Abstract
Immunoglobulin A nephropathy (IgAN) is the most common form of primary glomerulonephritis worldwide and an important cause of kidney disease in young adults. Highly variable clinical presentation and outcome of IgAN suggest that this diagnosis may encompass multiple subsets of disease that are not distinguishable by currently available clinical tools. Marked differences in disease prevalence between individuals of European, Asian, and African ancestry suggest the existence of susceptibility genes that are present at variable frequencies in these populations. Familial forms of IgAN have also been reported throughout the world but are probably underrecognized because associated urinary abnormalities are often intermittent in affected family members. Of the many pathogenic mechanisms reported, defects in IgA1 glycosylation that lead to formation of immune complexes have been consistently demonstrated. Recent data indicates that these IgA1 glycosylation defects are inherited and constitute a heritable risk factor for IgAN. Because of the complex genetic architecture of IgAN, the efforts to map disease susceptibility genes have been difficult, and no causative mutations have yet been identified. Linkage-based approaches have been hindered by disease heterogeneity and lack of a reliable noninvasive diagnostic test for screening family members at risk of IgAN. Many candidate-gene association studies have been published, but most suffer from small sample size and methodological problems, and none of the results have been convincingly validated. New genomic approaches, including genome-wide association studies currently under way, offer promising tools for elucidating the genetic basis of IgAN.
Electronic supplementary material
The online version of this article (doi:10.1007/s00467-010-1500-7) contains supplementary material, which is available to authorized users.
Keywords: IgA nephropathy, Genetics, Hereditary disease, IgA1 glycosylation, Genome-wide association study
Introduction
Primary immunoglobulin A nephropathy (IgAN) is a complex trait [1] and a significant cause of renal insufficiency in young adults [2–5]. Complex diseases refer to disorders with a genetic basis that do not obey single-gene Mendelian inheritance patterns. They are typically determined by the action of multiple genes and environmental factors acting independently or through more complex gene–gene and gene–environment interactions. The environmental risk factors for IgAN remain poorly defined. Observational studies associate male gender and mucosal infections with increased risk of primary IgAN, but the causal mechanisms underlying these observations are not clear. The arguments in support of genetic factors include profound differences in prevalence among different ethnicities, familial clustering of IgAN, and interindividual variation in disease course and prognosis. For instance, Asians (Chinese and Japanese) have a relatively high prevalence of IgAN compared with Caucasians, whereas the disease is infrequently diagnosed in individuals of African ancestry [6]. High frequency of IgAN has also been reported in biopsy series for Native Americans and Oceanians [7–11]. Extended kindreds with familial IgAN have been reported throughout the world, including the USA [12], France [13], Canada [14], Italy [15], Australia [10], and Lebanon [16]. Familial disease accounts for 10–15% of all cases in regions such as northern Italy, France, or eastern Kentucky in the USA, where thorough surveys of relatives have been performed [13, 17–20].
Recognition of familial disease has many clinical implications, particularly for selecting donors for transplantation. In most reported families, segregation of IgAN is consistent with autosomal dominant transmission with incomplete penetrance (not all obligate carriers develop the disease), though more complex genetic models cannot be excluded. The incomplete penetrance is likely explained by the requirement of additional genetic or environmental factors for clinical manifestation of the disease and is also consistent with a complex disease model. Gene-mapping studies of traits with complex determination are difficult, and thus far, no single mutation has been conclusively demonstrated to cause IgAN. In this review, we concentrate on the approaches to genetic studies of IgAN; we summarize the studies of the last 10–15 years, review the most recent work, and attempt to project future directions in the field.
What do we know about genetics of IgAN?
Until now, two basic approaches have been used in genetic studies of IgAN: linkage studies and candidate-gene association studies. Linkage studies involve recruiting families with multiple affected individuals. In a typical whole-genome linkage scan, up to 400 microsatellites, or equivalently ∼10,000 single nucleotide polymorphism (SNP) markers, equally spaced across the genome, are typed in families to interrogate marker cosegregation with a disease phenotype. The advantage of genome-wide linkage studies is that they do not require a priori assumptions about disease pathogenesis. Unfortunately, these studies are very sensitive to phenotype misspecification, and their power is limited to detecting rare genetic variants with a relatively large effect on the risk of disease. The LOD score (logarithm of the ratio of odds) is used to determine whether a given genomic locus is linked with a disease trait. An LOD score of 3 indicates 1,000:1 odds that linkage between the marker and the disease locus exists and is generally accepted as significant in genome-wide linkage studies.
Linkage studies of IgAN are faced with multiple challenges. Familial forms of IgAN are frequently underrecognized because the associated urinary abnormalities in affected family members are often mild or intermittent. Moreover, once familial disease is documented, systematic screening by renal biopsy cannot be justified among asymptomatic at-risk relatives, necessitating reliance on less accurate phenotypes, such as microscopic hematuria, to diagnose affection. Additionally, IgAN has been observed to co-occur in families with thin basement membrane disease (TBMD), an autosomal dominant disease caused by heterozygous mutations in the collagen type IV genes (COL4A3/COL4A4) [21]. Short of kidney biopsy or direct sequencing of the very large collagen genes, TBMD cannot be reliably excluded among relatives of IgAN patients. Finally, because urinary abnormalities may manifest intermittently, one also cannot unequivocally classify at-risk relatives as unaffected, necessitating affected-only linkage analysis. The inability to classify affected and unaffected individuals accurately is commonly encountered in linkage studies of complex traits, leading to decreased study power. Increasing sample size by including additional families is also not necessarily helpful in these situations because the diagnosis of IgAN likely encompasses several disease subsets, such that expansion to larger sample size can paradoxically reduce analytic power due to increased heterogeneity [22–25].
To date, three genome-wide linkage studies of familial IgAN have been reported [14, 26, 27]. Families in these studies have all been ascertained via at least two cases with biopsy-documented IgAN, with additional family members diagnosed as affected based on clinical evidence (renal failure or multiple documentation of hematuria/proteinuria). In the first study, 30 families with two or more affected members were examined [26]; multipoint linkage analysis under the assumption of genetic heterogeneity yielded a peak LOD score of 5.6 on chromosome 6q22-23 (locus named IGAN1), with 60% of families linked. The remainder of families linked to chromosome 3p24-23 with a suggestive LOD of 2.8. This study demonstrated IgAN is genetically heterogeneous but argued for the existence of a single locus with a major effect in some families. Another genome-wide linkage study involved 22 families that replicated linkage to chromosome 6q22-23 (nominal p = 0.01 at IGAN1 locus) but also detected two suggestive signals on chromosome 4q26-31 (LOD 1.8) and 17q12-22 (LOD 2.6) [27]. The most recent linkage scan was based on a uniquely large pedigree with 14 affected relatives (two individuals with biopsy-defined diagnosis, and 12 with hematuria/proteinuria on urine dipstick) [14]. Linkage to chromosome 2q36 was detected with a maximal multipoint LOD of 3.47. Most linkage intervals reported did not contain obvious candidate genes, but the 2q36 locus encompasses the COL4A3 and COL4A4, which are mutated in TBMD. Together with the high penetrance of hematuria, this finding suggests that affected individuals in the 2q36-linked family may belong to an IgAN subtype that overlaps with TBMD. To date, none of the genes underlying these linkage loci has been identified. The underlying reasons are numerous, including the phenotyping difficulties discussed above; the presence of locus heterogeneity, which limits the ability to precisely map the disease interval and find additional linked families to refine loci; or contribution from noncoding susceptibility alleles (e.g. point mutations or structural genomic variants within intronic or promoter regions), which usually escape detection if mutational screening is confined to exonic regions. It is expected that the availability of inexpensive Next-Gen sequencing will enable comprehensive interrogation of linkage intervals, facilitating identification of disease-risk alleles.
Genetic association studies typically involve a collection of sporadic cases and a group of unrelated controls. Association studies are predicated on the premise that human populations share susceptibility alleles inherited from remote ancestors and that these alleles were not purified out because they individually confer a small excess risk of disease, resulting in a relatively high frequency in human populations (common variant/common disease hypothesis). Consistent with this starting premise, association studies are limited to detecting common disease-contributing genetic variants (i.e. population frequency typically >5%). The association approach can thus identify variants with moderate to relatively small effects and, compared with linkage scans, may be less sensitive to locus heterogeneity or phenotype misspecification. On the other hand, these studies are very sensitive to population stratification, i.e. undetected population mismatches between cases and controls that create spurious associations. As with linkage studies, association studies can now be carried out on a genome-wide scale [Genome-Wide Association Studies (GWAS)], providing an unbiased examination of the genome and the ability to detect and correct for population stratification. In addition, current standards necessitate replication of findings in independent cohorts to declare true associations.
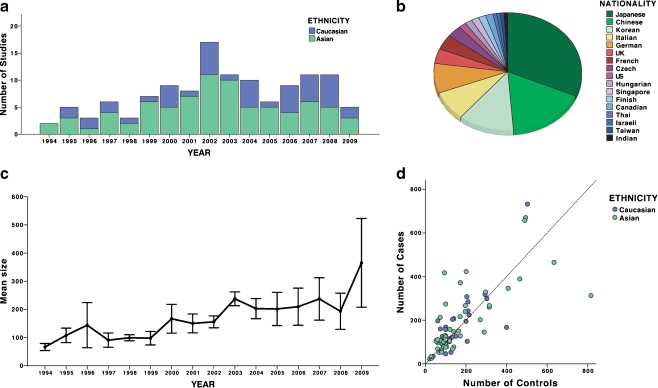
In contrast to the GWAS approach, candidate-gene association studies examine polymorphisms in only specific genes that are selected based on a priori assumptions about their involvement in the disease pathogenesis, and they are highly sensitive to population stratification, multiple testing, and reporting bias. As a result, most candidate-gene association studies in the literature have not been replicated [28]. Not surprisingly, candidate-gene studies for IgAN have also been largely unrevealing. Many candidates have been proposed, but most were studied in the context of IgAN progression rather than causality. In addition, in part due to our lack of knowledge about disease pathogenesis, most candidates were predicated on sparse a priori evidence for involvement in IgAN. Over the last 15 years, there were 123 candidate-gene association studies for IgAN published in the English literature and indexed on PubMed (Fig. 1, listed in the Supplemental Table 1S). Of these, 39 (31%) studies examined genetic polymorphisms in association with susceptibility to IgAN, 40 (32%) examined an association with disease severity, progression, or complications, and 44 (35%) examined both susceptibility and risk of progression. Approximately one third of all studies involved polymorphisms in the renin–angiotensin system (RAS). The quality of most studies was astoundingly poor. In general, they were severely underpowered, thus negative findings were almost universally inconclusive. Overall, the average size of case–control cohorts per study was 182 cases (range 23–916) and 171 controls (range 21–816), although a recent trend for increasing size of IgAN cohorts is notable (Fig. 1c, d). The lower average number of controls is a reflection of poor emphasis on the proper assembly of control groups. Many studies used ad hoc controls derived from unscreened blood donors who were poorly matched to the cases in terms of ancestry and geography. The potential impact of confounding by population stratification was ignored by a majority of studies, including very recent ones, despite the fact that the tools for quantification of this problem have been developed. Additionally, many studies tested several hypotheses (either multiple polymorphisms, multiple phenotypes, or multiple genetic models) and did not adequately correct for multiple testing. A very few studies performed permutation testing to derive empiric p values in the face of multiple, nonindependent tests. Other major problems included inadequate or variable SNP coverage of candidate genomic areas, with several studies examining only a single polymorphism. Thus far, only one group attempted to survey the entire genome, albeit in a severely underpowered cohort and with inadequate coverage of ∼80,000 SNPs [29, 30]. The results have not been replicated, and because these efforts do not pass current standards for genome-wide association studies, they remain inconclusive and difficult to interpret. Moreover, 77% of all published candidate-gene studies reported positive findings, an observation that is likely explained by a combination of high rate of false positives and a strong publication bias. Another silent problem in the literature relates to the fact that same patient cohorts are being tested for new polymorphisms without accounting for their use in prior publications. Most findings were not reproduced in other populations. None of the above problems is unique to the field of IgAN [31], and for these reasons, new general guidelines aimed at improving the design and execution of genetic association studies have recently been formulated (please refer to the STROBE [32] and STREGA [33] statements for more detailed discussion of these issues).
Fig. 1.
An overview of trends in the published genetic association studies of sporadic immunoglobulin A nephropathy (IgAN): a Trends in the numbers of genetic association studies by publication year and ethnicity (data from 1994 to mid-2009); b proportions of published genetic associations by nationality of study cohorts; c trends in the average size of IgAN cohorts by publication year (mean ± standard error); and d number of cases and controls per study by ethnicity. Only studies that use DNA-based genotyping are included
New approaches and ongoing studies: genetics of IgA1 glycosylation abnormalities
The requirement for a kidney biopsy for diagnosing IgAN is a major obstacle for family studies and a limiting step in the assembly of large case groups for genetic association studies. Serum IgA levels, though elevated in a significant portion of IgAN patients, lack the sensitivity and specificity required for a clinically useful diagnostic test. Fortunately, recent studies of glycosylation abnormalities of IgA1 offer prospects for a more reliable diagnostic biomarker for IgAN. In humans, IgA1 represents one of the two structurally and functionally distinct subclasses of IgA. Unlike IgA2, IgM, and IgG, IgA1 has heavy chains that contain a unique hinge-region segment between the first and second constant-region domains, which is the site of attachment of three to five O-linked glycan chains. O-glycans on circulatory IgA1 consist of N-acetylgalactosamine (GalNAc) with a β1,3-linked galactose; both residues may be sialylated. Carbohydrate composition of O-linked glycans on normal serum IgA1 is variable. Prevailing forms include the galactose-GalNAc disaccharide and its mono- and disialylated forms. Galactose-deficient variants with terminal GalNAc or sialylated GalNAc are more common in IgAN patients. These aberrantly glycosylated galactose-deficient forms predominate in glomerular immunodeposits and circulating complexes in IgAN [34–36].
O-linked glycans on circulatory IgA1 are synthesized in a step-wise manner, beginning with attachment of GalNAc to serine or threonine of the hinge region catalyzed by GalNAc transferases (Fig. 2). The O-glycan chain is then extended by attachment of galactose followed by addition of sialic acid residues to the GalNAc or galactose or both. Addition of galactose is catalyzed by Core-1-beta-1,3-galactosyltransferase (C1GalT1), and its stability is mediated by a molecular chaperone, Cosmc. The glycan structure is completed by the alpha-2,6-sialyltransferase II (ST6GalNAcII) and alpha-2,3-sialyltransferasaes (ST3Gal) that attach sialic acid to the GalNAc and galactose residues, respectively. Alternatively, ST6GalNAcII can add sialic acid to terminal GalNAc, a step that blocks any subsequent modifications and thus is a terminal step of O-glycan synthesis [37, 38]. Delineation of the IgA1 glycosylation pathway provided new logical candidates for genetic association studies. Genetic variations in genes encoding C1GalT1 (C1GALT1, chr. 7p13-14), Cosmc (C1GALT1C1, chr. Xq24), and ST6GalNAcII (ST6GALNAC2, chr. 17q25.1) were recently examined in a large cohort of 670 Chinese IgAN cases and 494 controls [39, 40], as well as in a smaller Italian study [41]. These studies identified risk haplotypes in ST6GALNAC2 and C1GALT1 and suggest a genetic interaction between these haplotypes. Similar to all other candidate studies, these results are preliminary and require validation.
Fig. 2.
Immunoglobulin A1 (IgA1) glycosylation pathway. Hinge region of human IgA1 contains serine (Ser) and threonine (Thr) residues, and some of them become O-glycosylated in B-cells’ Golgi apparatus. The predominating configuration of IgA1 glycans contains galactose (Gal) residues. Circulating IgA1 from IgA nephropathy (IgAN) patients is, to a large degree, galactose deficient and contains terminally sialylated N-acetylgalactosamine (GalNAc). GalNAcT2 UDP-GalNAc-transferase 2, C1GalT1 Core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, Cosmc C1GALT1-specific chaperone 1, ST6GalNAc II N-acetylgalactosaminide alpha-2,6-sialyltransferase II, NeuAc N-acetylneuraminic acid (sialic acid)
Critically, recent studies have shown that IgA1-producing cells are the source of elevated serum galactose-deficient IgA1 levels in patients with IgAN [42]. Aberrantly glycosylated IgA1 can be detected within these cells, and serum levels of galactose-deficient IgA1 correlated extremely well with levels of this immunoglobulin in the supernatant of cultured IgA1-producing cells isolated from peripheral blood of the same individual. These data indicate that aberrant glycosylation is not a secondary phenomenon attributable to modification of IgA1 during formation of immune complexes but, rather, reflect a specific defect originating in IgA1-producing cells. Additional studies did not reveal any specific defects in individual IgA1 glycosylation enzymes but found that each enzymatic step in the glycosylation pathway is shifted toward increased production of galactose-deficient IgA1, with elevated content of sialic acid on GalNAc [42]. Moreover, expression and enzymatic activity of C1GalT1 appear to be dissociated from O-glycosylation in IgAN [43], and aberrant glycosylation affects solely IgA1 and not other glycoproteins with O-linked glycans, such as IgD [44]. Taken together, these observations isolate the IgA1 glycosylation defect to a specific cell type and indicate that the primary disturbance likely originates in an upstream regulatory pathway(s) of these cells rather than in the specific glycosylation enzymes.
Recently, a reliable lectin-based enzyme-linked immunosorbent assay (ELISA) for determining serum galactose-deficient IgA1 has been developed. It utilizes a naturally occurring HAA lectin isolated from the Helix aspersa snail to detect circulating galactose-deficient O-linked glycans. In a large cohort of Caucasians from the southeastern USA, this test had 90% specificity and 76% sensitivity for diagnosing sporadic IgAN [45]. Similarly, ELISA-determined serum galactose-deficient IgA1 was elevated in 77% of pediatric patients with IgAN [46]. This simple and inexpensive serum ELISA holds much promise for providing the first noninvasive screening test for IgAN.
Our group used this assay to investigate the inheritance of galactose-deficient IgA1 in familial and sporadic forms of IgAN [47]. A high serum galactose-deficient IgA1 level was present in the majority of index cases, as well as among their parents (39%), siblings (28%), and children (30%), providing further support for a major dominant effect. Levels in spouses were indistinguishable from controls, ruling out an environmental effect. Heritability of galactose-deficient IgA1 (the proportion of a trait’s variation explained by inherited factors) was therefore statistically significant and estimated at 54%. Segregation analysis of galactose-deficient IgA1 suggested inheritance of a major dominant gene with an additional polygenic component. We further examined galactose-deficient IgA1 levels in relatives after stratification for galactose-deficient IgA1 levels in the index case. Among relatives of IgAN patients with high galactose-deficient IgA1 values, 33% of individuals also had high values. In contrast, relatives of IgAN patients with normal galactose-deficient IgA1 levels had levels that were indistinguishable from controls. These data strongly argue that galactose-deficient IgA1 values can identify distinct subpopulations among IgAN patients, which may differ in the underlying disease pathogenesis. Inheritance of galactose-deficient IgA1 has been confirmed in Chinese patients with familial and sporadic adult IgAN [48, 49] and recently extended to pediatric IgAN and Henoch-Schönlein purpura with nephritis (HSPN) [50]. Thus, aberrant IgA1 glycosylation is a common inherited defect that provides a unifying link in the pathogenesis of HSPN, and familial and sporadic IgAN among many populations across the globe. Furthermore, these data demonstrate that an elevated serum galactose-deficient IgA1 level is antecedent to disease but, because most family members with elevated levels are asymptomatic, IgA1 glycosylation abnormalities are not sufficient to produce IgAN, and additional cofactors must trigger formation of immune complexes.
A recent study by Suzuki et al. offered some insights into the additional cofactors required to form immune complexes and their deposition in glomeruli [51]. Molecular characterization of IgG autoantibodies that recognize abnormally glycosylated IgA1 molecules (specifically, galactose-deficient GalNAc-containing epitopes in the hinge region of IgA1) revealed a specific amino-acid substitution in the variable region of the IgG1 heavy chain that is more frequent in IgAN cases compared with controls. This substitution greatly enhances IgG1 binding to the galactose-deficient IgA1 molecules. It is likely that this substitution arises as a somatic mutation in IgG1-producing cells, which is then positively selected in the course of immune response to yet unidentified antigens. The triggering antigens may include viral or bacterial pathogens (supported by frequent synpharyngitic disease exacerbation) or possibly by ingested food epitopes (supported by clinical overlap of IgAN with celiac disease). These important observations established antiglycan IgG1 antibodies as at least one additional risk factor, or a “second hit”, which predisposes to disease development.
These data enable formulation of a working hypothesis on the pathogenesis of IgAN (Fig. 3). Analogous to elevated serum cholesterol level, which in conjunction with additional risk factors (such as smoking or hypertension) leads to the development of coronary heart disease, elevated galactose-deficient IgA1 levels constitute a genetic risk factor that, in combination with a second independent hit (the production of antiglycan antibodies), leads to formation of circulating immune complexes that deposit in the glomerulus, producing inflammation and kidney damage. This simple model can potentially explain familial or sporadic occurrence of IgAN. Whereas in the majority of cases elevated galactose-deficient IgA1 appears to be an inherited risk factor [47], one can surmise that the propensity to produce antiglycan antibodies results from an independent, stochastic process such as a somatic mutation or an environmental insult (e.g. a viral infection). Thus, the occurrence of this second hit in an individual with genetically elevated galactose-deficient IgA1 levels would result in sporadic IgAN. One can also hypothesize that in some cases, the propensity to produce antiglycan antibodies is by itself genetically determined, such that cosegregation of risk alleles for galactose-deficient IgA1 and antiglycan antibody production in the same family would produce the pattern of familial IgAN. Because the two mutations would be independently inherited, only a small number of family members would carry both risk alleles, resulting in the pattern of variable penetrance and small number of affected individuals typical of familial IgAN. The relative prevalence of such risk alleles among different populations could also explain geographic variation in IgAN prevalence. Most importantly, the pathogenesis model depicted in Fig. 3 predicts that interventions that decrease levels of galactose-deficient IgA1 or anti-glycan antibodies would reduce formation of immune complexes and positively impact the course of IgAN. Thus, identification of genes and pathways that specifically affect each side of the equation would provide targets for therapeutic intervention in IgAN. Such interventions would be analogous to administration of cholesterol-lowering drugs to reduce the risk of coronary heart disease.
Fig. 3.
The model of immunoglobulin A nephropathy (IgAN) pathogenesis in patients with high levels of galactose-deficient IgA1. Inherited defect of IgA1 glycosylation is not sufficient to cause the disease. Additional environmental or genetic factors are probably required for renal injury, which is likely mediated by antiglycan antibody production and immune complex formation
Future outlook: genomic approaches to IgA nephropathy
The availability of assays for circulating galactose-deficient IgA1 and antiglycan antibody production will likely accelerate gene-mapping studies. For example, profiling serum galactose-deficient IgA1 levels can now be used to reduce heterogeneity in linkage scans for IgAN. Additionally, galactose-deficient IgA1 can be used more directly, as an endophenotype (i.e. a measurable intermediate component of the causal pathway between genotype and disease) in genetic linkage studies of IgAN. Quantitative endophenotypes are frequently preferred in genetic studies of a complex disease, because they may more closely reflect specific pathogenic processes and provide more statistical power to detect genotypic correlations. Moreover, statistical methods used in quantitative linkage analysis are less sensitive to a trait’s heterogeneity. In parallel to linkage approaches, quantitative endophenotypes can also be used to enhance genetic association studies. This is best exemplified by quantitative studies of serum IgE levels that identified novel susceptibility loci for asthma using both genome-wide linkage and association approaches [52, 53]. Quantitative gene-mapping studies of serum galactose-deficient IgA1 levels in IgAN are underway.
The field of complex disease genetics is also evolving rapidly, and a new generation of genetic studies is soon to emerge for IgAN. First, rapid and cost-effective screening of the human genome with >1 million polymorphisms is now possible, enabling efficient execution of high-resolution genome-wide genetic association studies. In a GWAS design, a dense map of SNPs is surveyed for association with a trait of interest. Typically, large and well-phenotyped case–control groups (>1,000 individuals per group) are required to discover genetic variants, and independent cohorts are needed to replicate findings [54]. The main issues facing GWAS approaches include the analytical challenge of multiple hypotheses testing and accounting for population stratification. Both of these issues have been addressed in the genetics community, with rigorous standards now widely accepted [55, 56]. GWAS is also inherently limited to detecting susceptibility alleles that are relatively common. To date, most GWAS-discovered variants reside in noncoding segments of the genome and impart a small effect on disease, presumably via a regulatory role on expression of neighboring genes (odds ratios 1.15–1.35). Variants with even smaller effects can be detected with larger sample sizes [57]. Small effects of common susceptibility variants are hypothesized to be the consequence of long-standing purifying selection against alleles that produce major alterations in the encoded protein and may impair reproductive fitness. There are, however, exceptions to this rule in which common alleles impart relatively large effects on disease risk. Two examples are diseases of very late onset in which reproductive fitness is not impaired (e.g. common allele ε4 at APOE locus with large effect on the risk of Alzheimer’s disease [58]), and diseases in which recent changes in the environment may have resulted in alleles that were once neutral or favored now becoming disease contributing. Many diseases of the immune system may fit into the latter category, including type I diabetes mellitus, macular degeneration, systemic lupus erythematosus (SLE), inflammatory bowel disease, and celiac disease. In each case, common variants had moderate to large effect sizes, facilitating their detection by GWAS [59–64]. IgAN is also likely to fall into this category; thus, the GWAS design for IgAN may represent a powerful approach.
In addition to GWAS, several other genomic approaches are likely to be integrated for the purpose of gene identification (Table 1). These include analyses of structural rearrangements, genome-wide expression, and deep-sequencing data. Copy-number variants (CNVs) have been recognized as an important source of genetic variation in humans [65]. Comprehensive surveys of small insertions, deletions, and segmental duplications across the genome can be efficiently accomplished with modern SNP genotyping platforms [66]. This is relevant to IgAN because structural variants have been implicated in several immune-mediated traits, such as psoriasis or SLE [67–69]. We anticipate that genome-wide CNV analyses are likely to appear alongside the first GWAS for IgAN. Genome-wide expression profiling is another approach that can be helpful in dissecting the genetic basis of IgAN. Recent data suggest that tissue-specific patterns of gene expression are highly inherited, and expression quantitative trait loci (eQTL) mapping has been used to define loci that control transcription efficiency [70]. The combination of gene mapping and gene-expression profiling provides a powerful tool for understanding complex traits, as these techniques generate independent information that allows reciprocal prioritization of candidate genes. Such information can be integrated with the GWAS approach to better define functional defects responsible for disease susceptibility [71]. Moreover, novel integrative genomic approaches enable joint analysis of genetic, gene-expression, and biochemical phenotypes to derive pathways and molecular networks driving disease pathogenesis [72]. Application of these methods in humans is challenging because most tissues of interest cannot be readily accessed, and sampled specimens (such as kidney) are composed of heterogeneous cell types with distinct gene-expression profiles. However, this approach would be ideally suited for genetic studies of IgAN because expression studies can be performed in IgA1-secreting cells, enabling interrogation a single cell type and the specific biochemical phenotype of defective IgA1 glycosylation [42]. Lastly, high-throughput sequencing technology is evolving rapidly, and cost-effective whole-genome deep sequencing is now becoming feasible. Direct sequence analysis offers hope of detecting important rare genetic variants that contribute to the pathogenesis of common complex diseases [73–76]. The recent feasibility of exome sequencing (sequencing of all the exons of the genome in one shot) now also enables detection of mutations underlying oligogenic traits [77, 78]. This approach is quite promising, as variants identified via GWAS collectively account for a small fraction of the heritability of phenotypes studied, suggesting major contributions from rare sequence variants to disease pathogenesis. General approach and statistical tools for this approach are still under development, but this type of study is expected to provide a new wave of findings in the genetic determination of complex traits.
Table 1.
Genetic approaches to studies of immunoglobulin A nephropathy (IgAN) in humans
| Family-based approaches: |
| 1. Family aggregation studies (numerous reports) |
| 2. Traditional whole-genome linkage scans (3 reported) |
| 3. Quantitative whole-genome linkage scans for Gd-IgA1 and related phenotypes (in progress) |
| Population-based approaches: |
| 1. Candidate gene associations for susceptibility and progression of IgAN (over 120 reported) |
| 2. Genome-wide case–control association studies of IgAN (in progress) |
| 3. Genome-wide quantitative association studies of Gd-IgA1 and related phenotypes (in progress) |
| Anticipated future approaches: |
| 1. Studies of copy number variants (CNVs) in IgAN |
| 2. Population-based and family-based studies of genome-wide gene expression in IgAN |
| 3. Integrative genomics approaches to derive disturbed molecular networks in IgAN |
| 4. Whole-genome sequencing to discover rare variants in IgAN |
Summary and clinical implications
Similar to other immune-mediated disorders, IgAN is a genetically complex trait. Although there is a clear contribution of genetic factors to IgAN susceptibility, specific genes have not yet been identified. Gene-mapping studies for complex traits are challenging, but successful discovery of IgAN susceptibility genes will have major impact and far-reaching implications worldwide. Such findings may open doors to novel targeted therapeutic approaches. Clinical applications may involve genetic screening and diagnosis, improved risk stratification, or selection of suitable kidney transplant donors among related individuals based on genetic testing. Discovery of IgAN biomarkers combined with the availability of novel genomic technology are likely to transform the approach of genetic studies of IgAN. However, it is now clear that much larger cohorts of patients will be required than have been previously studied. Referral of patients for genetic research studies and systematic biobanking of blood, serum, and kidney tissue will be critical for execution of such studies. International and interdisciplinary collaborations will likely be required for successful identification of specific genetic causes of IgAN.
Below is the link to the electronic supplementary material.
Index of published genetic association studies in IgA nephropathy sorted by publication year (data from 1994 to 09/01/2009) (DOC 3642 kb)
Acknowledgements
Krzysztof Kiryluk is supported by the Daland Fellowship from the American Philosophical Society and Grant Number KL2 RR024157 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Bruce A. Julian, Robert J. Wyatt, Francesco Scolari, Jan Novak, and Ali G. Gharavi are supported by Grant Number DK082753 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The authors also acknowledge other grants from NIDDK supporting their research of IgAN: DK078244, DK080301, DK075868, DK071802, and DK077279. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of NCRR, NIDDK, or NIH.
Conflicts of Interest None
Questions
(Answers appear following the reference list)
- Which is true about familial forms of IgAN?
- Familial forms of IgAN have only been observed in genetically isolated populations
- Patients with familial IgAN should never be transplanted because of high risk of recurrence
- Patients with familial IgAN can be identified based on their characteristic presenting symptoms
- Most familial IgAN displays autosomal dominant inheritance
- b and d
- Which is true about glycosylation defects of IgA1 in IgAN?
- It is associated with a generalized defect in glycosylation of most circulating immunoglobulins
- The abnormally glycosylated IgA1 has a higher propensity for immune complex formation and deposition in mesangium
- Elevated levels of galactose-deficient IgA1 correlate with symptoms and prognosis in IgAN
- Elevated levels of galactose-deficient IgA1 are observed in large proportion of family members of IgAN patients
- b and d
- Which is true about genetic studies of IgAN?
- Consistent association of IgAN with cytokine haplotypes have been identified
- Multiple different genes can cause familial IgAN because different families demonstrate linkage to different segments of the genome
- Glycosylation defects in IgAN are caused by mutations in the C1GALT1 (beta-1,3 galatosylatransferase gene)
- Genome-wide association studies will be able to detect rare genes with small effect that contribute to IgAN
- b and d
- Which is true about serum levels of galactose-deficient IgA1:
- Elevated serum level of galactose-deficient IgA1 is sufficient to make the diagnosis of IgAN
- Normal serum level of galactose-deficient IgA1 is sufficient to exclude the diagnosis of IgAN
- Elevated serum level of galactose-deficient IgA1 is required for the development and progression of IgAN
- In the populations studied to date, inherited factors are estimated to account for approximately 50% of the total variation in galactose-deficient IgA1 levels
- None of the above
- Which is NOT true about genetic studies of IgAN:
- Genetic heterogeneity decreases power of linkage studies of familial IgAN
- The results of genetic association studies may be biased if cases and controls are derived from heterogenous populations
- Most candidate gene associations in sporadic IgAN have not been replicated
- Galactose-deficient IgA1 level represents a promising endophenotype for genetic linkage and association studies
- Several common copy-number polymorphisms have been consistently associated with IgAN
Footnotes
Answers:
1. d
2. e
3. b
4. d
5. e
References
- 1.Beerman I, Novak J, Wyatt RJ, Julian BA, Gharavi AG. The genetics of IgA nephropathy. Nat Clin Pract Nephrol. 2007;3:325–338. doi: 10.1038/ncpneph0492. [DOI] [PubMed] [Google Scholar]
- 2.D’Amico G (1987) The commonest glomerulonephritis in the world; IgA nephropathy. Q J Med 64:709–727 [PubMed]
- 3.Donadio JV, Grande JP. IgA nephropathy. N Engl J Med. 2002;347:738–748. doi: 10.1056/NEJMra020109. [DOI] [PubMed] [Google Scholar]
- 4.Barratt J, Feehally J. IgA nephropathy. J Am Soc Nephrol. 2005;16:2088–2097. doi: 10.1681/ASN.2005020134. [DOI] [PubMed] [Google Scholar]
- 5.D'Amico G, Imbasciati E, Barbiano Di Belgioioso G, Bertoli S, Fogazzi G, Ferrario F, Fellin G, Ragni A, Colasanti G, Minetti L, Ponticelli C. Idiopathic IgA mesangial nephropathy. Clinical and histological study of 374 patients. Medicine. 1985;64:49–60. [PubMed] [Google Scholar]
- 6.Hall YN, Fuentes EF, Chertow GM, Olson JL. Race/ethnicity and disease severity in IgA nephropathy. BMC Nephrol. 2004;5:10. doi: 10.1186/1471-2369-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoy WE, Hughson MD, Smith SM, Megill DM. Mesangial proliferative glomerulonephritis in southwestern American Indians. Am J Kidney Dis. 1993;21:486–496. doi: 10.1016/s0272-6386(12)80394-5. [DOI] [PubMed] [Google Scholar]
- 8.Smith SM, Harford AM. IgA nephropathy in renal allografts: increased frequency in Native American patients. Ren Fail. 1995;17:449–456. doi: 10.3109/08860229509037608. [DOI] [PubMed] [Google Scholar]
- 9.Hughson MD, Megill DM, Smith SM, Tung KS, Miller G, Hoy WE. Mesangiopathic glomerulonephritis in Zuni (New Mexico) Indians. Arch Pathol Lab Med. 1989;113:148–157. [PubMed] [Google Scholar]
- 10.O'Connell PJ, Ibels LS, Thomas MA, Harris M, Eckstein RP. Familial IgA nephropathy: a study of renal disease in an Australian aboriginal family. Aust NZ J Med. 1987;17:27–33. doi: 10.1111/j.1445-5994.1987.tb05045.x. [DOI] [PubMed] [Google Scholar]
- 11.Casiro OG, Stanwick RS, Walker RD. The prevalence of IgA nephropathy in Manitoba Native Indian children. Can J Public Health. 1988;79:308–310. [PubMed] [Google Scholar]
- 12.Julian BA, Quiggins PA, Thompson JS, Woodford SY, Gleason K, Wyatt RJ. Familial IgA nephropathy. Evidence of an inherited mechanism of disease. N Engl J Med. 1985;312:202–208. doi: 10.1056/NEJM198501243120403. [DOI] [PubMed] [Google Scholar]
- 13.Levy M. Familial cases of Berger's disease and anaphylactoid purpura more frequent than previously thought. Am J Med. 1989;87:246–248. doi: 10.1016/S0002-9343(89)80720-X. [DOI] [PubMed] [Google Scholar]
- 14.Paterson AD, Liu XQ, Wang K, Magistroni R, Song X, Kappel J, Klassen J, Cattran D, St George-Hyslop P, Pei Y. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol. 2007;18:2408–2415. doi: 10.1681/ASN.2007020241. [DOI] [PubMed] [Google Scholar]
- 15.Scolari F, Amoroso A, Savoldi S, Mazzola G, Prati E, Valzorio B, Viola BF, Nicola B, Movilli E, Sandrini M, Campanini M, Maiorca R. Familial clustering of IgA nephropathy: further evidence in an Italian population. Am J Kidney Dis. 1999;33:857–865. doi: 10.1016/S0272-6386(99)70417-8. [DOI] [PubMed] [Google Scholar]
- 16.Karnib HH, Sanna-Cherchi S, Zalloua PA, Medawar W, D'Agati VD, Lifton RP, Badr K, Gharavi AG. Characterization of a large Lebanese family segregating IgA nephropathy. Nephrol Dial Transplant. 2007;22:772–777. doi: 10.1093/ndt/gfl677. [DOI] [PubMed] [Google Scholar]
- 17.Johnston PA, Brown JS, Braumholtz DA, Davison AM. Clinico-pathological correlations and long-term follow-up of 253 United Kingdom patients with IgA nephropathy. A report from the MRC Glomerulonephritis Registry. Q J Med. 1992;84:619–627. [PubMed] [Google Scholar]
- 18.Rambausek M, Hartz G, Waldherr R, Andrassy K, Ritz E. Familial glomerulonephritis. Pediatr Nephrol. 1987;1:416–418. doi: 10.1007/BF00849246. [DOI] [PubMed] [Google Scholar]
- 19.Schena FP, Scivittaro V, Ranieri E, Sinico R, Benuzzi S, Cillo M, Aventaggiato L. Abnormalities of the IgA immune system in members of unrelated pedigrees from patients with IgA nephropathy. Clin Exp Immunol. 1993;92:139–144. doi: 10.1111/j.1365-2249.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schena FP, Scivittaro V, Ranieri E. IgA nephropathy: pros and cons for a familial disease. Contrib Nephrol. 1993;104:36–45. doi: 10.1159/000422394. [DOI] [PubMed] [Google Scholar]
- 21.Frasca GM, Soverini L, Gharavi AG, Lifton RP, Canova C, Preda P, Vangelista A, Stefoni S. Thin basement membrane disease in patients with familial IgA nephropathy. J Nephrol. 2004;17:778–785. [PubMed] [Google Scholar]
- 22.Durner M, Greenberg DA, Hodge SE. (1992) Inter- and intrafamilial heterogeneity: effective sampling strategies and comparison of analysis methods. Am J Hum Genet. 1992;51:859–870. [PMC free article] [PubMed] [Google Scholar]
- 23.Durner M, Greenberg DA. Effect of heterogeneity and assumed mode of inheritance on lod scores. Am J Med Genet. 1992;42:271–275. doi: 10.1002/ajmg.1320420302. [DOI] [PubMed] [Google Scholar]
- 24.Cavalli-Sforza LL, King MC. Detecting linkage for genetically heterogeneous diseases and detecting heterogeneity with linkage data. Am J Hum Genet. 1986;38:599–616. [PMC free article] [PubMed] [Google Scholar]
- 25.Ott J. The number of families required to detect or exclude linkage heterogeneity. Am J Hum Genet. 1986;39:159–165. [PMC free article] [PubMed] [Google Scholar]
- 26.Gharavi AG, Yan Y, Scolari F, Schena FP, Frasca GM, Ghiggeri GM, Cooper K, Amoroso A, Viola BF, Battini G, Caridi G, Canova C, Farhi A, Subramanian V, Nelson-Williams C, Woodford S, Julian BA, Wyatt RJ, Lifton RP. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22-23. Nat Genet. 2000;26:354–357. doi: 10.1038/81677. [DOI] [PubMed] [Google Scholar]
- 27.Bisceglia L, Cerullo G, Forabosco P, Torres DD, Scolari F, Perna M, Foramitti M, Amoroso A, Bertok S, Floege J, Mertens PR, Zerres K, Alexopoulos E, Kirmizis D, Ermelinda M, Zelante L, Schena FP, European IgAN Consortium Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet. 2006;79:1130–1134. doi: 10.1086/510135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 29.Obara W, Iida A, Suzuki Y, Tanaka T, Akiyama F, Maeda S, Ohnishi Y, Yamada R, Tsunoda T, Takei T, Ito K, Honda K, Uchida K, Tsuchiya K, Yumura W, Ujiie T, Nagane Y, Nitta K, Miyano S, Narita I, Gejyo F, Nihei H, Fujioka T, Nakamura Y. Association of single-nucleotide polymorphisms in the polymeric immunoglobulin receptor gene with immunoglobulin A nephropathy (IgAN) in Japanese patients. J Hum Genet. 2003;48:293–299. doi: 10.1007/s10038-003-0027-1. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo S, Iida A, Nitta K, Tanaka T, Yamada R, Ohnishi Y, Maeda S, Tsunoda T, Takei T, Obara W, Akiyama F, Ito K, Honda K, Uchida K, Tsuchiya K, Yumura W, Ujiie T, Nagane Y, Miyano S, Suzuki Y, Narita I, Gejyo F, Fujioka T, Nihei H, Nakamura Y. Association of a single-nucleotide polymorphism in the immunoglobulin mu-binding protein 2 gene with immunoglobulin A nephropathy. J Hum Genet. 2005;50:30–35. doi: 10.1007/s10038-004-0214-8. [DOI] [PubMed] [Google Scholar]
- 31.(1999) Freely associating. Nat Genet 22:1–2 [DOI] [PubMed]
- 32.Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370:1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 33.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N. STrengthening the REporting of Genetic Association Studies (STREGA)-an extension of the STROBE statement. Genet Epidemiol. 2009;33:581–598. doi: 10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
- 34.Hiki Y, Odani H, Takahashi M, Yasuda Y, Nishimoto A, Iwase H, Shinzato T, Kobayashi Y, Maeda K. Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int. 2001;59:1077–1085. doi: 10.1046/j.1523-1755.2001.0590031077.x. [DOI] [PubMed] [Google Scholar]
- 35.Tomana M, Matousovic K, Julian BA, Radl J, Konecny K, Mestecky J. Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int. 1997;52:509–516. doi: 10.1038/ki.1997.361. [DOI] [PubMed] [Google Scholar]
- 36.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J. Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest. 1999;104:73–81. doi: 10.1172/JCI5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raska M, Moldoveanu Z, Suzuki H, Brown R, Kulhavy R, Hall S, Vu HL, Carlsson F, Lindahl G, Tomana M, Julian BA, Wyatt RJ, Mestecky J, Novak J. Identification and characterization of CMP-NeuAc:GalNAc-IgA1 α2, 6-sialyltransferase in IgA1-producing cells. J Mol Biol. 2007;369:69–78. doi: 10.1016/j.jmb.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, Julian BA, Tomana M, Wyatt RJ, Edberg JE, Alarcón GS, Kimberly RP, Tomino Y, Mestecky J, Novak J. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li GS, Zhang H, Lv JC, Shen Y, Wang HY. Variants of C1GALT1 gene are associated with the genetic susceptibility to IgA nephropathy. Kidney Int. 2007;71:448–453. doi: 10.1038/sj.ki.5002088. [DOI] [PubMed] [Google Scholar]
- 40.Zhu L, Tang W, Li G, Lv J, Ding J, Yu L, Zhao M, Li Y, Zhang X, Shen Y, Zhang H, Wang H. Interaction between variants of two glycosyltransferase genes in IgA nephropathy. Kidney Int. 2009;76:190–198. doi: 10.1038/ki.2009.99. [DOI] [PubMed] [Google Scholar]
- 41.Pirulli D, Crovella S, Ulivi S, Zadro C, Bertok S, Rendine S, Scolari F, Foramitti M, Ravani P, Roccatello D, Savoldi S, Cerullo G, Lanzilotta SG, Bisceglia L, Zelante L, Floege J, Alexopoulos E, Kirmizis D, Ghiggeri GM, Frasca G, Schena FP, Amoroso A. Genetic variant of C1GalT1 contributes to the susceptibility to IgA nephropathy. J Nephrol. 2009;22:152–159. [PubMed] [Google Scholar]
- 42.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, Julian BA, Tomana M, Wyatt RJ, Edberg JC, Alarcon GS, Kimberly RP, Tomino Y, Mestecky J, Novak J. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck KS, Smith AC, Molyneux K, El-Barbary H, Feehally J, Barratt J. B-cell O-galactosyltransferase activity, and expression of O-glycosylation genes in bone marrow in IgA nephropathy. Kidney Int. 2008;73:1128–1136. doi: 10.1038/sj.ki.5002748. [DOI] [PubMed] [Google Scholar]
- 44.Smith AC, Wolff JF, Molyneux K, Feehally J, Barratt J. O-glycosylation of serum IgD in IgA nephropathy. J Am Soc Nephrol. 2006;17:1192–1199. doi: 10.1681/ASN.2005101115. [DOI] [PubMed] [Google Scholar]
- 45.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 2007;71:1148–1154. doi: 10.1038/sj.ki.5002185. [DOI] [PubMed] [Google Scholar]
- 46.Lau KK, Wyatt RJ, Moldoveanu Z, Tomana M, Julian BA, Hogg RJ, Lee JY, Huang WQ, Mestecky J, Novak J. Serum levels of galactose-deficient IgA in children with IgA nephropathy and Henoch-Schonlein purpura. Pediatr Nephrol. 2007;22:2067–2072. doi: 10.1007/s00467-007-0623-y. [DOI] [PubMed] [Google Scholar]
- 47.Gharavi AG, Moldoveanu Z, Wyatt RJ, Barker CV, Woodford SY, Lifton RP, Mestecky J, Novak J, Julian BA. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J Am Soc Nephrol. 2008;19:1008–1014. doi: 10.1681/ASN.2007091052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin X, Ding J, Zhu L, Shi S, Jiang L, Zhao M, Zhang H. Aberrant galactosylation of IgA1 is involved in the genetic susceptibility of Chinese patients with IgA nephropathy. Nephrol Dial Transplant. 2009;24:3372–3375. doi: 10.1093/ndt/gfp294. [DOI] [PubMed] [Google Scholar]
- 49.Tam KY, Leung JC, Chan LY, Lam MF, Tang SC, Lai KN. Macromolecular IgA1 taken from patients with familial IgA nephropathy or their asymptomatic relatives have higher reactivity to mesangial cells in vitro. Kidney Int. 2009;75:1330–1339. doi: 10.1038/ki.2009.71. [DOI] [PubMed] [Google Scholar]
- 50.Kiryluk K, Moldoveanu Z, Sanders JT, Suzuki H, Julian BA, Mesteky J, Novak J, Gharavi AG, Wyatt RJ (2009) Aberrant glycosylation of IgA1 is inherited in pediatric Henoch-Schönlein nephritis. J Am Soc Nephrol 20:435A (abstract PO1408) [DOI] [PMC free article] [PubMed]
- 51.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J. Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest. 2009;119:1668–1677. doi: 10.1172/JCI38468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, Gohlke H, Wagenpfeil S, Ollert M, Ring J, Behrendt H, Heinrich J, Novak N, Bieber T, Kramer U, Berdel D, Berg A, Bauer CP, Herbarth O, Koletzko S, Prokisch H, Mehta D, Meitinger T, Depner M, Mutius E, Liang L, Moffatt M, Cookson W, Kabesch M, Wichmann HE, Illig T. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, Sayers I. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 56.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 58.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 59.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wellcome Trust Case Control Consortium Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapää-Dahlqvist S, Petri M, Manzi S, Seldin MF, Rönnblom L, Syvänen AC, Criswell LA, Gregersen PK, Behrens TW. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 64.International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 66.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaschl H, Aitman TJ, Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, Smith A, Blakemore AI, Froguel P, Owen CJ, Pearce SH, Teixeira L, Guillevin L, Graham DS, Pusey CD, Cook HT, Vyse TJ, Aitman TJ. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, Escaramis G, Ballana E, Martin-Ezquerra G, Heijer M, Kamsteeg M, Joosten I, Eichler EE, Lazaro C, Pujol RM, Armengol L, Abecasis G, Elder JT, Novelli G, Armour JA, Kwok PY, Bowcock A, Schalkwijk J, Estivill X. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat Genet. 2002;32(Suppl):522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- 71.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 73.Li B, Leal SM. Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 2009;5:e1000481. doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 75.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Index of published genetic association studies in IgA nephropathy sorted by publication year (data from 1994 to 09/01/2009) (DOC 3642 kb)