Abstract
Many carbohydrate-active enzymes have complex architectures comprising multiple modules that may be involved in catalysis, carbohydrate binding, or protein-protein interactions. Carbohydrate-binding modules (CBMs) are a common ancillary module whose function is to promote the adherence of the complete enzyme to carbohydrate substrates. CBM family 32 has been proposed to be one of the most diverse CBM families classified to date, yet all of the structurally characterized CBM32s thus far recognize galactose-based ligands. Here we report a unique binding specificity and mode of ligand binding for a family 32 CBM. NagHCBM32-2 is one of four CBM32 modules in NagH, a family 84 glycoside hydrolase secreted by Clostridium perfringens. NagHCBM32-2 has the β-sandwich scaffold common to members of the family; however, its specificity for N-acetylglucosamine is unusual among CBMs. X-ray crystallographic analysis of the module at resolutions from 1.45–2.0 Å and in complex with disaccharides reveals that its mode of sugar recognition is quite different from that observed for galactose specific CBM32s. This study continues to reveal the diversity of CBMs found in family 32 and how these CBMs might impact the carbohydrate-binding specificity to the extracellular glycoside hydrolases in C. perfringens.
Keywords: NagH, Clostridium perfringens, carbohydrate-binding module, glycan, N-acetylglucosamine, glycoside hydrolase
INTRODUCTION
Carbohydrate specific antibodies, lectins and solute-binding proteins are three major classes of proteins whose study has helped shape our understanding of protein-carbohydrate interactions. A fourth category of carbohydrate binding proteins, carbohydrate-binding modules (CBMs), has recently emerged as a biologically important class of carbohydrate recognizing protein that has remarkable diversity of structure and function 1. CBMs are non-catalytic carbohydrate recognizing entities that are found as independently folding domains in multi-modular carbohydrate-active enzymes. To date, the large majority of studied CBMs are polysaccharide specific modules whose primary role appears to be to target the entire enzyme to specific carbohydrate structures in the plant cell wall 1. This keeps the enzyme in proximity to polysaccharide substrates thus increasing the enzyme’s efficiency. The importance of carbohydrate-active enzymes and their cognate CBMs in the biological recycling of photosynthetically fixed carbon and their potential applications in controlled bioconversion strategies has, in part, driven CBM research. This has resulted in the current classification of 53 CBM families, which are defined on the basis of primary structure similarity and whose individual members have binding specificities that span a wide array of plant cell wall polysaccharides, storage polysaccharides and other glycans 1,2.
CBMs are now increasingly being found in carbohydrate-active enzymes from bacterial pathogens and bacteria involved in commensal relationships with human hosts. Characterized members of families 32 3–5, 40 4,6, 41 7, 47 8, 48 9, and 51 10 have demonstrated specificity for human glycans such as terminally galactosylated or sialylated glycans, the LewisY antigen, the blood group A/B-antigens, and glycogen. Of these families, CBM32 stands out as one of the largest and most diverse families whose members are often found associated with bacterial enzymes that are likely to interact with human glycans 11. Indeed, CBM32s are found extensively in the glycoside hydrolases of pathogenic species of Bacteroides, Clostridium, and Streptococcus 11. Clostridium perfringens is a particularly interesting example that features prominently when considering CBM32s in bacterial pathogens. C. perfringens is a prolific producer of toxins and secreted virulence factors, all of which contribute to its prowess as a pathogen that causes gangrene, necrotic enteritis and gastroenteritis12,13. In addition to its major toxins, C. perfringens produces a battery of additional toxins/virulence factors. Of the eight known additional toxins, three are glycoside hydrolases (enzymes that cleave glycosidic bonds by hydrolysis reactions): the sialidases (NanI and NanJ) and a hyaluronidase (μ-toxin or NagH)14–19. The sialidase NanJ is known to contain one galactose specific CBM32 4 while NagH contains four putative CBM32s 3. In addition to the sialidases and the NagH, the recently released genome sequences of 3 isolates of C. perfringens (ATCC 13124, 13, and SM101) have revealed a large number of open reading-frames encoding additional putative glycoside hydrolases 20,21. In C. perfringens ATCC 13124, thirteen putative glycoside hydrolases are predicted to be secreted into the extracellular milieu while six more are predicted to be cell-wall attached through a sortase-mediated process. These enzymes, whose predicted specificities are consistent with activity on human glycans, contain twenty-six putative CBM32s. Five more putative CBMs are found in hypothetical C. perfringens proteins of unknown function bringing the total number of putative CBM32s in the C. perfringens (ATCC 13124) genome to 31. Significantly, a phylogenetic analysis of all CBM32s, which are often annotated as FA5/8C modules, and the C. perfringens subset of CBM family 32 reveals that modules in this family display a high degree of sequence divergence, hinting at the possible variability of ligands other than galactose recognized by CBM32s 11.
All of the CBM32s that have been structurally characterized to date recognize galactose-based ligands; these are either terminal galactose residues or polymers of galacturonic acid 3–5,22. However, the focus of these studies has been mainly on a relatively small and closely related region of the CBM32 phylogenetic tree and likely does not reflect the potential diversity of the CBM32 binding specificity hypothesized by Abbott et al. 11. Here we focus on a branch of the CBM32 phylogenetic tree that is quite distantly related to the cluster of galactose binding CBM32s, leading us to surmise that modules on this branch may have a specificity for non-galactose based ligands. Members of this branch are found exclusively in family 84 exo-β-D-N-acetylglucosaminidases, which are homologs of C. perfringens NagH. Indeed, the second of the four CBM32s from NagH, here referred to as NagHCBM32-2, is found in this cluster. Given a tenet of CBM research - CBM binding specificity will more often than not match the specificity of the catalytic module 1 - we hypothesized that this putative CBM will recognize a ligand containing N-acetylglucosamine (glcNAc). In this work, we provide support for this hypothesis by quantifying the interaction of NagHCBM32-2 with glycans bearing terminal N-acetylglucosamine residues and presenting the structural basis of this interaction through high-resolution X-ray crystallographic analysis. Not only does this work provide compelling support for the proposed ligand diversity of family 32 CBMs but also it continues to illuminate the possible role of CBM32s, and CBMs in general, in host-pathogen recognition.
RESULTS AND DISCUSSION
NagHCBM32-2 is an N-acetylglucosamine binding CBM. NagH from C. perfringens (ATCC 13124) is a large protein (1627 amino acids) with a complex modular architecture 3,23,24 (Figure 1a). Roughly in the middle of this protein, amino acids ~660–1160, are four contiguous modules that are classified as family 32 CBMs. Phylogenetic analysis and amino acid sequence comparisons with galactose binding CBM32s identified the second module of this quartet, NagHCBM32-2, as potentially having unique binding properties. The domain boundaries of this particular module were more accurately predicted by combining secondary structure analysis with sequence alignments. The gene fragment encoding NagHCBM32-2 was then cloned and the polypeptide overproduced in E. coli for subsequent study.
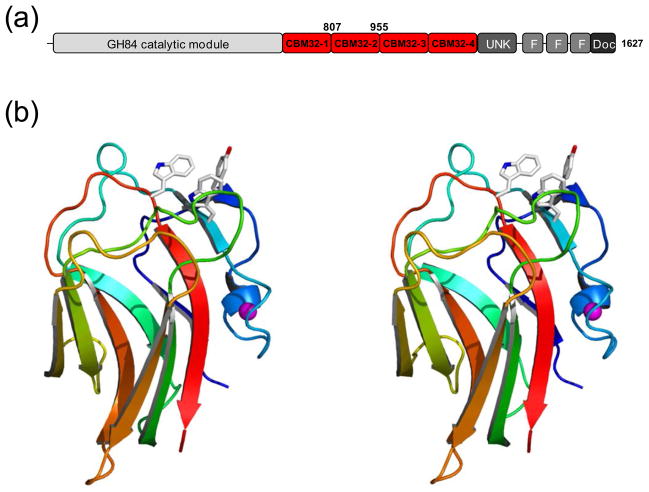
Figure 1.
Structural features of C. perfringens NagH. (a) Schematic of the modular architecture of NagH. The catalytic module is shown in grey and red indicates family 32 carbohydrate-binding modules. CBM32-2 corresponds to NagHCBM32-2 whose amino acid boundaries are indicated above the schematic. Other module designations are F, found in various architectural regions (FIVAR) modules; Doc, dockerin modules; and UNK, module of unknown function. (b) Divergent stereo color ramped cartoon representation of the high-resolution X-ray crystal structure of NagHCBM32-2. Residues in the carbohydrate-binding site are shown in stick representation. The bound metal ion is shown as a magenta sphere.
The carbohydrate-binding specificity of NagHCBM32-2 was initially screened by glycan array screening through Core H of the Consortium for Functional Glycomics but, unfortunately, the results of this approach were not conclusive. We attributed the poor binding of NagHCBM32-2 on the arrays to the low binding affinity of the module in its isolated monomeric form, a property that appears to be characteristic of this family of CBMs 3,4,25. Given the common occurrence of chromophoric amino acid side chains in the binding sites of CBM32s, which in other cases have been shown to be sensitive to ligand binding 3,4,22,25, we turned to examining the carbohydrate binding properties of NagHCBM32-2 by UV difference analysis. UV difference scans of NagHCBM32-2 obtained the presence of a wide variety of monosaccharides, including glucose, mannose, galactose, N-acetylglucosamine (glcNAc), N-acetylgalactosamine (galNAc), fucose, and sialic acid, yielded a characteristic UV difference spectrum only when glcNAc was added to the protein, indicating binding to this sugar (Supplementary Figure S1). Quantitative UV difference titrations and isothermal titration calorimetry (ITC) of glcNAc titrated into NagHCBM32-2 gave association constants (Ka) of 2.22 (± 0.25) × 103 M−1 and 1.88 (± 0.02) × 103 M−1, respectively, which were in good agreement with one another and consistent with the affinities of other family 32 CBMs for monosaccharides 3,4,25 (Supplementary Figure S1).
The majority of family 32 CBMs studied to date recognize non-reducing terminal galactose residues. Though NagHCBM32-2 displays a different specificity we conjectured that NagHCBM32-2 might specifically bind non-reducing terminal glcNAc residues. In initial qualitative binding analyses, NagHCBM32-2 showed no capacity to bind lacNAc (galβ1-4glcNAc) but did bind glcNAcβ1-3galNAc suggesting a requirement for a non-reducing terminal glcNAc, which was subsequently confirmed by the determination of the structure of NagHCMB32-2 in complex with glcNAcβ1-3galNAc (see below). Thus, we focused our attention on potential ligands possessing non-reducing terminal glcNAc residues.
GlcNAc is typically found in the human body as components of complex N-linked glycans and O-linked glycans. In these glycans, glcNAc is found at the “non-reducing” position in a handful of disaccharide motifs where glcNAc is attached to mannose, galNAc, galactose, or glcNAc with a variety of linkages (Supplementary Figure S2). We sought to investigate the interaction of NagHCBM32-2 with some of these disaccharides to determine if an additional sugar unit attached to the glcNAc is a better ligand and if it might discriminate between motifs found in N-linked vs. O-linked glycans. The Ka of NagHCBM32-2 for chitobiose (glcNAcβ1-4glcNAc), a motif found at the core of glycans N-linked to proteins, was determined by UV difference titrations to be 1.25 (± 0.13) × 103 M−1 and thus not substantially different from the CBM’s affinity for the monosaccharide glcNAc. GlcNAcβ1-2mannose was bound more tightly with a Ka of 7.86 (± 0.37) × 103 M−1, also determined by UV difference titrations and roughly 4-fold higher than the affinity for glcNAc. ITC analysis of glcNAcβ1-3galNAc binding yielded a Ka of 5.91 (± 0.33) × 103 M−1, approximately 3 fold higher than the affinity for glcNAc. NagHCBM32-2 appears, therefore, to have a preference for disaccharides; however, it does display some promiscuity with respect to the sugar linked to the glcNAc. Indeed, the CBM was able to bind with very similar affinities to disaccharide motifs found in both N-linked (glcNAcβ1-2mannose) and O-linked (glcNAcβ1-3galNAc) glycans. To further probe the limited specificity of the putative secondary subsite, we also tested the binding to glcNAcβ1-3mannose, which is a biologically uncommon motif and thus instructive in probing the non-specific recognition of the reducing sugar. The affinity for this sugar was determined by UV difference titrations to be 3.98 (±1.08) × 103 M−1, only slightly lower than the values determined for the other two disaccharides but still higher than for a glcNAc monosaccharide. Taken together, these results indicate that NagHCBM32-2 requires a glcNAc as the primary recognition determinant but binding can be enhanced when a sugar residue is linked to the reducing end of glcNAc.
Structural insights into N-acetylglucosamine recognition
The ligand binding properties of NagHCBM32-2 are unprecedented for a CBM32, and CBMs in general. To further explore the molecular basis of NagHCBM32-2’s carbohydrate-binding specificity we determined its three-dimensional structure by X-ray crystallography. Initially, the uncomplexed structure of NagHCBM32-2 was determined to 1.45 Å by single isomorphous replacement using seleno-methionine substituted protein. Like other CBM32s NagHCBM32-2 adopts a β-sandwich comprising a 5-stranded anti-parallel β-sheet packed against a 4-stranded anti-parallel β-sheet (Figure 1b). Also in accordance with other CBM32s, NagHCBM32-2 coordinates a single metal ion, which has been modeled as a Ca2+ ion on the basis of coordination geometry (bond lengths of ~2.2 Å), coordination chemistry (only oxygen ligands), and B-factor analysis. This atom is distal to the binding site and is unlikely to play a role in carbohydrate recognition (Figure 1b).
To obtain insight into carbohydrate recognition by NagHCBM32-2 we co-crystallized the module with glcNAcβ1-3galNAc and glcNAcβ1-2mannose. Determination of these structures yielded clear electron density for sugars bound to each of the four molecules of NagHCBM32-2 in the asymmetric unit for both of the carbohydrate ligands (Figure 2). The binding site is at the tip of the β-sandwich amongst the loops that link the two β-sheets (Figure 1b), a location that appears to be conserved in family 32 CBMs and several other CBM families 3,8,26,27. The binding site is remarkably shallow giving the impression that the sugar is sitting flat on the surface of the protein (Figure 3a). Despite its shallow nature, it is clear from the topography of the binding site around the non-reducing end of the glcNAc residue that this protein can accommodate only terminal glcNAc residues as additional sugar residues on the non-reducing terminus would be sterically legislated against (Figure 3a). In contrast, a second subsite at the reducing end of the glcNAc clearly accepts an additional sugar. The reducing end of the sugar in the second subsite extends into solvent suggesting that this protein could bind longer sugars unhindered but would be unlikely to make any additional interactions with it. Thus, the general features of the binding site indicate that NagHCBM32-2 binds the terminal glcNAc moieties of glycans that are soluble, attached to macromolecules such as proteins or lipids, or possibly to the non-reducing ends of glcNAc containing polysaccharides such as glycosaminoglycans.
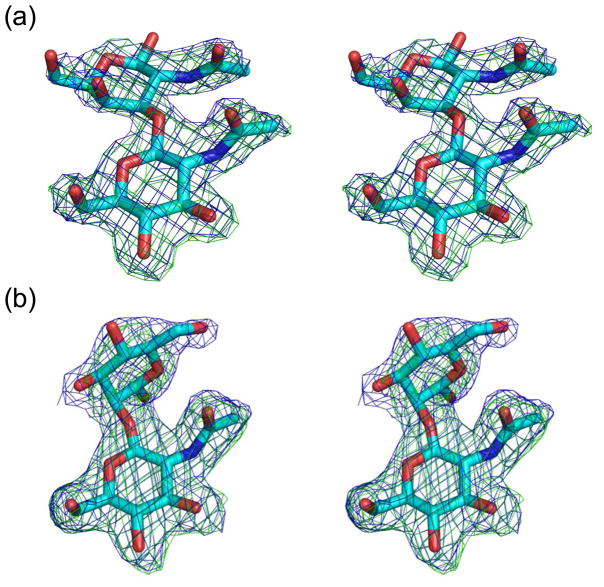
Figure 2.
Electron density of (a) glcNAcβ1-3galNAc and (b) glcNAcβ1-2mannose bound to NagHCBM32-2. Sugars are shown in cyan stick representation. Blue mesh represents the maximum-likelihood44/σa-weighted52 2Fobs-Fcalc electron density maps contoured at 1σ (0.41 electrons/Å3 for glcNAcβ1-3galNAc and 0.31 electrons/Å3 for glcNAcβ1-2mannose). Green mesh represents the maximum-likelihood/σa-weighted Fobs-Fcalc electron density maps obtained from refinements with the sugar atoms omitted and contoured at 3σ (0.25 electrons/Å3 for glcNAcβ1-3galNAc and 0.18 electrons/Å3 for glcNAcβ1-2mannose). For simplicity, only the sugars bound to monomer A (of four monomers) in the asymmetric unit are shown. Images are shown in divergent stereo.
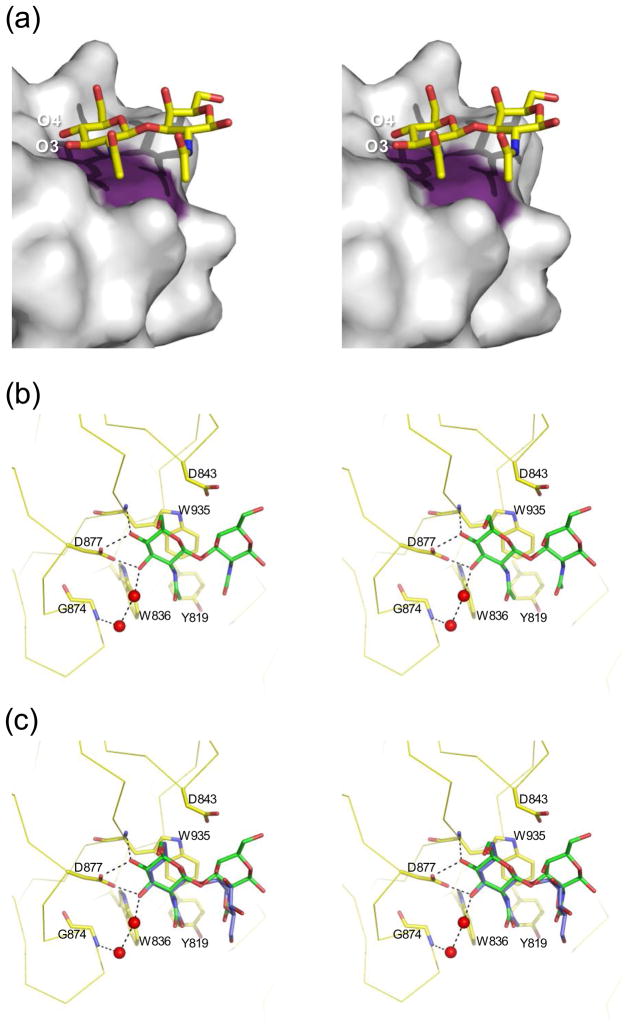
Figure 3.
Structural properties of ligand binding by NagHCBM32-2. (a) Surface properties of the NagHCBM32-2 binding site. The solvent accessible surface of NagHCBM32-2 is shown in grey with the surface contributed by a Trp935 colored in purple. The sugar is shown in yellow stick representation. O3 and O4 of the non-reducing end are labeled for reference. (b) Representation of the interactions made in the active site. Residues in the active site involved in binding glcNAcβ1-3galNAc are shown in yellow stick representation and labeled with the backbone of the protein shown in ribbon representation. The sugar is shown in green stick representation. Possible hydrogen bonds are shown as black dotted lines. Relevant waters are shown as red spheres. These interactions are identical for the four molecules in the active site. (c) Overlap of glcNAcβ1-3galNAc (green) with glcNAcβ1-2mannose (blue; only from monomer A). Only the sidechains from the glcNAcβ1-3galNAc complex are shown as the conformations of the sidechains in both complexes were nearly identical. All panels are shown in divergent stereo.
The primary interactions between the sugar and the protein occur in the glcNAc-binding subsite and these interactions were virtually identical for the two carbohydrate ligands (Figures 3b and c). Trp935 acts as a hydrophobic platform that lies parallel to the surface of the protein and interacts with the B-face of the glcNAc sugar ring. The O4 of glcNAc makes two hydrogen bonds, one with the main chain amide nitrogen of Trp935 and one with the carboxylate group of Asp877. These interactions select for the equatorial O4 hydroxyl of glcNAc allowing the protein to discriminate against galacto-configured sugars. Asp877 also makes a hydrogen bond with the O3 of glcNAc. Finally, one indirect interaction is mediated by two water molecules between Gly874 and the O3 of glcNAc. Selectivity for glcNAc necessarily involves its defining acetamido group. The equatorial acetamido group of glcNAc is oriented so that it, and particularly its C8 methyl group, tucks into a shallow hydrophobic pocket created by Tyr819, Trp836 and Trp935 thus providing substantial additional van der Waals interactions (Figures 3b and c). This interaction likely provides the selective binding of glcNAc over glucose.
Non-selective binding at a secondary subsite
While the primary subsite imparts specificity for non-reducing glcNAc residues, the secondary subsite appears much less selective. With respect to occupation of galNAc in the secondary subsite, this residue of glcNAcβ1-3galNAc makes no direct or indirect hydrogen bonds in the secondary subsite. However, the C4-C5-C6 edge of the galNAc pyranose ring lies against the planar arm of Asp843, resulting in several van der Waals interactions (Figures 3b). Likewise, the acetamido group of galNAc nestles against a hydrophobic surface provided by the aromatic residues Tyr819 and Trp935 as well as packing against the acetamido group of the glcNAc in the primary subsite (Figures 3b). These additional interactions likely explain the enhanced affinity of NagHCBM32-2 for glcNAcβ1-3galNAc relative to glcNAc.
The affinity of NagHCBM32-2 for glcNAcβ1-2mannose is less well explained by the structure. The axial configuration of the mannose O2, which is involved in the glycosidic linkage, disposes the plane of the mannose at roughly right angles to that of the glcNAc residue in the primary binding site (or the galNAc residue in the glcNAcβ1-3galNAc ligand) (Figure 3c). This results in limited additional van der Waals interactions between Trp 935 and C1 of the mannose residue. However, the conformation of the mannose residue and surrounding sidechains varied slightly for the sugars bound to each of the four NagHCBM32-2 monomers in the asymmetric unit (not shown). In monomer A, no direct or indirect hydrogen bonds are made with mannose (Figure 3c). However, in monomer B and C a water mediated hydrogen bond network, involving two water molecules, bridges Asp843 and O1 of mannose. Finally, in monomer D, Tyr 819 makes a water mediated hydrogen bond to the O6 of mannose. This variability between the interactions observed in the four monomers may reflect transient binding conformations of the mannose; however, we cannot rule out the influences of crystal packing. The recognition of mannose in the secondary subsite seems to be particularly plastic as a limited number of apparently transient interactions might be made. Nevertheless, the few additional interactions between the sugar in the secondary subsite and the protein appear to be sufficient to improve the binding energy of the disaccharides relative to the monosaccharide glcNAc. Overall, the recognition of the sugar residues in the CBM’s secondary subsite appears to be structurally non-specific as it accommodates mannose and galNAc (and tolerates glcNAc). This appears to be the result of very few specific interactions made between sugars in this subsite and the protein.
Carbohydrate recognition in family 32 CBMs
The CBM32s whose carbohydrate binding properties have been studies at the structural level all bind ligands containing galacto-configured sugars. With the exception of YeCBM32 from Yersinia enterocolitica, which binds polymers of galacturonate 22, these CBM32s bind to non-reducing terminal galactose residues through a structurally conserved arrangement of residues in the active site 3–5. The CBM32 from C. perfringens GH84C (NagJ), here referred to as NagJCBM32, possesses this galactose binding motif and its ligand binding has been studied in detail through high resolution structural studies 3. This CBM32 provides the most suitable reference point for comparison with NagHCBM32-2.
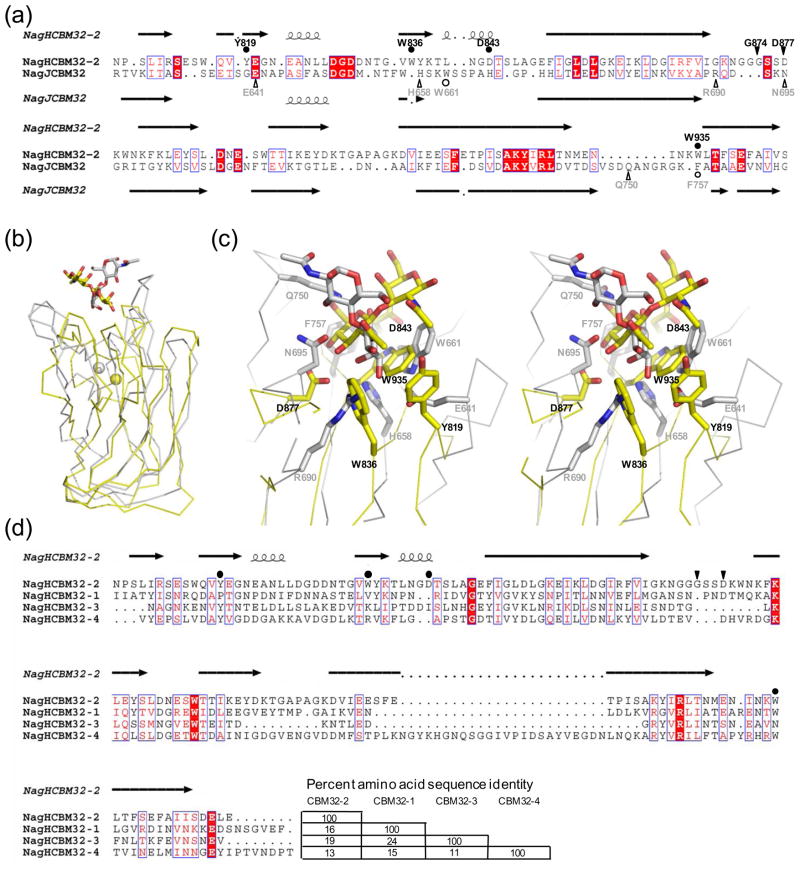
A structure based alignment of NagHCBM32-2 and NagJCBM32 using the secondary structure matching algorithm 28 (as implemented in COOT29) yields an overall low amino acid sequence identity of 15%, though the sequence identity is considerably better within the secondary structure elements that define the β-sandwich core of the module (Figure 4a). This structural alignment gives an r.m.s.d. of 2.7 Å over 122 matched amino acid residues indicating the structural relatedness of these proteins, which is visually evident when they are overlapped (Figure 4b). Indeed, the general location of the carbohydrate binding sites is also conserved. Quite striking at the primary structure level, however, is the complete lack of conserved residues involved in ligand recognition (Figure 4a and c). The only apparently semi-conserved residues are Trp935 of NagHCBM32-2 and Phe757 in NagJCBM32 (Figure 4a), yet these are not well conserved in three-dimensional space (Figure 4c). Trp935 of NagHCBM32-2 plays a key role in ligand recognition by packing against the pyranose ring of glcNAc and is only loosely conserved in three dimensions with Trp661 of NagJCBM32, which plays a similar role in galactose recognition by this CBM (Figure 4c). Likewise, Asp877 of NagHCBM32-2 and Asn695 of NagJCBM32 both engage in hydrogen bonds with their respective ligands and are approximately conserved in the structure (Figure 4c). However, they are involved in interactions with quite different parts of the ligand: Asp877 with O4 and O3 of glcNAc and Asn695 with O1 of galactose. These observations highlight that even at the tertiary structure level there is extremely little similarity in binding sites of NagHCBM32-2 and NagJCBM32.
Figure 4.
Comparison of NagHCBM32-2 with other family 32 carbohydrate-binding modules. (a) Amino acid sequence alignment of NagHCBM32-2 with the CBM, NagJCBM32, from C. perfringens NagJ based on a structural overlay produced by secondary structure matching 28. Secondary structures for NagHCBM32-2 and NagJCBM32 are shown above and below the alignment, respectively. Residues in NagHCBM32-2 involved in hydrogen bonds with the carbohydrate are indicated by filled triangles and labeled. Residues making primarily van der Waals interactions are indicated with filled circles. Residues performing these functions in NagJCBM32 are indicated below the alignment in open symbols. (b) Structural overlay of NagHCBM32-2 (yellow) and NagJCBM32 (grey). Bound sugars are shown in stick representation and metal ions as spheres. (c) The overlapped active sites of NagHCBM32-2 (yellow) and NagJCBM32 (grey) shown in divergent stereo. Bound ligands and residues that interact with these are shown in stick representation. Residues in NagHCBM32-2 are labeled in black and those in NagJCBM32 labeled in grey. (d) Amino acid alignment of NagHCBM32-2 with the other putative CBM32s in NagH. This alignment was produced using ClustalW 53. The secondary structure and functional residues of NagHCBM32-2 are indicated as in panel (a). Amino acid sequence identities of the modules are shown at the end of the alignment. The alignments in panels (a) and (d) were displayed using ESPript 54.
Where examined, CBM families that have diverse binding specificities typically have quite strong conservation of key functional amino acids with variation in a secondary group of functional amino acids that provides differences in ligand specificity 25,26,30–32. The closest to diverging from this observation is the family 51 CBMs; however, even with these CBMs there is quite good functional conservation of residues involved in recognizing a critical galacto-configured portion of their ligands 10. Thus, the comparison of NagHCBM32-2 with NagJCBM32 illustrates the unique and extreme binding site plasticity that may be found in family 32 CBMs. Nevertheless, this assertion must be tempered by the observation that family 32 is also currently the most diverse family at the primary amino acid sequence level with known or putative CBM32s on distant branches of the phylogenetic tree that may show as little as 10% sequence identity to one another11. Under normal circumstances sequence identity this low would be sufficient to classify new CBM families. However, the unique evolutionary structure of this family results in a wide and continuous spectrum of sequence variability making it impossible to divide CBM family 32 into separate families. For example, a putative CBM32 from another C. perfringens enzyme shows ~30% amino acid sequence identity to NagJCBM32, a known galactose binding CBM, which clusters with the canonical galactose binding CBM32s. This level of identity is sufficient to justify the sequence-based membership of the putative CBM32 in the family with NagJCBM32. Likewise, NagHCBM32-2 shows ~30% amino acid sequence identity to the putative CBM, which rationalizes NagHCBM32-2s classification as a family 32 CBM, yet NagHCBM32-2 only has ~15% amino acid sequence identity with NagJCBM32. The fascinating result of this “one big family” organization is the large structural and functional divergence that we have observed here. It is likely that we are yet to fully appreciate the functional variability in this family of CBMs.
NagH and the recognition of human glycans
Our results provide compelling evidence that NagHCBM32-2 binds non-reducing terminal glcNAc residues. The affinity for this motif is increased when it is β-1,3-linked to galNAc or β-1,2-linked to mannose and unaffected when it is β-1,4-linked to a second glcNAc. Precisely why a β-1,4-linked glcNAc provides no binding advantage is presently unclear but we can speculate that 180° relative rotation of the glcNAc residues in chitobiose may put the acetamido group of the reducing end sugar in a position such that it makes unfavorable interactions that compensate for any energetic gains made by the presence of the second sugar. Nevertheless, the β-linkage does appear to be generally selected for as it allows the pyranose ring of the non-reducing end glcNAc to pack against the indole ring of Trp935 in a parallel orientation. Though we were unable to test disaccharides of glcNAc α-linked to a second sugar, the structures of NagHCBM32-2 in complex with disaccharides suggest that the α-linkage would result in steric clashes with Trp935 and likely prevent the recognition of α-linked glcNAc residues. However, on the basis of the loose specificity of the protein and further examination of the X-ray crystal structure, it is entirely plausible that NagHCBM32-2 also binds to the other glcNAc containing motifs in N- or O-linked glycans: glcNAcβ1-3galactose, glcNAcβ1-4mannose, glcNAcβ1-6mannose, and glcNAcβ1-6galNAc (See Supplementary Figure S2). Furthermore, NagHCBM32-2 may also have the capacity to bind non-reducing glcNAc residues that might be found in glycosaminoglycans such as keratan and hyaluronate (See Supplementary Figure S2). Indeed, this latter possibility would be in keeping with NagH’s functional assignment as a hyaluronidase 19. However, we believe it improbable that NagH is a hyaluronidase and therefore unlikely that NagHCBM32-2’s biological function is to bind hyaluronate.
Recent structural and biochemical characterization of four homologs of NagH, all from glycoside hydrolase family 84 and including NagJ from C. perfringens, have revealed these proteins to be exo-β-D-N-acetylglucosaminidases, not hyaluronidases 33–36. This family of glycoside hydrolases uses a substrate assisted catalytic mechanism that requires the acetamido group of the glcNAc substrate and uses catalytic residues that are invariant in the family 33,36. Thus, NagH could not be active on the glucosamine found in hyaluronate (or the galactose in keratan). Furthermore, NagH and its composite modules contain no unique primary structure properties to suggest the protein has anything other than the exo-β-D-N-acetylglucosaminidase activity found in other family members (not shown). Though NagH may cleave the non-reducing end glcNAc residues of hyaluronate, C. perfringens does not appear to possess the β-glucuronidase that it would require to continue the depolymerizaton of hyaluronate using exo-glycosidases. On this basis, it seems implausible that the biological substrate of NagH is hyaluronate and suggests that the μ-toxin hyaluronidase of C. perfringens is an as yet unidentified protein. It is more likely that NagH is indeed an exo-β-D-N-acetylglucosaminidase involved in depolymerizing N- and O-linked glycans, like its homologs. Such glycans would have to be uncapped by additional enzymes, for example sialidases and β-galactosidases, all of which C. perfringens appears to possess, to expose β-linked non-reducing terminal glcNAc residues. This is consistent with the specificity of NagHCBM32-2 for β-linked non-reducing terminal glcNAc residues and in keeping with the proposed targeting function of CBMs 1.
NagH also contains three additional predicted CBM32s (Figure 1a). These three putative CBMs show little amino acid sequence identity to NagHCBM32-2 or to each other (Figure 4d). Furthermore, these all appear to lack any apparent conservation of residues that are involved in either glcNAc (Figure 4d) or galactose recognition (not shown); this is perhaps consistent with their positions on separate and distant branches of the CBM32 phylogenetic tree 11. The carbohydrate binding function, if there is any, for NagHCBM32-3 and NagCBM32-4 is unknown but we have evidence that the first CBM in the quartet, NagHCBM32-1, is able to bind galactose and galNAc (E. Ficko-Blean, unpublished results), a property that is also conspicuously at odds with NagH’s proposed hyaluronidase activity. This suggests that NagH also has the capacity to adhere to glycans terminating in sugars other than glcNAc. While this is inconsistent with the assumed principle that CBM specificity matches that of its cognate catalytic module, this does appear to be an emerging theme with Clostridial CBM32s where galactose binding CBM32s have been found appended to an exo-β-D-N-acetylglucosaminidase (NagJ)3 and a sialidase (NanJ)4 and thus mismatched with the activity of the catalytic modules. In these enzymes, the CBMs would appear to play a general role in adhering the enzyme to glycosylated molecules, rather than targeting a particular glycan. In NagH this may also be the case where the presence of multiple CBMs with differing specificities simply increases the likelihood of an interaction with a glycan bearing molecule or surface. Alternatively, the presence of several CBMs may promote avid binding to glycosylated surfaces displaying multiple glycans terminating in a variety of sugar residues. This type of avid binding using mismatched binding specificities might also provide greater specificity for glycan clusters that simultaneously display particular combinations of terminal sugars. The possibility for avid binding and its effect on specificity becomes particularly salient in light of NagH’s recently demonstrated capacity to form non-covalent complexes with other CBM32-containing glycoside hydrolases from C. perfringens, including NagJ and NanJ 23,24. This creates considerable potential for diverse and high-affinity binding of these molecular complexes to human glycans.
Conclusion
CBM family 32 is a large family of modules that displays a great deal of amino acid sequence divergence. On the basis of this sequence diversity we proposed that the family likely harbors modules with a wide range of carbohydrate-binding specificities. Centeno et al. reported the weak binding of a family 32 CBM from Cellvibrio mixtus to β-1,3-glucans supporting this theory of diversity 37. The determination of NagHCBM32-2’s binding to terminal glcNAc residues, which is unique among CBMs, provides additional support for the purported ligand diversity in family 32 CBMs 11. Furthermore, the structural analysis of NagHCBM32-2’s binding site and its comparison with canonical galactose binding CBM32s provides persuasive evidence for the evolutionary plasticity of CBM32 binding sites and their concomitant binding specificities.
The CBM32s from C. perfringens provides a unique snapshot of the diversity found in the family. These CBMs also have the interesting potential role of involvement in the organism’s recognition of host glycans. The characterization of NagHCBM32-2 now reveals that NagH is capable of adhering to terminal N-acetylglucosamine. The repeated presence of CBM32s, as single modules or in multiples, throughout C. perfringens glycoside hydrolases 3,24, combined with the potentially varied binding specificities of the CBMs, paints a picture of this bacterium’s enzymes, and possibly the bacterium itself, having a fascinatingly extensive capacity to recognize human glycans.
EXPERIMENTAL PROCEDURES
Carbohydrates
Monosaccharides were purchased from Sigma while all other glycans were purchased from Toronto Research Chemicals.
Cloning. C. perfringens
ATCC 13124 (Sigma) genomic DNA was used as a template to PCR amplify the gene fragment of the nagH gene encoding NagHCBM32-2. Primers were designed with engineered restriction sites for recombinant cloning purposes. The forward primer: CATATGGCTAGCAATCCAAGTTTAATAAGAAGTGAATCTTGGCAAGTT contains a NheI restriction endonuclease site. The reverse primer: GAATTCGGATCCTTACTCTTTATTTCCTGCATTTTCTAATTCATCACT contains a BamHI restriction endonuclease site. The gene fragment (nucleotides 2419–2865 encoding amino acids 807–955) was cloned into pET28a(+) (Invitrogen) via the NheI and BamHI restriction sites generating pCBM32-2. The recombinant polypeptide has an N-terminal six-histidine tag followed by a thrombin protease cleavage site then the CBM.
Protein Production and Purification
For protein production pCBM32-2 was transformed into the expression strain Escherichia coli BL21 Star (DE3) (Novagen). Luria-Burtani broth (3L) containing 50 μg/mL kanamycin was inoculated with the transformed cells and incubated at 37°C until an OD~1 at 595 nm was reached whereupon protein production was induced using 0.5 mM isopropyl 1-thio-β-D-galactopyranoside. Incubation of the cultures was continued overnight at 37°C. Cells were harvested at 5180 × g for 10 minutes and lysed in Binding Buffer (20 mM Tris-HCl, pH 8.0, and 0.5 M NaCl) using a French Pressure Cell. The lysate was clarified by centrifugation at 48384 × g for 45 minutes and the supernatant loaded onto a Ni-NTA IMAC column (Amersham). Protein was eluted with binding buffer containing increasing concentration of imidazole (0–500mM). Samples from fractions were run using SDS PAGE and those fractions judged to have purity > 95% were pooled, concentrated and buffer exchanged into 20 mM Tris-HCl, pH 8.0, using a stirred cell concentrator (Amicon).
Protein to be used for crystallography was digested with thrombin (Novagen) overnight to remove the N-terminal six-histidine tag. The sample was further purified by size exclusion chromatography using a Sephacryl S-200 (GE Biosciences). Fractions containing NagHCBM32-2 were pooled and concentrated in a stirred cell concentrator.
In order to produce seleno-methionine labeled NagHCBM32-2 the methionine auxotrophic strain E. coli 834 (DE3) (Novagen) was transformed with pCBM32-2. Seleno-methionine minimal media (AthenaES) containing 40 mg of L-seleno-methionine and kanamycin to 50 μg/mL was inoculated with the transformed E. coli 834 (DE3) cells. Protein production and purification continued as described for unlabeled NagHCBM32-2.
Determination of protein concentration
The concentration of purified protein was determined by UV absorbance at 280 nm using the calculated molar extinction coefficient of 36130 M−1cm−1 38.
Crystallization and data collection
The hanging drop vapour diffusion method was used at 18 °C for all crystallization experiments. The optimized condition that gave diffraction quality crystals for the native and seleno-methionine preparation was 0.1M Bis-Tris, pH 6.0, 0.2 M MgCl2, 25% polyethylene glycol (PEG) 2000 MME. The crystallization solution supplemented with 10 % ethylene glycol was used as a cryoprotectant. NagHCBM32-2 was co-crystallized in complex with 5 mM glcNAcβ1-3galNAc or 5 mM glcNAcβ1-2mannose in 0.1 M Tris-HCl, pH 8.5, 0.2M sodium acetate, 30 % PEG 4000. In this case the crystallization solution containing 15% ethylene glycol was used as a cryoprotectant. In both cases crystals were cryo-cooled directly in a nitrogen stream at 113 K. Data was collected with a Rigaku R-AXIS IV++ area detector coupled to an MM-002 X-ray generator with Osmic “blue” optics and an Oxford Cryostream 700. Data was processed with Crystal Clear/d*trek 39. Data collection statistics are given in Table 1.
Table 1.
X-ray data collection and refinement statistics.
| NagHCBM32-2 | Selenium derivative | glcNAcβ1-2mannose | glcNAcβ1-3galNAc | |
|---|---|---|---|---|
| Data collection statistics | ||||
| Space Group | P212121 | P212121 | P3121 | P3121 |
| Unit Cell (α, β, γ)(Å) | 56.1, 61.5, 83.0 | 56.3, 61.9, 83.0 | 91.3, 91.3, 132.7 | 91.2, 91.2, 132.6 |
| Asymmetric Unit Contents |
2 molecules | 2 molecules | 4 molecules | 4 molecules |
| Resolution Range | 49.39-1.60 (1.64-1.60) | 20.00-1.45 (1.49-1.45) | 39.52-2.03 (2.10-2.03) | 20.00-2.00 (2.05-2.00) |
| R-merge (%) | 3.9 (25.6) | 5.7 (37.5) | 9.4 (33.9) | 9.9 (30.3) |
| Completeness (%) | 93.8 (86.9) | 99.1 (99.9) | 99.7 (100.0) | 98.6 (99.9) |
| I/σI | 19.2 (3.9) | 15.7 (3.2) | 11.6 (5.1) | 10.4 (4.6) |
| Redundancy | 3.5 (2.7) | 7.1 (4.2) | 7.0 (7.2) | 7.0 (6.7) |
| Total Reflections | 125786 | 773883 | 291472 | 299940 |
| Unique Reflections | 36193 | 109314 | 41797 | 43009 |
| Refinment statistics | ||||
| R-Value (%) | 17.5 | 20.1 | 19.4 | 16.9 |
| R-free Value (%) | 21.1 | 24.4 | 26.0 | 21.3 |
| Model stereochemistry | ||||
| bond lengths (Å) | 0.015 | 0.010 | 0.010 | 0.015 |
| bond angles (deg.) | 1.469 | 1.243 | 1.299 | 1.508 |
| chiral centres (Å3) | 0.107 | 0.094 | 0.079 | 0.111 |
| Average B-factors (Å2) | ||||
| Overall | 18.19 | 17.79 | 27.93 | 19.44 |
| Protein | 14.71 (A); 16.92 (B); | 14.28 (A); 16.61 (B) | 32.68 (A); 24.39 (B); 24.48 (C); 27.80 (D) | 21.19 (A); 17.18 (B); 17.07 (C); 17.41 (D) |
| Water molecules | 29.55 | 28.50 | 32.44 | 26.73 |
| Ligands | 13.48 (Ca); 14.79(Na); 12.55 (BTB) | 13.53 (Ca); 16.51 (Na); 11.53 (BTB) | 45.53 (A); 34.36 (B); 26.00 (C); 34.73 (D); 24.40 (Ca) | 30.74 (A); 23.39 (B); 16.41 (C); 24.65 (D); 16.5 (Ca); 32.2 (ACT) |
| Number of Atoms | ||||
| Total | 2720 | 2739 | 5092 | 5331 |
| Protein atoms | 1121 (A); 1106 (B) | 1127 (A); 1095 (B); | 1100 (A); 1161 (B); 1117 (C); 1128 (D); | 1100 (A); 1153 (B); 1109 (C); 1136 (D) |
| Waters | 475 | 499 | 478 | 704 |
| Ligand atoms | 2 (Ca); 2 (Na); 14 (BTB) | 2 (Ca); 2 (Na) 14 (BTB) | 26 (A); 26 (B); 26 (C); 26 (D); 4 (Ca) | 30 (A); 34 (B); 34 (C); 34 (D); 4 (Ca); 3 (ACT) |
| Ramachandran statistics | ||||
| Most favored | 88.0 | 88.8 | 88.2 | 89.3 |
| Additional allowed | 12.0 | 11.2 | 11.8 | 10.7 |
| Disallowed | 0 | 0 | 0 | 0 |
| PDB code | 2w1q | 2w1s | 2wdb | 2w1u |
Structure determination
ShelxC/D 40 was used to determine selenium substructure using isomorphous differences between the native and seleno-methionine derivative datasets. Two selenium sites were located, which were then used for single isomorphous replacement phasing with SHARP 41. Refinement of the two sites resulted in a phasing powers to 1.45 Å of 0.59 and 0.79 for centric and acentric reflections, respectively. Solvent flattening using DM 42 with a solvent content of 50% resulted in a figure-of-merit of 0.84. ARP/wARP 43 was able to build nearly complete models of the two molecules in the asymmetric unit. These were then completed manually using successive rounds of refinement and model building using the programs COOT 29 and REFMAC 44.
NagHCBM32-2 in complex with sugars was solved by molecular replacement using MOLREP 45 with the native structure of NagHCBM32-2 as a search model to find the four monomers in the asymmetric unit. These structures and their carbohydrate ligands were successive rounds of model building using COOT and refinement using REFMAC.
In all cases, water molecules were added using the REFMAC implementation of ARP/wARP and inspected visually prior to deposition. Five percent of the observations were flagged as “free” and used to monitor refinement procedures 46. Model validation was performed with SFCHECK 47 and PROCHECK 48. Final model statistics are given in Table 1.
UV difference experiments
UV difference scans were performed by taking a baseline absorbance scan on protein in solution (~30 μM, 20 mM Tris-HCl pH 8.0) between 270–300 nm. A second scan was performed after the addition of excess solid monosaccharide such as D-glucose, glcNAc (N-acetyl-D-glucosamine), L-fucose, D-galactose, D-galNAc (N-acetyl-D-galactosamine), and D-mannose. The baseline scan was subtracted from the second scan resulting in a difference scan. The difference scan was examined for the characteristic peaks and troughs associated with binding 49,50.
UV difference titrations were done as described previously 3,4. GlcNAc (25 mM) was titrated into NagHCBM32-2 (33.5 μM in 20 mM Tris-HCl, pH 8.0). GlcNAcβ1-3mannose (25 mM) and glcNAcβ1-2mannose (20 mM) were titrated into NagHCBM32-2 (28.5 μM in 20 mM Tris-HCl, pH 8.0). Absorbance was measured between 270 nm and 300 nm. Peak to trough differences were determined by subtracting trough from peak within the spectra for three wavelength pairs (292.1 nm, 278.4 nm; 284.4 nm, 278.4 nm; 292.1 nm, 289.1 nm). This value was plotted as a function of ligand concentration. Data was analyzed using MicroCal Origin software (version 7.0) using a one site binding model. Data is reported as an average of independent experiments and error reported is the standard deviation.
Isothermal titration calorimetry
ITC Calorimetry was performed as described previously 3 using a VP-ITC (MicroCal, Northampton, MA). Protein was dialyzed in 20 mM Tris-HCl, pH 8.0. Ligand was prepared by weight in buffer saved from dialysis. 25 injections of glcNAcβ1-3galNAc (2.75 mM) were titrated in 10 μL aliquots into NagHCBM32-2 (200.28 μM). Similarly, 25 injections of glcNAc (17.5 mM) were titrated in 10 μL aliquots into NagHCBM32-2 (200.28 μM). Heats of dilution, determined by titration of ligand into buffer, were subtracted from the appropriate experimental run. Due to the low affinities, and thus low C-values 51, the stoichiometries (n) were fixed at 1, justified by the crystal data which shows one binding site in the CBM. Data was fit with a one site binding model using MicroCal Origin software (version 7.0).
Supplementary Material
Acknowledgments
We are grateful to Core H of the Consortium for Functional Glycomics for their array screening efforts. The resources and collaborative efforts provided by The Consortium for Functional Glycomics were funded by NIGMS - GM62116. This work was supported by a Canadian Institutes of Health Research Operating Grant. EF-B is supported by doctoral fellowships from the Natural Sciences and Engineering Research Council of Canada and the Michael Smith Foundation for Health Research (MSFHR). ABB is a Canada Research Chair in Molecular Interactions and a MSFHR Career Scholar.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors have been deposited with the PDB codes of 2w1q (native), 2w1s (Se-met derivative), 2wdb (glcNAcβ1-2mannose complex), and 2w1u (glcNAcβ1-3galNAc complex).
References
- 1.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine tuning polysaccharide recognition. Biochem J. 2004;382:769–782. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2008;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ficko-Blean E, Boraston AB. The interaction of carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor. J Biol Chem. 2006;281:37748–57. doi: 10.1074/jbc.M606126200. [DOI] [PubMed] [Google Scholar]
- 4.Boraston AB, Ficko-Blean E, Healey M. Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens. Biochemistry. 2007;46:11352–60. doi: 10.1021/bi701317g. [DOI] [PubMed] [Google Scholar]
- 5.Newstead SL, Watson JN, Bennet AJ, Taylor G. Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. Acta Crystallogr D Biol Crystallogr. 2005;61:1483–91. doi: 10.1107/S0907444905026132. [DOI] [PubMed] [Google Scholar]
- 6.Moustafa I, Connaris H, Taylor M, Zaitsev V, Wilson JC, Kiefel MJ, von Itzstein M, Taylor G. Sialic acid recognition by Vibrio cholerae neuraminidase. J Biol Chem. 2004;279:40819–26. doi: 10.1074/jbc.M404965200. [DOI] [PubMed] [Google Scholar]
- 7.van Bueren AL, Higgins M, Wang D, Burke RD, Boraston AB. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol. 2007;14:76–84. doi: 10.1038/nsmb1187. [DOI] [PubMed] [Google Scholar]
- 8.Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–71. doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
- 9.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–62. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Gregg KJ, Finn R, Abbott DW, Boraston AB. Divergent modes of glycan recognition by a new family of carbohydrate-binding modules. J Biol Chem. 2008;283:12604–13. doi: 10.1074/jbc.M709865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott DW, Eirin-Lopez JM, Boraston AB. Insight into ligand diversity and novel biological roles for family 32 carbohydrate-binding modules. Mol Biol Evol. 2008;25:155–67. doi: 10.1093/molbev/msm243. [DOI] [PubMed] [Google Scholar]
- 12.Petit L, Gibert M, Popoff MR. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104–10. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 13.Rood JI. Virulence genes of Clostridium perfringens. Annu Rev Microbiol. 1998;52:333–60. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 14.Rood JI, Wilkinson RG. Relationship between hemagglutinin and sialidase from Clostridium perfringens CN3870: chromatographic characterization of the biologically active proteins. J Bacteriol. 1976;126:831–44. doi: 10.1128/jb.126.2.831-844.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rood JI, Wilkinson RG. Relationship between hemagglutinin and sialidase from Clostridium perfringens CN3870: gel filtration of mutant and reverant activities. J Bacteriol. 1976;126:845–51. doi: 10.1128/jb.126.2.845-851.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roggentin P, Gutschker-Gdaniec GH, Hobrecht R, Schauer R. Early diagnosis of clostridial gas gangrene using sialidase antibodies. Clin Chim Acta. 1988;173:251–62. doi: 10.1016/0009-8981(88)90012-5. [DOI] [PubMed] [Google Scholar]
- 17.Roggentin P, Rothe B, Lottspeich F, Schauer R. Cloning and sequencing of a Clostridium perfringens sialidase gene. FEBS Lett. 1988;238:31–4. doi: 10.1016/0014-5793(88)80219-9. [DOI] [PubMed] [Google Scholar]
- 18.Traving C, Schauer R, Roggentin P. Gene structure of the ‘large’ sialidase isoenzyme from Clostridium perfringens A99 and its relationship with other clostridial nanH proteins. Glycoconj J. 1994;11:141–51. doi: 10.1007/BF00731154. [DOI] [PubMed] [Google Scholar]
- 19.Canard B, Garnier T, Saint-Joanis B, Cole ST. Molecular genetic analysis of the nagH gene encoding a hyaluronidase of Clostridium perfringens. Mol Gen Genet. 1994;243:215–24. doi: 10.1007/BF00280319. [DOI] [PubMed] [Google Scholar]
- 20.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–40. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott DW, Hrynuik S, Boraston AB. Identification and characterization of a novel periplasmic polygalacturonic acid binding protein from Yersinia enterolitica. J Mol Biol. 2007;367:1023–33. doi: 10.1016/j.jmb.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Adams JJ, Gregg K, Bayer EA, Boraston AB, Smith SP. Structural basis of Clostridium perfringens toxin complex formation. Proc Natl Acad Sci U S A. 2008;105:12194–9. doi: 10.1073/pnas.0803154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ficko-Blean E, Gregg KJ, Adams JJ, Hehemann JH, Czjzek M, Smith SP, Boraston AB. Portrait of an enzyme: A complete structural analysis of a multi-modular beta-N-acetylglucosaminidase from Clostridium perfringens. J Biol Chem. 2009 doi: 10.1074/jbc.M808954200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boraston AB, Notenboom V, Warren RA, Kilburn DG, Rose DR, Davies G. Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and “galactose-binding” domains. J Mol Biol. 2003;327:659–69. doi: 10.1016/s0022-2836(03)00152-9. [DOI] [PubMed] [Google Scholar]
- 26.Czjzek M, Bolam DN, Mosbah A, Allouch J, Fontes CM, Ferreira LM, Bornet O, Zamboni V, Darbon H, Smith NL, Black GW, Henrissat B, Gilbert HJ. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J Biol Chem. 2001;276:48580–48587. doi: 10.1074/jbc.M109142200. [DOI] [PubMed] [Google Scholar]
- 27.Jamal-Talabani S, Boraston AB, Turkenburg JP, Tarbouriech N, Ducros VM, Davies GJ. Ab initio structure determination and functional characterization of CBM36; a new family of calcium-dependent carbohydrate binding modules. Structure (Camb) 2004;12:1177–87. doi: 10.1016/j.str.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, Davies GJ. Differential oligosaccharide recognition by evolutionarily-related beta-1,4 and beta-1,3 glucan-binding modules. J Mol Biol. 2002;319:1143–1156. doi: 10.1016/S0022-2836(02)00374-1. [DOI] [PubMed] [Google Scholar]
- 31.Henshaw J, Horne A, van Bueren AL, Money VA, Bolam DN, Czjzek M, Ekborg NA, Weiner RM, Hutcheson SW, Davies GJ, Boraston AB, Gilbert HJ. Family 6 carbohydrate binding modules in beta -agarases display exquisite selectivity for the non-reducing termini of agarose-chains. J Biol Chem. 2006;281:17099–107. doi: 10.1074/jbc.M600702200. [DOI] [PubMed] [Google Scholar]
- 32.Henshaw J, Bolam DN, Pires VM, Czjzek M, Henrissat B, Ferreira LM, Fontes CM, Gilbert HJ. The family 6 carbohydrate-binding module CmCBM6-2 contains two ligand-binding sites with distinct specificities. J Biol Chem. 2004;279:21560–8. doi: 10.1074/jbc.M401620200. [DOI] [PubMed] [Google Scholar]
- 33.Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, Davies GJ. Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13:365–71. doi: 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- 34.Sheldon WL, Macauley MS, Taylor EJ, Robinson CE, Charnock SJ, Davies GJ, Vocadlo DJ, Black GW. Functional analysis of a group A streptococcal glycoside hydrolase Spy1600 from family 84 reveals it is a beta-N-acetylglucosaminidase and not a hyaluronidase. Biochem J. 2006;399:241–7. doi: 10.1042/BJ20060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, van Aalten DM. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–78. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–22. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 37.Centeno MS, Goyal A, Prates JA, Ferreira LM, Gilbert HJ, Fontes CM. Novel modular enzymes encoded by a cellulase gene cluster in Cellvibrio mixtus. FEMS Microbiol Lett. 2006;265:26–34. doi: 10.1111/j.1574-6968.2006.00464.x. [DOI] [PubMed] [Google Scholar]
- 38.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55 ( Pt 10):1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 40.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 41.Evans G, Bricogne G. Triiodide derivatization and combinatorial counter-ion replacement: two methods for enhancing phasing signal using laboratory Cu Kalpha X-ray equipment. Acta Crystallogr D Biol Crystallogr. 2002;58:976–91. doi: 10.1107/s0907444902005486. [DOI] [PubMed] [Google Scholar]
- 42.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–4. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 46.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 47.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 49.Boraston AB, Ghaffari M, Warren RA, Kilburn DG. Identification and glucan-binding properties of a new carbohydrate-binding module family. Biochem J. 2002;361:35–40. doi: 10.1042/0264-6021:3610035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boraston AB, Creagh AL, Alam MM, Kormos JM, Tomme P, Haynes CA, Warren RA, Kilburn DG. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry. 2001;40:6240–6247. doi: 10.1021/bi0101695. [DOI] [PubMed] [Google Scholar]
- 51.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 52.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst A. 1986;42:140–149. [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix. Nucl Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.