Abstract
This study demonstrates, by using neutral comet assay and pulsed field gel electrophoresis, that hyperosmotic stress causes DNA damage in the form of double strand breaks (dsb). Different solutes increase the rate of DNA dsb to different degrees at identical strengths of hyperosmolality. Hyperosmolality in the form of elevated NaCl (HNa) is most potent in this regard, whereas hyperosmolality in the form of elevated urea (HU) does not cause DNA dsb. The amount of DNA dsb increases significantly as early as 15 min after the onset of HNa. By using neutral comet and DNA ladder assays, we show that this rapid induction of DNA damage is not attributable to apoptosis. We demonstrate that renal inner medullary cells are able to efficiently repair hyperosmotic DNA damage within 48 h after exposure to hyperosmolality. DNA repair correlates with cell survival and is repressed by 25 μM LY294002, an inhibitor of DNA-activated protein kinases. These results strongly suggest that the hyperosmotic stress resistance of renal inner medullary cells is based not only on adaptations that protect cellular proteins from osmotic damage but, in addition, on adaptations that compensate DNA damage and maintain genomic integrity.
Cells adapt to osmotic stress by cell volume regulation and protection of protein structure, activity, and metabolism to maintain proper cell function. Such protective responses are necessary because osmotic stress alters cell volume, the concentration and stability of proteins, and the rate of biochemical reactions (1). Proteins are protected from osmotic stress by molecular chaperones and compatible organic osmolytes (2, 3). We recently showed that mouse renal inner medullary collecting duct (mIMCD3) cells respond to hyperosmotic stress by induction of cell cycle arrest, the tumor suppressor p53, and the growth arrest and DNA damage inducible proteins GADD45 and GADD153 (4, 5). These responses are known hallmarks of signaling pathways that counteract DNA damage in mammalian cells (6, 7). DNA damage is defined as an alteration of DNA structure capable of causing cellular injury and reduction of viability or reproductive fitness of the organism (8). Thus, an induction of DNA repair pathways may be required to confer the high osmotic stress tolerance that is characteristic of renal inner medullary cells. However, in contrast to the wealth of knowledge about osmotic effects on cell volume and protein stability and function, little is known about the consequences of osmotic stress on DNA integrity in mammalian cells. Studies on several mammalian cell lines, including CHO cells (9), PAP-HT25 cells (10), human peripheral lymphocytes (11), and V79 cells (12), indicate that osmotic stress can lead to chromosomal aberrations. Such aberrations may be the consequence of an increased frequency of DNA double strand breaks (dsb) or result from an inhibition of constitutive DNA-repair mechanisms. Hyperosmotic stress inhibits inducible DNA repair pathways that are activated in response to ionizing radiation in some mammalian cells (13, 14). However, it is not known whether osmotic stress by itself increases the amount of DNA dsb. We conducted this study to test the hypothesis that hyperosmotic stress causes DNA dsb and that such damage could be the trigger for the hyperosmotic induction of p53, GADD45, GADD153, and cell cycle arrest. Our results indicate that hyperosmolality because of elevated [NaCl] (HNa) but not from high [urea] (HU) significantly increases the number of DNA dsb in mIMCD3 cells. These data suggest that DNA dsb are not caused by hyperosmolality per se but by changes in ionic strength, cell volume, or macromolecular crowding. Thus, the accumulation of urea in the renal inner medulla may be the result of evolutionary selection based on a high compatibility of this abundant solute with genomic integrity during hyperosmotic stress.
Methods
Cell Culture and Osmotic Stress Experiments.
mIMCD3 cells were generously provided by Steven R. Gullans (Harvard University, Boston) and used for all experiments. They were grown in medium consisting of 45% DMEM (low glucose), 45% Coon's improved F-12 medium (Irvine Scientific), and 10% FBS (Life Technologies, Grand Island, NY). Osmolality of isosmotic control medium was 300 ± 5 milliosmol (mosmol)/kg H2O. Cells were osmotically stressed by instantly replacing control medium with hyperosmotic medium that was prepared by addition of NaCl, urea, or other solutes as indicated. For some experiments, the medium was supplemented with 25 μM LY294002, a specific inhibitor of DNA-activated protein kinases, including DNA-dependent protein kinase (DNA-PK), ataxia telangiectasia mutated (ATM), and ATM related (ATR).
Neutral Comet Assay.
Neutral comet assays were carried out as described previously (15, 16) with minor modifications. Briefly, cells were harvested by gentle trypsinization, resuspended in 0.5% low melting point agarose (FMC), and applied to microscopic slides that were precoated with agarose. Slides were incubated exactly 5 min in cold (4°C) neutral lysis buffer containing 2% SDS, 25 mmol/liter EDTA-Na2, 35 mmol/liter N-lauroyl sarcosine, and 10 mmol/liter Tris base (pH 8.0). Single cell electrophoresis was carried out by placing slides containing agarose-embedded, lysed cells in a horizontal electrophoresis tray (Owl Scientific, Woburn, MA) and applying 40 V for 4 min at 20°C. Slides were stained with SYBR Gold (Molecular Probes) and analyzed by using a Leitz DMRD/RB fluorescence microscope (Leica, Deerfield, IL). Images were taken with a color chilled 3CCD digital camera (Hamamatsu, Middlesex, NJ) and the comet moment quantified for at least 50 cells per sample by using quantity-one software (Bio-Rad). The comet moment is defined as the ratio of DNA in the comet tail versus DNA in the comet head (17).
Pulsed Field Gel Electrophoresis (PFGE).
Cell samples were prepared by gentle trypsinization followed by embedding 107 cells/ml in 0.75% PFGE grade agarose (Bio-Rad). Agarose blocks were digested for 3 days at 37°C in ESP solution (18). ESP solution contained 0.5 mol/liter EDTA-Na2, 1% N-lauroyl sarcosine, and 1 mg/ml proteinase K (Roche Molecular Biochemicals). After digestion, agarose blocks were stored for <1 wk in fresh ESP solution at 4°C. PFGE was carried out in a CHEF Mapper XA system (Bio-Rad) using the following conditions: 0.75% agarose, 200 s pulse time, 48–72 h run time, 2 V/cm voltage, 106° angle, 1× TAE (40 mmol/liter Tris base, 40 mmol/liter glacial acetic acid, 1 mmol/liter EDTA-Na2) running buffer, 10°C constant temperature. Gels were stained in 5 μg/ml ethidium bromide for 30 min and analyzed with a FluorS MultiImager and quantity-one software (Bio-Rad). Genomic DNA up to 10 Mb in size can be separated by PFGE (18). Because mouse chromosomes are larger than 10 Mb, intact DNA will stay in the wells and broken pieces migrate into the lane during PFGE. The amount of DNA dsb is proportional to the FAR (FAR = amount of DNA migrated into the lane vs. total DNA present in the lane plus well) (19). We used Saccharomyces cerevisiae chromosomal DNA (Bio-Rad) as size standards. All samples within the same experiment were treated absolutely identical with regard to proteinase K digestion and PFGE, and no data from different experiments are compared. The number of replicates refers to data from different dishes of cells within an experiment.
DNA Ladder Assays.
Genomic DNA was isolated from 5 × 106 cells per sample by using a DNA ladder kit according to manufacturer instructions (Roche Molecular Biochemicals, cat. no. 1835246). We modified the protocol slightly by adding an incubation of the samples in 2 μg/ml of RNase A/T1 (Ambion, Austin, TX) at room temperature. After isolation, 3 μg of genomic DNA per sample were loaded on 1% agarose gels and electrophoresed for 3 h at 50 V. Gels were stained with 5 μg/ml ethidium bromide, destained in deionized water, and the DNA bands visualized by using a FluorS MultiImager (Bio-Rad).
Cell Morphology and Survival.
Morphological characteristics of mIMCD3 cells were evaluated with an inverted phase contrast microscope (CK40; Olympus, New Hyde Park, NY). Photographs were taken at ×400 magnification of each cell culture dish by using a 35-mm camera (Yashica, Somerset, NY). Cell survival was measured as the number of cells left attached on a dish after exposing them to different treatments. Cell counts were performed after harvest by using a Hausser hemocytometer.
Statistics.
Data analysis was done by using STATMOST32 software. Time series effects were evaluated by analysis of variance and differences between values within a single series by Student–Newman–Keuls test. Differences between pairs of data for the same time point were analyzed by F test followed by either paired t test or Mann–Whitney test.
Results
Hyperosmotic Stress Causes DNA dsb.
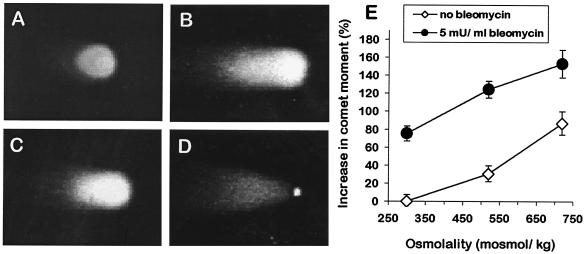
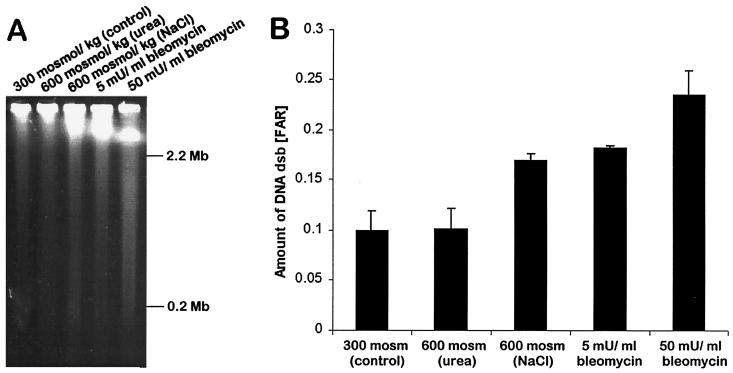
mIMCD3 cells respond to hyperosmotic stress by activation of growth arrest and DNA damage inducible proteins GADD45 and GADD153 and the tumor suppressor p53 (4, 5). Therefore, we investigated whether exposure of these cells to hyperosmotic stress leads to an increase in DNA dsb. When analyzed by neutral comet assay, mIMCD3 cells exposed to isosmotic medium have short comet tails, indicating that the number of constitutive DNA dsb is low (Fig. 1A). In contrast, comet tails are much larger after 60 min exposure of cells to 725 mosmol/kg HNa (Fig. 1B). Treatment of cells with 5 milliunit/ml bleomycin for 60 min also increases comet tails of mIMCD3 cells (Fig. 1C). Bleomycin was a positive control because it is a potent inducer of DNA dsb (20). These results suggest that DNA dsb increase rapidly during HNa. Indeed, the quantitative evaluation of comet tails from three independent experiments shows that DNA dsb increase significantly (P < 0.05) with increasing concentrations of NaCl in the medium (Fig. 1E). The amount of DNA damage during HNa is substantial because 60 min exposure of cells to 5 milliunit/ml bleomycin increases DNA dsb about as much as exposure to 600–650 mosmol/kg HNa (Fig. 1E). Effects of bleomycin and HNa are additive (Fig. 1E). We confirmed the hyperosmotic induction of DNA dsb with PFGE. Again, the amount of broken DNA is significantly higher in cells treated for 60 min with 600 mosmol/kg HNa than in isosmotic controls and comparable to that in cells treated with 5 milliunit/ml bleomycin for 60 min (Fig. 2). These data demonstrate that HNa leads to a significant and rapid increase in DNA dsb in mIMCD3 cells.
Figure 1.
Induction of DNA dsb in mIMCD3 cells by HNa. Neutral comet assay was used to measure DNA dsb. (A) Control cells kept in isosmotic medium have only a short comet tail. (B) The comet tail increases after exposure of cells for 1 h to 725 mosmol/kg HNa. (C) Exposure of cells for 1 h to 5 milliunit/ml bleomycin also increases the comet tail. (D) Appearance of apoptotic cells when analyzed by neutral comet assay. Note the highly condensed nucleus and the very high ratio of comet tail length and width to nuclear diameter. (E) Quantification of comet moment (fraction of DNA in the comet tail versus the comet head) in mIMCD3 cells. Data are means ± SEM (n = 3).
Figure 2.
HNa but not HU increases DNA dsb in mIMCD3 cells. PFGE was used to measure DNA dsb. (A) Representative PFGE gel illustrating the rate of migration of whole genomic DNA from cells exposed to isosmotic medium (300 mosmol/kg), 600 mosmol/kg HNa, 600 mosmol/kg HU, and 5 or 50 milliunit/ml bleomycin. (B) Quantification of DNA dsb by PFGE. The fraction of DNA recovered (FAR), which is the amount of DNA that has migrated from the well into the lane vs. total DNA in lane plus well, is plotted. Data shown are means ± SEM (n = 3).
DNA Damage Results from Elevated NaCl but Not Urea.
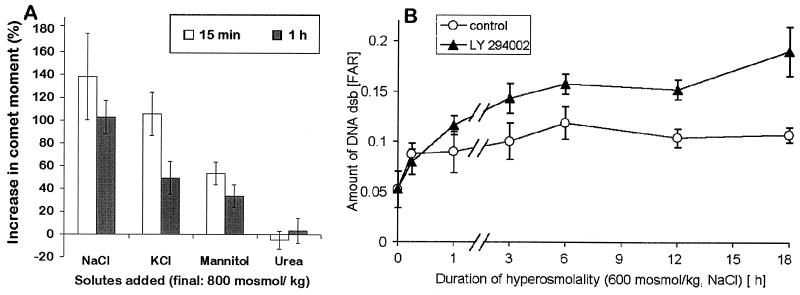
We investigated whether the hyperosmotic induction of DNA dsb is related to hyperosmolality per se or a result of the concentration increase of particular solutes. Interestingly, 60 min exposure of cells to 600 mosmol/kg HU did not significantly increase DNA dsb (Fig. 2). Urea penetrates mammalian cell membranes very well and does not disturb cell volume. In contrast to inorganic ions, urea is also a neutral solute. Thus, the different effects of HNa vs. HU on DNA dsb could be attributed to either (i) solute-specific mechanisms, or (ii) increases in intracellular ionic strength, macromolecular density, or cell/nuclear volume. To distinguish between these two possibilities, we exposed cells to hyperosmolality in the forms of elevated NaCl, KCl, mannitol, and urea. Within 15 and 60 min, all solutes except urea cause an increase in DNA dsb (Fig. 3A). At 60 min, this increase is significantly larger (P < 0.05) for NaCl compared with KCl and mannitol. At 15 min, the increase in DNA dsb was not significantly different between NaCl and KCl, but significantly smaller (P < 0.05) for mannitol (Fig. 3A). These data suggest that hyperosmotic induction of DNA dsb is partly a result of changes in intracellular ionic strength, cell/nuclear volume, or macromolecular density, and partly because of solute-specific effects. The significant differences between NaCl, KCl, and mannitol for inducing DNA dsb are difficult to explain because, in theory, all three solutes do not freely penetrate cell membranes and should lead to comparable increases of intracellular inorganic ions. It may be possible, however, that the intracellular Na+ concentration increases more during HNa as a consequence of steeper Na+ gradients for Na+-coupled cotransporters. If this is the case, our results suggest that Na+ is more potent for inducing DNA dsb than K+.
Figure 3.
Kinetics and solute-specificity of hyperosmotic induction of DNA dsb in mIMCD3 cells. (A) Cells were exposed to hyperosmolality of 800 mosmol/kg for 15 min and 1 h. Hyperosmolality was because of addition of various solutes to regular growth medium. All solutes except urea increase DNA dsb rapidly (within 15 min). (B) DNA dsb after exposure of mIMCD3 cells to 600 mosmol/kg HNa are elevated for at least 18 h. Inhibition of DNA-activated protein kinases by LY294002 during exposure of cells to 600 mosmol/kg HNa leads to a significantly larger increase in DNA dsb (P < 0.05), indicating that these kinases are important for preventing DNA dsb from occurring or for their repair. Data are means ± SEM (n = 3).
The Kinetics of DNA Damage During Hyperosmolality Is Rapid.
DNA dsb increase very rapidly after exposure of cells to severe, lethal HNa (800 mosmol/kg; Fig. 3A). Under these conditions, maximum damage is observed already after 15 min (data not shown). The kinetics of hyperosmotic induction of DNA dsb is slower (maximum at 3–6 h) when cells are exposed to lower, survivable levels of HNa (600 mosmol/kg; Fig. 3B). Interestingly, the amount of DNA damage remains elevated (but does not further increase) for at least 18 h (Fig. 3B). These kinetic data suggest that DNA dsb mainly occur in the early phase of HNa and that they occur more rapidly with increasing severity of HNa. The rate of DNA dsb increases rapidly within minutes to a few hours depending on the severity of HNa. It stays elevated but does not increase further for at least 18 h, indicating that the cells need longer than 18 h to fully match their DNA repair machinery to account for all of the damage. However, because DNA dsb remain elevated for many hours, either the rate of DNA dsb remains higher than normal even after the initial damage or DNA repair capacity is reduced in the first 24 h after HNa because of decreased DNA accessibility as a result of salt-induced chromatin compaction. Interestingly, during hyperosmolality in the form of high KCl DNA repair may be more efficient than during HNa (Fig. 3A).
DNA dsb Are Not Solely the Result of Apoptosis.
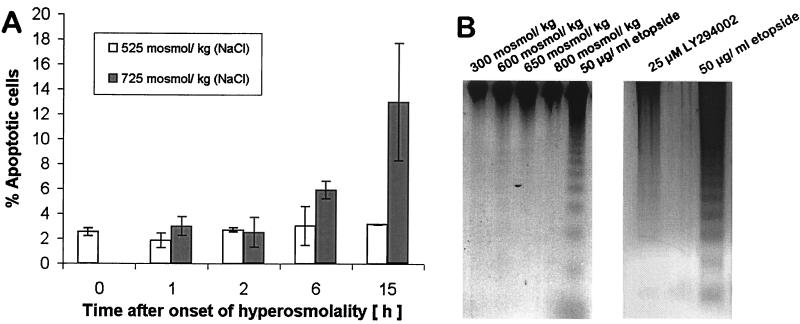
Many types of stress, including hyperosmotic stress (21, 22), induce apoptosis if the stress level exceeds the tolerance limit of cells. A characteristic end stage of apoptosis is degradation of genomic DNA into oligonucleosomal ladders (23). Thus, we can expect DNA dsb as a result of apoptosis when the osmotic tolerance threshold (600 mosmol/kg for mIMCD3 cells) is exceeded. However, increased DNA dsb observed rapidly after onset of HNa are not the result of apoptosis, even if HNa exceeds 600 mosmol/kg. This conclusion is based on four lines of evidence: (i) The kinetics of DNA dsb following HNa are too rapid to be accounted for by apoptosis, which generally needs several hours to more than a day to be apparent as DNA degradation; (ii) The size of DNA fragments resulting from the rapid hyperosmotic induction of DNA dsb is much larger (several Mb; Fig. 2A) than the oligonucleosome fragments generated by apoptosis; (iii) We have determined the number of apoptotic cells with the neutral comet assay that identifies apoptotic cells clearly on the basis of their highly condensed nuclei and very large comet tails (Fig. 1D) (24). Apoptotic cells do not increase significantly within 2 h of HNa, even if such stress exceeds the cellular tolerance threshold (Fig. 4A); and (iv) Oligonucleosomal ladders are not detectable in mIMCD3 cells exposed to hyperosmotic stress up to 800 mosmol/kg within 3 h of exposure (Fig. 4B). We conclude that increased DNA dsb occurring rapidly after the onset of HNa are not the result of apoptosis.
Figure 4.
Apoptosis cannot account for the increased DNA dsb during the early phase of hyperosmotic stress because its kinetics is slower than the induction of DNA dsb. (A) The percentage of apoptotic cells determined by neutral comet assay is low and not significantly different from isosmotic controls at all times when cells are exposed to 525 mosmol/kg HNa. Only after 15 h of exposure of cells to 725 mosmol/kg HNa does the number of apoptotic cells increase markedly. Data are means ± SEM (n = 3). (B) DNA ladder assay of mIMCD3 cells exposed to various degrees of HNa for 3 h. Equal amounts of whole genomic DNA were loaded in each well. Exposure of cells to 50 μg/ml etopside in serum-free medium served as a positive control for which the nucleosomal DNA ladder that is characteristic for apoptosis can be seen (last lane). DNA ladders are not present in genomic DNA of mIMCD3 cells exposed to HNa for 3 h, indicating that apoptosis is manifested at later times. Treatment with 25 μM LY294002 for 24 h did not induce apoptosis.
DNA-Activated Protein Kinases Confer Osmoresistance.
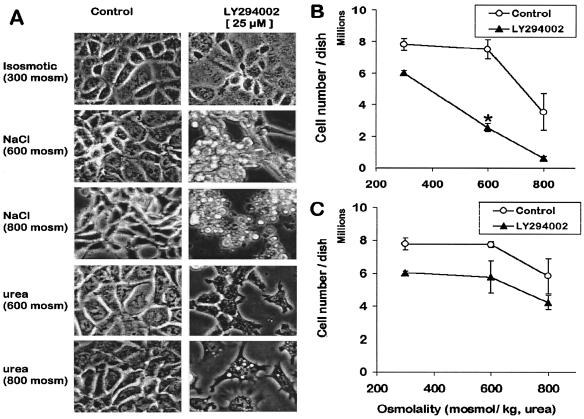
Ionizing radiation and chemical carcinogens that cause DNA dsb also induce DNA-activated protein kinases (DNA-PK, ATM, ATR) and p53 (25). Under these conditions, activation of DNA-activated protein kinases depends on the amount of DNA dsb and accounts for p53 activation and stress compensation (26). Because we have shown that HNa induces DNA dsb (this study) and p53 (5), we investigated whether DNA-activated protein kinases are necessary for cell survival during HNa. The synthetic compound LY294002 was used to inhibit DNA-activated protein kinases followed by analysis of cell morphology and survival after exposing cells to various osmolalities for 15 h. Inhibition of these kinases greatly alters cell morphology when cells are maintained in 600 and 800 mosmol/kg HNa (Fig. 5A). Specifically, LY294002 causes severe cell shrinkage and detachment (Fig. 5A). This is not the case with HU, although vacuolarization of some cells results under such conditions (Fig. 5A). Inhibition of DNA-activated protein kinases reduces cell survival by about 20% irrespective of osmolality, presumably because of inhibition of constitutive DNA repair (Fig. 5 B and C). However, cell survival at 600 mosmol/kg is only significantly reduced compared with isosmotic controls during HNa but not HU, and only if LY294002 is present (Fig. 5 B and C). These data indicate that DNA-activated protein kinases are necessary for cell survival during HNa but not HU.
Figure 5.
Solute-specific effects of inhibition of DNA-activated protein kinases on mIMCD3 cells exposed to hyperosmotic stress. (A) Morphology of mIMCD3 cells after growing them for 15 h in various media with or without 25 μM LY294002. (Magnification, ×400.) NaCl/urea 600 = 600 mosmol/kg HNa/HU, NaCl/urea 800 = 800 mosmol/kg HNa/HU. (B and C) Effect of LY294002 on number of cells left in a confluent 10-cm dish after 15 h culture in various osmolalities. (B) HNa. Data are means ± SEM (n = 3). Note that cell number drops significantly after exposure to 600 mosmol/kg with LY294002 but not without it. (C) HU. Data are means ± SEM (n = 3). Unlike with HNa, cell numbers are not significantly different between 300 and 600 mosmol/kg even in the presence of LY294002 during HU. When error bars are not visible, they are smaller than the corresponding symbol.
DNA Repair Requires DNA-Activated Protein Kinase Activity.
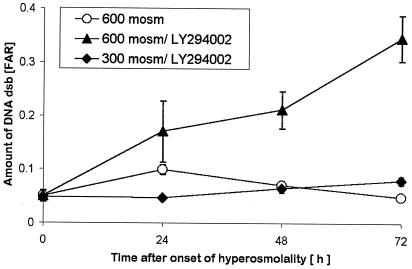
The dependence of cell survival on DNA-activated protein kinases during HNa but not HU suggests that they may be necessary for the repair of DNA dsb. We tested this notion by measuring DNA dsb in cells exposed to 600 mosmol/kg HNa. When DNA-activated kinases are inhibited by 25 μmol/liter LY294002, DNA dsb increase significantly more than without this inhibitor starting at 3 h (Fig. 3B). The effect of the inhibitor precedes apoptosis (Fig. 3B) but is greatest when cells are exposed to HNa for longer than 24 h (Fig. 6B). Whereas, in control cells, DNA dsb are fully repaired within 3 days, DNA dsb continue to increase to very high levels in the presence of 25 μmol/liter LY254002 (Fig. 6B). We conclude that DNA-activated protein kinases are necessary for DNA dsb repair during hyperosmotic stress.
Figure 6.
Suppression of cell growth and DNA repair by the DNA-activated protein kinase inhibitor LY294002. DNA dsb in mIMCD3 cells exposed to 600 mosmol/kg HNa are significantly increased at 24 h in isosmotic controls and in cells treated with 25 μM LY294002 plus HNa but not in cells treated with 25 μM LY294002 in isosmotic medium. DNA dsb continue to increase significantly when cells are left in HNa plus 25 μM LY294002, whereas they decrease to baseline by day 3 in HNa without LY294002. There is a small but significant increase in DNA dsb at 72 h when cells are exposed to 25 μM LY294002 in isosmotic medium. However, this increase is much smaller compared with the effect of LY294002 in HNa. DNA dsb were measured by PFGE; FAR = amount of DNA in lane/amount of DNA in (lane plus well). Data are means ± SEM (n = 3).
Discussion
We report that HNa but not HU causes DNA dsb in mammalian kidney inner medullary cells and that these cells have means to counteract such DNA damage. These results represent evidence for hyperosmotic stress-induced DNA damage. They provide a rationale for our previous observations of hyperosmotic induction of DNA damage pathways and checkpoints in mammalian kidney cells exposed to HNa (4, 5). The mechanism by which HNa causes DNA dsb is currently elusive. Only few DNA-damaging agents cause direct breakage of the DNA phosphodiester backbone, among them ionizing radiation, hydrogen peroxide, and bleomycin (17). The majority of genotoxic agents cause damage to nucleotide bases. Such damage is removed by nucleotide excision repair, which introduces DNA strand breakage as an intermediate stage (17). We currently do not know whether DNA dsb resulting from HNa are a direct result of breakage of the phosphodiester backbone or an indirect consequence of strand breakage during nucleotide excision repair. The fast kinetics of hyperosmotic DNA dsb seems to favor direct breakage of the phosphodiester backbone as the underlying mechanism. This, in turn, may explain the increased frequency of chromosomal aberrations in some mammalian cells exposed to hyperosmolality (9–12).
Several mechanisms by which DNA dsb could occur during hyperosmolality can be envisioned: First, increasing ionic strength, macromolecular crowding, or physical distortion of the nuclear matrix as a result of cell shrinkage may cause changes in DNA bending. This could translate into mechanical straining of DNA strands in areas where chromatin packaging of DNA is too rigid and enhance the likelihood of breakage in these regions. A certain number of DNA dsb occurs constitutively but such damage is readily compensated for by constitutive DNA repair. An increase of DNA dsb disturbs the steady state between constitutive DNA damage and repair with a net increase in DNA dsb. Because of the fast kinetics of hyperosmotic induction of DNA dsb, it is plausible that DNA regions prone to mechanical straining break rapidly during hyperosmotic stress as a direct consequence of cell/nuclear shrinkage, increased ionic strength, or macromolecular crowding, which are phenomena that are not evoked by HU. DNA repair (homologous recombination and/or nonhomologous end joining) seems to be a two-step process to: (i) prevent a further increase in DNA dsb and (ii) normalize the amount of DNA dsb.
The second scenario by which hyperosmotic stress could cause DNA dsb is by means of formation of free radicals. Although we are unaware of any direct evidence for an increase in free radicals during hyperosmotic stress, several indirect indications exist. For instance, several free radical scavenging enzymes, including catalase, superoxide dismutase, and gluthatione peroxidase, are activated during hyperosmotic stress in cells of the rat blood brain barrier (27). Furthermore, phospholipase A2 activity is rapidly osmoregulated in many cell types (28) leading to prostaglandin synthesis and production of free radicals by interconversion of prostaglandins (29). In addition, many compatible organic osmolytes protect cells against free radicals (30, 31).
DNA dsb may also be caused by changes of chromatin compactness and DNA accessibility during hyperosmotic stress. Such changes are likely because most forms of hyperosmolality lead to an increase of cationic counter ions that compete with histones and other positively charged nuclear proteins for binding to the negatively charged DNA phosphodiester backbone. Thus, an increase in these ions may replace histones from DNA and change chromatin structure. Such effects are commonly used in vitro for salt-extracting histones from chromatin (32), but it is currently uncertain to what extent they play a role in vivo. Changes in chromatin compactness lead to changes in DNA accessibility, and this may disturb the steady state of constitutive DNA repair vs. DNA damage or allow enhanced access of nucleases or radicals to certain regions of DNA. Alternatively, the stability of chromosomal sized DNA may suffer from replacement of histones by inorganic cations leading to an increased frequency of DNA dsb. These effects of inorganic ions are offset by accumulation of compatible organic osmolytes, which are more compatible with chromatin structure (33).
HU does not lead to increases in cationic counter ions because urea penetrates cell membranes relatively freely and causes no change in cell volume or ionic strength. This may explain the lack of increased DNA dsb during HU. Hyperosmolality because of increases of other solutes (KCl or mannitol) increases DNA dsb similar to HNa, presumably because they all result in changes of cell volume and ionic strength. However, KCl and mannitol are less potent for inducing DNA dsb than NaCl. This difference is likely a result of ion-specific effects. HNa may activate Na+-coupled cotransporters or increase passive leakage of NaCl into cells and thereby transiently increase the intracellular sodium concentration. However, it remains to be tested whether increasing intracellular Na+ is more potent than K+ for causing DNA dsb. Nevertheless, our data provide a rationale for why urea and not exclusively inorganic salts accumulate in several hyperosmotic tissues of vertebrates, including the mammalian kidney inner medulla and elasmobranch tissues. Although urea is highly proteotoxic, its availability as a nitrogeneous waste product and its compatibility with DNA structure may have been factors for its evolutionary selection as a major solute in hyperosmotic tissues of vertebrates. Urea is filtered into the nephron as a metabolic waste. However, it is not simply excreted but instead reabsorbed from the nephron into the inner medullary interstitium where it partly replaces NaCl. The reabsorption of urea into the renal inner medullary interstitium may be a result of evolutionary selection based on its benefit of being more compatible with genomic integrity of renal cells than NaCl during periods when the kidney has to adjust to producing highly concentrated urine.
Renal inner medullary cells tolerate large osmolality changes even if NaCl is increasing. Thus, they must have means to detect and repair DNA dsb under these conditions. Our data suggest that DNA-activated protein kinases such as DNA-PK, ATM, and/or ATR are necessary for detecting and/or repairing DNA dsb during hyperosmotic stress. These kinases are structurally similar to phosphatidylinositol-3-kinase (PI3K) and can be specifically inhibited by 25 μM LY 294002 (34, 35). However, our data do not exclude that PI3K, which is inhibited by 25 μM LY294003, or an unknown LY294002-sensitive protein kinase may mediate cell survival and DNA repair during hyperosmotic stress. Although plausible, we view the involvement of PI3K as less likely because DNA-activated protein kinases are directly activated by DNA dsb and participate in DNA damage pathways, whereas PI3K is not (35). Specifically, DNA-activated protein kinases phosphorylate and activate p53 on Ser15 (36), a phenomenon observed in mIMCD3 cells exposed to HNa but not HU (5).
In conclusion, our study indicates that HNa but not HU causes DNA dsb and that this DNA damage leads to activation of DNA-activated protein kinases that are necessary for DNA repair and cell survival during HNa. This, in turn, provides an explanation for the induction of the p53 pathway and growth arrest in mIMCD3 cells exposed to HNa.
Abbreviations
- dsb
double strand breaks
- PFGE
pulsed field gel electrophoresis
- DNA-PK
DNA-dependent protein kinase
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia mutated related
- HNa
hyperosmolality in the form of elevated NaCl
- HU
hyperosmolality in the form of elevated urea
- mosmol
milliosmol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Somero G N, Yancey P. In: Osmolytes and Cell Volume Regulation: Physiological and Evolutionary Principles. Hoffmann J F, Jamieson J D, editors. Washington, DC: Am. Physiol. Soc.; 1997. pp. 441–484. [Google Scholar]
- 2.Gullans S R, Cohen D M, Kojima R, Randall J, Brenner B M, Santos B, Chevaile A. Kidney Int. 1996;49:1678–1681. doi: 10.1038/ki.1996.245. [DOI] [PubMed] [Google Scholar]
- 3.Burg M B, Kwon E D, Kültz D. Annu Rev Physiol. 1997;59:437–455. doi: 10.1146/annurev.physiol.59.1.437. [DOI] [PubMed] [Google Scholar]
- 4.Kültz D, Madhany S, Burg M B. J Biol Chem. 1998;273:13645–13651. doi: 10.1074/jbc.273.22.13645. [DOI] [PubMed] [Google Scholar]
- 5.Dmitrieva N, Kültz D, Michea L, Ferraris J D, Burg M B. J Biol Chem. 2000;275:18243–18247. doi: 10.1074/jbc.M000522200. [DOI] [PubMed] [Google Scholar]
- 6.Hollander M C, Fornace A J., Jr . In: Cell DNA Repair Mechanisms: Impact on Human Diseases and Cancer. Vos J-M H, editor. New York: Verlag; 1995. pp. 219–237. [Google Scholar]
- 7.Kaufmann W K, Kies P E. Mutat Res. 1998;400:153–167. doi: 10.1016/s0027-5107(98)00041-4. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann W K, Paules R S. FASEB J. 1996;10:238–247. doi: 10.1096/fasebj.10.2.8641557. [DOI] [PubMed] [Google Scholar]
- 9.Galloway S M, Deasy D A, Bean C L, Kraynak A R, Armstrong M J, Bradley M O. Mutat Res. 1987;189:15–25. doi: 10.1016/0165-1218(87)90029-2. [DOI] [PubMed] [Google Scholar]
- 10.Uchida S, Green N, Coon H, Triche T, Mims S, Burg M. Am J Physiol. 1987;253:C230–C242. doi: 10.1152/ajpcell.1987.253.2.C230. [DOI] [PubMed] [Google Scholar]
- 11.Kalweit S, Nowak C, Obe G. Mutat Res. 1990;245:5–9. doi: 10.1016/0165-7992(90)90017-e. [DOI] [PubMed] [Google Scholar]
- 12.Nowak C. Mutat Res. 1990;230:227–234. doi: 10.1016/0027-5107(90)90060-h. [DOI] [PubMed] [Google Scholar]
- 13.Kosaka T, Kaneko I, Koide F. Int J Radiat Biol. 1990;58:417–425. doi: 10.1080/09553009014551771. [DOI] [PubMed] [Google Scholar]
- 14.Okayasu R, Iliakis G. Radiat Res. 1993;136:262–270. [PubMed] [Google Scholar]
- 15.Östling O, Johanson K J. Biochem Biophys Res Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 16.Bauch T, Bocker W, Mallek U, Muller W U, Streffer C. Strahlenther Onkol. 1999;175:333–340. doi: 10.1007/s000660050020. [DOI] [PubMed] [Google Scholar]
- 17.Green M H, Lowe J E, Delaney C A, Green I C. Methods Enzymol. 1996;269:243–266. doi: 10.1016/s0076-6879(96)69026-0. [DOI] [PubMed] [Google Scholar]
- 18.Maule J. Mol Biotechnol. 1998;9:107–126. doi: 10.1007/BF02760813. [DOI] [PubMed] [Google Scholar]
- 19.Prise K M, Ahnstrom G, Belli M, Carlsson J, Frankenberg D, Kiefer J, Lobrich M, Michael B D, Nygren J, Simone G, Stenerlow B. Int J Radiat Biol. 1998;74:173–184. doi: 10.1080/095530098141564. [DOI] [PubMed] [Google Scholar]
- 20.Ordoukhanian P, Taylor J S. Bioconjugate Chem. 2000;11:94–103. doi: 10.1021/bc9900993. [DOI] [PubMed] [Google Scholar]
- 21.Santos B C, Chevaile A, Hebert M J, Zagajeski J, Gullans S R. Am J Physiol. 1998;274:F1167–F1173. doi: 10.1152/ajprenal.1998.274.6.F1167. [DOI] [PubMed] [Google Scholar]
- 22.Michea L, Ferguson D R, Peters E M, Andrews P M, Kirby M R, Burg M B. Am J Physiol Renal Physiol. 2000;278:F209–F218. doi: 10.1152/ajprenal.2000.278.2.F209. [DOI] [PubMed] [Google Scholar]
- 23.Kohn K W, O'Connor P M, Pommier Y. In: Programmed Cell Death. Vos J-M H, editor. Austin, TX: R. G. Landes; 1995. pp. 248–283. [Google Scholar]
- 24.Fairbairn D W, Carnahan K G, Thwaits R N, Grigsby R V, Holyoak G R, O'Neill K L. FEMS Microbiol Lett. 1994;115:341–346. doi: 10.1111/j.1574-6968.1994.tb06661.x. [DOI] [PubMed] [Google Scholar]
- 25.Lakin N D, Jackson S P. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 26.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 27.Shukla A, Shukla R, Dikshit M, Srimal R C. Free Radical Biol Med. 1993;15:97–100. doi: 10.1016/0891-5849(93)90128-h. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann E K, Pedersen S F. Contrib Nephrol. 1998;123:50–78. doi: 10.1159/000059921. [DOI] [PubMed] [Google Scholar]
- 29.Bagchi D, Wetscher G J, Bagchi M, Hinder P R, Perdikis G, Stohs S J, Hinder R A, Das D K. Chem Biol Interact. 1997;104:65–85. doi: 10.1016/s0009-2797(97)03766-6. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Ogasawara M, Koyama I, Nemoto M, Yoshida T. Biol Pharm Bull. 1993;16:970–972. doi: 10.1248/bpb.16.970. [DOI] [PubMed] [Google Scholar]
- 31.Hong Z, Lakkineni K, Zhang Z, Verma D P. Plant Physiol. 2000;122:1129–1136. doi: 10.1104/pp.122.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Holt C, Brandt W F, Greyling H J, Lindsey G G, Retief J D, Rodrigues J D, Schwager S, Sewell B T. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 33.Buche A, Colson P, Houssier C. J Biomol Struct Dyn. 1990;8:601–618. doi: 10.1080/07391102.1990.10507831. [DOI] [PubMed] [Google Scholar]
- 34.Izzard R A, Jackson S P, Smith G C. Cancer Res. 1999;59:2581–2586. [PubMed] [Google Scholar]
- 35.Smith G C, Divecha N, Lakin N D, Jackson S P. Biochem Soc Symp. 1999;64:91–104. [PubMed] [Google Scholar]
- 36.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]