenotype [44] and the lasR quorum sensing-deficient phenotype [9], [15] (Figure 3). Even inactivation of MexZ was not linked to antibiotic resistance in our collection of isolates. This last observation results intriguing, since inactivation of this gene leads to the overproduction of the MexXY efflux system which is one of the most frequently reported mechanisms of acquiring resistance to aminoglycoside and other drugs among CF isolates [26] (Figure 3). Our results suggest that antibiotic resistance due to overproduction of MexXY may require the existence of additional regulatory mechanisms other than the inactivation of mexZ [15], [48], [49]. Thus, it is possible to suggest that those mutations linked to hypermutability, but not the inactivation of mucA/lasR/mexZ, may confer a level of antibiotic resistance detectable by disk diffusion, which is the methodology used in this study.
In a previous recent work, we determined in vitro that MRS-deficiency accelerates the acquisition of mucA [30] and lasR [31], [32] mutations, which led us to the hypothesis that such an association may also be occurring in vivo. In line with this, Waine et al [23] described a link between the hypermutator and mucoid phenotypes in P. aeruginosa isolates obtained from CF patients. However, in the present study, by scoring for mutations in hypermutator and nonmutator CF isolates of our collection, no association could be established between hypermutability and mutagenesis, not only for mucA but also for the lasR and mexZ mutations. A possible explanation for this discrepancy is that in Waine et al., the phenomenon was not characterized at the genetic level, and that the reversion phenomenon of the mucoid to a nonmucoid phenotype, which is highly frequent in isolates recovered from CF patients [38], was not evaluated. In fact, this analysis on our collection showed that 37.5% of the non-mucoid isolates harbored mutations in mucA, indicating that they had previously been mucoid before reverting to the phenotype. Furthermore, contrary to our isolates which were clonally different, almost half of the isolates in Waine et al. corresponded to epidemic clones, a feature that increases dissimilarities among collections.
It has been recently shown that hypermutator strains accelerate the mutagenic genetic adaptation thus enhancing the accumulation of new mutations [22]. However, no differences were observed between hypermutators and nonmutators isolates with respect to the distribution of mutations among several sequenced genes, suggesting that the hypermutability had a widespread effect on genes during the adaptation process in the lung, but was not linked to any specific adaptive trait, even to antibiotic resistance. [22]. Consistent with this antecedent, we observed similar mutational spectra in the mucA, lasR and mexZ genes of hypermutator and nonmutator isolates. However and most interestingly, these mutational spectra vary in a gene dependant manner, with mucA being dominated by small deletions (1 bp), lasR by base substitutions and mexZ by large deletions (>4 bp). This observation is intriguing, since it suggests the involvement of different mechanisms of mutagenesis, possibly operating simultaneously, during the process of CF chronic infection. mucA and lasR, but not mexZ mutational spectra coincide well not only with the spectrum generated by a MRS deficiency [39], [50] but also with the activity of SOS-inducible error prone polymerases (i.e., Pol IV) [51]. Therefore, it can be postulated that the induced hypermutability due to the stressful milieu of the CF lung plays an important role in mutagenesis, thereby contributing to the genetic adaptation of P. aeruginosa to this environment. Consistent with this hypothesis, we have recently shown that Pol IV activity is an essential ingredient to establish mucA as the main target for mutagenesis in mucoid conversion, with this factor having a prominent role in the generation of -1 deletion in a monomeric simple sequence repeats of five Gs (G5-SSR426) one of the most prevalent mutations among CF mucoid isolates [30]. In fact, elimination of G5SSR426 by site directed mutagenesis not only significantly reduces the mutation frequency in mucA, but also makes mucA to be no longer the major pathway for mucoid conversion [52]. In a very recent work, carried out at the same time as our study, a lack of association between stable hypermutability and mucA and lasR mutations was also found [53]. According to this work, mucoid isolates as well as lasR-deficient isolates occur prior to the appearance of hypermutator strains, which had previously been shown to accumulate at later stages of the infection [6], [47]. Thus, it is possible that mutations in these genes might be more important for survival at the earlier stages of chronic infection, when the effect of hypermutators is still poor. Taken all together, our results clearly suggest that mutations which inactivate mucA, lasR and mexZ arise by different mechanisms in each gene, with these being independent of MRS-deficiency. However, further research is required to achieve a better understanding of the complex and variable mutagenic strategies used by P. aeruginosa to continuously adapt to the environment of the CF lung.
Materials and Methods
P. aeruginosa isolates from CF patients
Thirty-eight P. aeruginosa isolates, representing different colony morphotypes (with respect to mucoidy, iridescence, colony size and pigmentation), were collected from sputum samples of 26 Argentinean CF patients with chronic P. aeruginosa lung infection (with at least four years of continuous isolation of P. aeruginosa). These patients were either children/teenagers or adults, who had been treated at two local Hospitals: Hospital de Niños Santísima Trinidad (Córdoba city) (12 patients; age range, 5 to 20 years; mean age, 13±3.5) or Hospital Alemán (Buenos Aires city) (14 patients; age range, 15 to 43 years; mean age, 27±7). The P. aeruginosa reference strain PAO1 and hypermutable strains MPAOMS and MPAOML were used as controls (Table 4). All strains were maintained and stored at −70°C in glycerol stocks. For the different assays, strains were routinely subcultured on Luria Bertani (LB) agar plates from the frozen glycerol stocks and the inoculums were prepared using overnight cultures in LB broth at 37°C with appropriate aeration.
Table 4. Bacterial strains, plasmids and primers used in this study.
| Genotype, relevant characteristics, or sequence (5′-3′) | Source or reference | |
| Strains | ||
| P. aeruginosa | ||
| PAO1 | wild-type; phototropic | [60] |
| MPAOMS | mutS::ISlacZA/hah (MPA32417); Tcr | [61] |
| MPAOML | mutL::ISlacZA/hah (MPA46306); Tcr | [61] |
| Plasmids | ||
| pMC5-mutS | pBBR1MCS-5 carrying P.aeruginosa mutS; Gmr | [56] |
| pMC5-mutL | pBBR1MCS-5 carrying P.aeruginosa mutL; Gmr | [62] |
| Primers | ||
| mucA-for | GAAGCCTGACACAGCGGCAAATGC | |
| mucA-rev | CCTCAGCGGTTTTCCAGGCTGGCTGC | |
| lasR-for | CACGGGCGCATGCGCCTC | |
| lasR-rev | AAGCTTCTATATAGAAGGGCA | |
| mexZ-for | CGCGACAGTAGCATATAATC | |
| mexZ-rev | TACATCGACGGCAAGCGCCT | |
| mutS-for1 | GCCCGTATGACCGACCTCT | |
| mutS-rev1 | CCGAGTCGCGATCGAAGT | |
| mutS-for2 | CCGCGCGCCATGGGACTTCGAT | |
| mutS-rev2 | TTCGGCGAGTTCGGGATA | |
| mutS-for3 | CACCACCATCGGCACCTAT | |
| mutS-rev3 | GTTGGCCACGAACGGTGT | |
| mutS-for4 | TGGTCGAGCAGGTGCTGG | |
| mutS-rev4 | ATTCTAGCAGCTTGTGCGG | |
| mutL-for1 | ACAGCCTGTCCAGCGACAAC | |
| mutL-rev1 | CTCGTCTCGCGCCTCGTGCA | |
| mutL-for2 | TGCACGAGGCGCGAGACGAG | |
| mutL-rev2 | TAACGGCGCGAAGTAGGCCTT | |
| mutL-for3 | AAGGCCTACTTCGCGCCGTTA | |
| mutL-rev3 | AGGCAGGGAAGACATCGGAAC |
Tcr, tetracycline resistance; Gmr, gentamicin resistance.
Ethics Statement
The P. aeruginosa isolates were obtained from the Bacteriological Laboratories of the Hospital de Niños Santísima Trinidad (Córdoba) and Hospital Alemán (Buenos Aires), Argentina, as byproducts of the routine established for bacterial typing and antimicrobial susceptibility testing. In this sense, sputa sampling was not performed with the aim to fulfill the study described in this work, and isolates recovered from these sputa were just a derivative of the habitual CF patient therapeutic controls. Thus, we obtained a collection of 38 P.aeruginosa isolates which was characterized in this work. It is important to clarify that bacterial isolates were processed anonymously, there was no contact with the patients or access to medical records, and that the therapeutic treatments of the patients were not modified in any case as a consequence of the results obtained in this study. The research protocols followed in this study were approved and reviewed by the Institutional Review Board of the Biological Chemistry Research Center of Córdoba (Centro de Investigaciones en Química Biológica de Córdoba, CIQUIBIC), which waived the need for a written consent from the patients involved.
Genotyping by Pulse Field Gel Electrophoresis (PFGE)
All isolates were genotyped by PFGE using the SpeI enzyme as described elsewhere [54]. The clonal relatedness was determined according to Tenover et al. [55], who established that isolates with PFGE patterns which differed: by 1–3 bands are closely related clones; by 4–6 bands are possibly related clones; and by ≥7 bands are considered to belong to different strains.
Determination of mutation frequency of P. aeruginosa CF isolates
Mutation rates determined via fluctuation analysis can provide good estimations of mutagenesis [53]. However, since our interest was merely to identify hypermutators in the collection of P. aeruginosa CF isolates, we performed the quantification of rifampin-resistant mutant frequencies which is considered to be proportional to the estimation of mutation frequencies. Mutation frequencies were determined as previously described [56]. Briefly, three bacterial colonies of each P. aeruginosa isolate were cultured in LB medium at 37°C for 24 h with shaking performed at 225 r.p.m. Appropriate dilutions of the cultures were plated on LB agar plates to determine the total number of viable cells, or on LB agar supplemented with 100 µg ml−1 rifampin to count the number of rifampin-resistant cells, following incubation overnight at 37°C. Then, the mutation frequency was determined as the ratio of the number of rifampin–resistant cells and the number of viable cells. Strains were considered hypermutators when their mutation frequencies were ≥2×10−7, which corresponds to a mutation frequency approximately 20 fold higher than that obtained from the reference P. aeruginosa PAO1 strain (2.4±1.3×10−8) and which has been shown to score for strong MRS-deficient mutators [19]. Determinations were carried out in duplicate for two independent experiments, and the results were expressed as means with their standard deviations.
Sequence analysis of mucA, lasR, mexZ, mutS and mutL genes
Primers for PCR amplification and DNA sequencing of mucA, lasR, mexZ, mutS and mutL genes are shown in Table 4. PCR amplifications were performed under the following conditions: 8 min at 95°C, 33 cycles of 1 min at 94°C, 1 min 20 sec at 60°C (for mucA, mexZ, mutS and mutL) or at 50°C (lasR) 2 min at 72°C, and a final extension of 10 min at 72°C. The PCR products were cleaned with a Gel Purification kit (QIAGEN) and both strands were directly sequenced by using their respective PCR primers (Macrogene Corp., USA). To score for mutations within the genes, the sequencing results were compared with the corresponding gene sequences of the PAO1 strain (www.pseudomonas.com) by using the BLAST program of the NCBI database (www.ncbi.nlm.nih.gov/blast/).
The analyses of the linkage between hypermutability and mutations observed in mucA, lasR and mexZ included only nonsynonymous mutations, which are expected to cause partial or complete loss of function in the genes by generation of premature stop codons, shifts of reading frames or deletions/insertions in the coding sequence. To predict the effect of nonsynonymous (missense) mutations on protein function, the SIFT software was [57] used with default parameters (median conservation of sequences = 3; removal of sequences >90% identical to the query). Mutations were considered as “not tolerated” when P<0.05.
Complementation assays with the mutS and mutL genes
To investigate the genetic basis for the hypermutator phenotype, the hypermutator isolates were complemented with the cloned P. aeruginosa wild-type mutS and mutL genes (Table 4). Vector pBBR1MC-5 [58], which harbors mutS or mutL genes, was successively transferred into the different hypermutator P. aeruginosa CF isolates by electroporation as described by Choi and Schweizer [59]. The transformants were selected on LB agar plates supplemented with 100 µg of gentamicin ml−1, and the complementation was checked by the rifampin test as described above. For each isolate, complementation was verified for three independent transformant colonies.
Antibiotic susceptibility tests and resistant mutant subpopulation quantification
The inhibition-zone diameters of the thirty eight P. aeruginosa CF isolates were determined for imipenem, meropenem, ceftazidime, ciprofloxacin and tobramycin in Mueller-Hilton (MH) agar plates by using conventional disks (Rosco). Approximately 0.5 McFarland standard suspensions were used for the inocula standardization of each isolate, and the diameters of the inhibition zones were measured after 24 h and 48 h of incubation at 37°C. The relative number (<10, 10 to 100, >100) of resistant mutant subpopulations growing within the inhibition zones was determined for each antibiotic after 48 h of incubation at 37°C, as described previously [43]. P. aeruginosa PAO1 strain and the hypermutable MPAOMS and MPAOML strains (Table 4) were used as controls. All assays were performed in duplicate.
Antibiotic multiresistance was defined as resistance to at least three antibiotics.
Statistical analysis
Statistical analyses were performed using Fisher's exact test, considering a P value of >0.05 as significant.
Acknowledgments
We would like to thank Dr. Paul Hobson, native speaker, for revision of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Agencia Nacional de Promoción Científica y Tecnológica (ANPCYT) Grants PICT-2005-33589 and PICT-2007-00687 (http://www.agencia.gov.ar/), and Secretaría de Ciencia y Técnica (SECYT-UNC) Grant 05/C486 (http://www.secyt.unc.edu.ar/) supported this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin C, Ichou MA, Massicot P, Goudeau A, Quentin R. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J Clin Microbiol. 1995;33:1461–1466. doi: 10.1128/jcm.33.6.1461-1466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen D, Singh PK. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci U S A. 2006;103:8305–8306. doi: 10.1073/pnas.0602526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, et al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, et al. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luzar MA, Thomassen MJ, Montie TC. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth AL, Pitt TL. Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J Clin Microbiol. 1995;33:37–40. doi: 10.1128/jcm.33.1.37-40.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 15.Vogne C, Aires JR, Bailly C, Hocquet D, Plésiat P. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother. 2004;48:1676–1680. doi: 10.1128/AAC.48.5.1676-1680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciofu O, Riis B, Pressler T, Poulsen HE, Høiby N. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob Agents Chemother. 2005;49:2276–2282. doi: 10.1128/AAC.49.6.2276-2282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maciá MD, Blanquer D, Togores B, Sauleda J, Pérez JL, et al. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother. 2005;49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montanari S, Oliver A, Salerno P, Mena A, Bertoni G, et al. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology. 2007;153:1445–1454. doi: 10.1099/mic.0.2006/003400-0. [DOI] [PubMed] [Google Scholar]
- 19.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 20.Oliver A, Baquero F, Blázquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol Microbiol. 2002;43:1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- 21.Hogardt M, Schubert S, Adler K, Götzfried M, Heesemann J. Sequence variability and functional analysis of MutS of hypermutable Pseudomonas aeruginosa cystic fibrosis isolates. Int J Med Microbiol. 2006;296:313–320. doi: 10.1016/j.ijmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waine DJ, Honeybourne D, Smith EG, Whitehouse JL, Dowson CG. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J Clin Microbiol. 2008;46:3491–3493. doi: 10.1128/JCM.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenna DT, Doherty CJ, Foweraker J, Macaskill L, Barcus VA, et al. Hypermutability in environmental Pseudomonas aeruginosa and in populations causing pulmonary infection in individuals with cystic fibrosis. Microbiology. 2007;153:1852–1859. doi: 10.1099/mic.0.2006/005082-0. [DOI] [PubMed] [Google Scholar]
- 25.Ferroni A, Guillemot D, Moumile K, Bernede C, Le Bourgeois M, et al. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr Pulmonol. 2009;44:820–825. doi: 10.1002/ppul.21076. [DOI] [PubMed] [Google Scholar]
- 26.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother. 2007;51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver A, Levin BR, Juan C, Baquero F, Blázquez J. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob Agents Chemother. 2004;48:4226–4233. doi: 10.1128/AAC.48.11.4226-4233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plasencia V, Borrell N, Maciá MD, Moya B, Pérez JL, et al. Influence of high mutation rates on the mechanisms and dynamics of in vitro and in vivo resistance development to single or combined antipseudomonal agents. Antimicrob Agents Chemother. 2007;51:2574–2581. doi: 10.1128/AAC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez O, Juan C, Pérez JL, Oliver A. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob Agents Chemother. 2004;48:3573–3575. doi: 10.1128/AAC.48.9.3573-3575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moyano AJ, Luján AM, Argarana CE, Smania AM. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol. 2007;64:547–559. doi: 10.1111/j.1365-2958.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 31.Luján AM, Moyano AJ, Segura I, Argaraña CE, Smania AM. Quorum-sensing-deficient (lasR) mutants emerge at high frequency from a Pseudomonas aeruginosa mutS strain. Microbiology. 2007;153:225–237. doi: 10.1099/mic.0.29021-0. [DOI] [PubMed] [Google Scholar]
- 32.Smania AM, Segura I, Pezza RJ, Becerra C, Albesa I, et al. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology. 2004;150:1327–1338. doi: 10.1099/mic.0.26751-0. [DOI] [PubMed] [Google Scholar]
- 33.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 34.LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 35.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 36.Pezza RJ, Villarreal MA, Montich GG, Argaraña CE. Vanadate inhibits the ATPase activity and DNA binding capability of bacterial MutS. A structural model for the vanadate-MutS interaction at the Walker A motif. Nucleic Acids Res. 2002;30:4700–4708. doi: 10.1093/nar/gkf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boucher JC, Yu H, Mudd MH, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, et al. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology. 2008;154:103–113. doi: 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- 39.Levy DD, Cebula TA. Fidelity of replication of repetitive DNA in mutS and repair proficient Escherichia coli. Mutat Res. 2001;474:1–14. doi: 10.1016/s0027-5107(00)00169-x. [DOI] [PubMed] [Google Scholar]
- 40.Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, et al. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics. 2002;162:557–566. doi: 10.1093/genetics/162.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taddei F, Matic I, Godelle B, Radman M. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol- 1997;5:427–428; discussion 428–429. doi: 10.1016/S0966-842X(97)01157-8. [DOI] [PubMed] [Google Scholar]
- 42.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, et al. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 43.Maciá MD, Borrell N, Pérez JL, Oliver A. Detection and susceptibility testing of hypermutable Pseudomonas aeruginosa strains with the Etest and disk diffusion. Antimicrob Agents Chemother. 2004;48:2665–2672. doi: 10.1128/AAC.48.7.2665-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978;4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- 45.Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 2010;6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandsberg LF, Ciofu O, Kirkby N, Christiansen LE, Poulsen HE, et al. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob Agents Chemother. 2009;53:2483–2491. doi: 10.1128/AAC.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogardt M, Hoboth C, Schmoldt S, Henke C, Bader L, et al. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis. 2007;195:70–80. doi: 10.1086/509821. [DOI] [PubMed] [Google Scholar]
- 48.Sobel ML, McKay GA, Poole K. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2003;47:3202–3207. doi: 10.1128/AAC.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, et al. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaaper RM, Dunn RL. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol. 2006;188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moyano AJ, Smania AM. Simple sequence repeats and mucoid conversion: biased mucA mutagenesis in mismatch repair-deficient Pseudomonas aeruginosa. PLoS One. 2009;4:e8203. doi: 10.1371/journal.pone.0008203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 54.Römling U, Tümmler B. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:464–465. doi: 10.1128/jcm.38.1.464-465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pezza RJ, Smania AM, Barra JL, Argaraña CE. Nucleotides and heteroduplex DNA preserve the active conformation of Pseudomonas aeruginosa MutS by preventing protein oligomerization. Biochem J. 2002;361:87–95. doi: 10.1042/0264-6021:3610087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 59.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 61.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacquelín DK, Filiberti A, Argaraña CE, Barra JL. Pseudomonas aeruginosa MutL protein functions in Escherichia coli. Biochem J. 2005;388:879–887. doi: 10.1042/BJ20042073. [DOI] [PMC free article] [PubMed] [Google Scholar]