Summary
Mutations in a number of genes encoding voltage-gated sodium channels cause a variety of epilepsy syndromes in humans, including genetic (generalized) epilepsy with febrile seizures Plus (GEFS+) and Dravet syndrome (DS, severe myoclonic epilepsy of infancy). The vast majority of these mutations are in the SCN1A gene, and all are dominantly inherited. Most of the mutations that cause DS result in loss of function, whereas all of the known mutations that cause GEFS+ are missense, presumably altering channel activity. Family members with the same GEFS+ mutation often display a wide range of seizure types and severities, and at least part of this variability likely results from variation in other genes. Many different biophysical effects of SCN1A-GEFS+ mutations have been observed in heterologous expression systems, consistent with both gain and loss of channel activity. However, results from mouse models suggest that the primary effect of both GEFS+ and DS mutations is to decrease the activity of GABAergic inhibitory neurons. Decreased activity of the inhibitory circuitry is thus likely to be a major factor contributing to seizure generation in patients with GEFS+ and DS, and may be a general consequence of SCN1A mutations.
Keywords: GEFS, Dravet syndrome, Genetics, Knock-in mice, Knockout mice
Introduction
The epilepsies are a diverse collection of neurological disorders involving recurrent and unprovoked seizures that are the manifestation of abnormal electrical activity in the central nervous system (CNS), and they represent one of the most common neurological diseases, with a lifetime incidence of up to 3% in the general population (Hauser et al., 1993). Although many epilepsies are secondary to injury or another disease, approximately 40% are idiopathic, meaning that the underlying cause is unknown (Steinlein, 2002). It is presumed that most of the idiopathic epilepsies result from genetic abnormalities, with the majority likely caused by mutations in multiple currently unidentified genes. Nevertheless, research has uncovered a growing number of single-gene mutations that cause epilepsy (Noebels, 2003). The majority of these genes encode ion channels or receptors, including voltage-gated sodium, potassium, calcium, and chloride channels and receptors for acetylcholine and γ-amino butyric acid (GABA). Even in these cases, though, the mechanisms by which the defects cause epilepsy are not well understood.
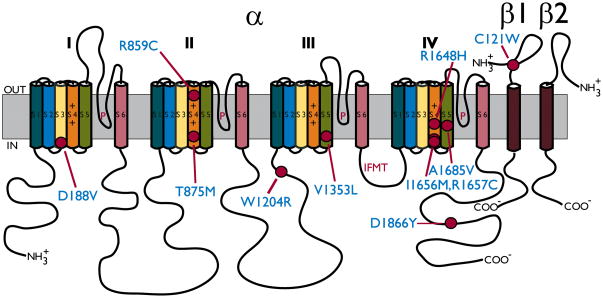
It seems logical that mutations in voltage-gated sodium channels would cause epilepsy, because these channels are in part responsible for controlling electrical excitability. Membrane depolarization activates the channel, causing a voltage-dependent conformational change that increases the permeability to sodium ions, further depolarizing the cell. This is followed by inactivation, in which the channel closes and the permeability to sodium ions decreases, allowing the membrane potential to return to its resting level. The sodium channel protein consists of a highly processed approximately 260-kDa α subunit that comprises four homologous domains termed I–IV (Figure 1). Within each domain, there are six transmembrane segments called S1–S6. A hairpin-like P-loop between S5 and S6 forms part of the channel pore, and the intracellular loop that connects domains III and IV forms the inactivation gate (Catterall, 2000). Channels in the adult CNS are associated with accessory β1, β2, β3, or β4 subunits. The β subunits are expressed in a complementary fashion, so that α subunits in the CNS are associated with either β1 or β3, and β2 or β4 (Isom et al., 1994; Morgan et al., 2000; Yu et al., 2003). Each β subunit consists of a single transmembrane segment, an extracellular IgG-like loop, and an intracellular C-terminus (Figure 1). The β2 or β4 subunit is covalently linked to the α subunit by a disulfide bond (Messner & Catterall, 1985), and the β1 or β3 subunit is noncovalently attached. The voltage dependence, kinetics, and localization of the α subunits are modulated by interactions with the β subunits.
Figure 1. Epilepsy-Causing Mutations in SCN1A.
Schematic diagram of the Nav1.1 sodium channel α subunit and associated β1 and β2 subunits. The Nav1.1 α subunit is associated non-covalently with β1 or β3, and it is associated via a disulfide linkage with either β2 or β4. The α subunit consists of four homologous domains (I–IV), each containing six transmembrane segments (S1–S6). The voltage-sensing S4 segment has multiple positively charged amino acids (+), and the intracellular loop between domains III and IV functions as the inactivation gate, containing four residues (IFMT) that interact with the inactivation docking site. The GEFS+ mutations that are mentioned in this review are indicated by filled circles. A complete list of the epilepsy-causing mutations in SCN1A can be found at http://web.scn1a.info (Lossin, 2009) and http://www.molgen.ua.ac.be/SCN1AMutations/(Claes et al., 2009).
The mammalian voltage-gated sodium channel α subunits are encoded by nine genes, termed SCN1A through SCN11A (SCN6A and SCN7A represent the same gene that encodes a non-voltage-gated sodium channel). The genes encode nine distinct isoforms, termed Nav1.1 through Nav1.9 (Goldin et al., 2000; Goldin, 2002). Four isoforms are expressed at high levels in the CNS (Nav1.1, Nav1.2, Nav1.3 and Nav1.6), and four are expressed at high levels in the peripheral nervous system (Nav1.6, Nav1.7, Nav1.8 and Nav1.9). The remaining two isoforms are expressed predominantly in adult skeletal muscle (Nav1.4) and in embryonic skeletal muscle and heart muscle (Nav1.5). The isoforms in the CNS are present at different times in development and in different locations (Trimmer & Rhodes, 2004). Nav1.1 becomes detectable shortly after birth and increases until adulthood (Beckh et al., 1989). Nav1.2 becomes detectable during embryonic development and reaches maximal levels during adulthood (Beckh et al., 1989). Nav1.3 is often considered an embryonic isoform, because its expression peaks at birth and it is usually undetectable in adult rodents (Beckh et al., 1989), but it has been detected at low levels in adulthood in humans (Whitaker et al., 2001). Nav1.3 expression has also been observed in the adult rat nervous system after dorsal root ganglion injury (Black et al., 1999; Hains et al., 2003) and in some epileptic rodent models (Yu et al., 2006; Guo et al., 2008). Nav1.6 is expressed during late embryonic and early postnatal periods (Felts et al., 1997; Schaller & Caldwell, 2000) and is also present at high levels during adulthood (Schaller et al., 1995).
In the adult CNS, Nav1.1 is the predominant channel in the caudal regions and the spinal cord, whereas levels of Nav1.2 are highest in the rostral regions (Gordon et al., 1987; Beckh et al., 1989). Nav1.1 is detected at high levels in the dendrites and cell bodies (Westenbroek et al., 1989; Gong et al., 1999), and it is also present in the axon initial segment of fast-spiking parvalbumin positive neurons (Ogiwara et al., 2007). Nav1.2 is observed at high levels in the dendrites and unmyelinated axons (Gong et al., 1999). Nav1.6 is present in both sensory and motor pathways, and its subcellular distribution includes axons, dendrites, cell bodies, and pre- and post-synaptic sites (Whitaker et al., 1999; Tzoumaka et al., 2000; Caldwell et al., 2000). Nav1.6 is the main sodium channel at the nodes of Ranvier (Krzemien et al., 2000; Caldwell et al., 2000).
Because of their distinct subcellular distribution and functional properties, each sodium channel isoform most likely plays a unique role in the initiation and propagation of action potentials. For example, action potentials are initiated preferentially at the distal end of the axon initial segment compared to the proximal end. This preference may reflect the high density of Nav1.6 channels in the distal end compared to the high density of Nav1.2 channels in the proximal end, as Nav1.6 channels have a lower threshold for activation (Hu et al., 2009). Because Nav1.1 is clustered at the axon initial segments of parvalbumin-positive basket cells (Ogiwara et al., 2007), which project their terminal axonal arbors around the soma and proximal dendrites of excitatory neurons, it may play an important role in modulating excitability of the network.
Epilepsy Syndromes Caused by SCN1A Mutations
Mutations in three of the sodium channel α subunit genes that are mainly expressed in the CNS have been shown to cause epilepsy in humans. Mutations in SCN1A (Nav1.1) and SCN2A (Nav1.2) cause several subtypes of dominant idiopathic generalized epilepsy. Mutations in both genes lead to genetic epilepsy with febrile seizures plus (GEFS+; MIM 604233) (Escayg et al., 2000; Escayg et al., 2001; Sugawara et al., 2001; Wallace et al., 2001b). This syndrome is also caused by mutations in the SCN1B gene encoding the sodium channel β1 subunit (Wallace et al., 1998; Wallace et al., 2002) and two GABAA receptor genes: GABRG2 encoding the γ2 subunit (Baulac et al., 2001; Wallace et al., 2001a; Harkin et al., 2002) and GABRD encoding the δ subunit (Dibbens et al., 2004). SCN1A mutations are the main cause of both Dravet syndrome (DS, MIM 607208) and intractable childhood epilepsy with generalized tonic-clonic seizures, also known as severe idiopathic generalized epilepsy of infancy (Mulley et al., 2005; Fujiwara, 2006). SCN2A mutations cause benign familial neonatal-infantile seizures (BFNIS; MIM 607745) and some cases of DS (Kamiya et al., 2004; Ogiwara et al., 2009; Shi et al., 2009), and a mutation in SCN1B has also been identified in a patient with DS (Patino et al., 2009). One mutation in SCN3A (Nav1.3) has been reported in a patient with partial epilepsy (Holland et al., 2008). Interestingly, mutations in SCN9A, which is considered to be expressed predominantly in the peripheral nervous system, were recently identified in patients with febrile seizures. SCN9A might also serve as a genetic modifier of DS (Singh et al., 2009).
GEFS+ is an autosomal dominant disorder with a complex and heterogeneous clinical presentation (Scheffer & Berkovic, 1997; Singh et al., 1999). The syndrome was originally called generalized epilepsy with febrile seizures plus, which is misleading because the clinical phenotype also includes partial epilepsy. Therefore, the term autosomal dominant epilepsy with febrile seizures plus was proposed by Ito et al. (2002), and more recently Scheffer et al. (2009; Ottman et al., 2010) proposed the alternative nomenclature genetic epilepsy with febrile seizures plus. Febrile seizures occur in approximately 5% of all children under the age of six years and typically are not associated with increased risk of epilepsy in adolescence and adulthood (Nelson & Ellenberg, 1976; Commission on Classification and Terminology of the International League Against Epilepsy, 1989). In contrast, patients with GEFS+ demonstrate febrile seizures that persist beyond six years of age and are associated with generalized or partial epilepsies, such as absence epilepsy, myoclonic seizures, atonic seizures, and myoclonic-astatic epilepsy (Scheffer & Berkovic, 1997; Singh et al., 1999). Affected individuals within GEFS+ families often display a wide variety of epilepsy subtypes with markedly different ages of onset and severity, suggesting the action of genetic and/or environmental modifiers.
DS is a catastrophic early-life epilepsy disorder in which the seizures are usually refractory to treatment and are associated with intellectual disability. This syndrome includes classical DS (severe myoclonic epilepsy of infancy) and borderline DS (severe myoclonic epilepsy of infancy borderline), in which patients show only a subset of clinical features (Scheffer et al., 2009; Mullen & Scheffer, 2009; Ottman et al., 2010). DS is characterized by febrile hemiclonic seizures or generalized status epilepticus starting at approximately six months of age, with other seizure types including partial, absence, atonic, and myoclonic seizures occurring after one year. In classical DS, development is delayed and patients often experience motor impairment, including spasticity and ataxia (Scheffer & Berkovic, 2003).
Genetics of SCN1A Mutations
Mutations in SCN1A account for approximately 10% of GEFS+ cases and mutations or deletions of SCN1A account for approximately 85% of DS cases (reviewed in Lossin, 2009; Claes et al., 2009). Updated lists of these mutations can be found at http://web.scn1a.info and http://www.molgen.ua.ac.be/SCN1AMutations/. Approximately 30 SCN1A-GEFS+ mutations have been identified to date. These have all been missense mutants (amino acid substitutions) that presumably alter but do not abolish SCN1A activity. In contrast, approximately half of the more than 600 SCN1A mutations in DS patients result from frameshift, nonsense, and splice site mutations (reviewed in Lossin, 2009; Claes et al., 2009). Several of these mutations occur very early in the sequence of SCN1A, strongly suggesting that expression of the mutant allele is reduced or that a non-functional protein product is generated from the mutant allele, demonstrating haploinsufficiency of SCN1A (Meisler & Kearney, 2005; Catterall et al., 2008).
A large number of SCN1A missense mutations have also been identified in DS patients. It is likely that many of these also abolish channel function, possibly by altering the properties of the channel, trafficking or subcellular localization, or interactions with other molecules. Unlike GEFS+ mutations that segregate within affected families, most DS mutations are de novo in the affected child. In a recent study, Heron et al. (2009) demonstrated that these de novo mutations most frequently originate on the paternal chromosome. The greater number of mitoses during spermatogenesis compared with oogenesis and the susceptibility of the methylated DNA of sperm cells to acquire mutations likely account for this observation.
Adding to the complexity of untangling the molecular mechanisms involving SCN1A mutations, microdeletions within SCN1A, and sometimes extending to include as many as 49 contiguous genes, including SCN2A, SCN3A, and SCN9A, have also been found in approximately 2–3% of DS patients (Suls et al., 2006; Wang et al., 2008; Davidsson et al., 2008; Marini et al., 2009). Although some of these patients display additional dysmorphic features (Davidsson et al., 2008), their clinical presentation is often surprisingly indistinguishable from DS patients in whom only SCN1A is affected. This suggests that haploinsufficiency for the adjacent genes may not contribute significantly to the disease presentation. Partial duplication/amplification of SCN1A has also been identified in three DS patients (Marini et al., 2009). Because these copy number variations are not detected by standard polymerase chain reaction screening, the percentage of DS cases resulting from SCN1A mutations is most likely an underestimate. Marini et al. (2009) estimated a 2–3% frequency of chromosomal abnormalities causing DS, which still leaves approximately 15% of cases in which no abnormality of SCN1A has been identified. Only a small number of DS cases have been shown to be due to mutations in SCN1B, SCN2A or GABRG2 (Harkin et al., 2002; Kamiya et al., 2004; Ogiwara et al., 2009; Shi et al., 2009; Patino et al., 2009), so the unexplained cases may be due to mutations in additional genes or may have polygenic etiologies (Marini et al., 2009).
Somatic and Germline Mosaicism
While the majority of DS patients carry de novo SCN1A mutations, some cases have been described in which the mutation was inherited from an asymptomatic or mildly affected parent, suggesting that the expression of the mutation in the transmitting parent was modified by environmental and/or genetic factors or was the result of somatic mosaicism. Somatic and germline SCN1A mosaicism has been reported in a small number of cases (Gennaro et al., 2006; Morimoto et al., 2006; Depienne et al., 2006; Marini et al., 2006). The possibility of SCN1A mosaicism in the parents of children with DS should therefore be considered when performing genetic counseling and determining recurrent risk.
Genetic Modifiers in GEFS+ and DS
One of the characteristics of GEFS+ is that family members with the same SCN1A mutation often display a wide range of seizure types and severities, suggesting that additional environmental or genetic factors likely influence clinical presentation. As mentioned earlier, similar factors may also alter the severity of SCN1A-DS mutations that are sometimes inherited from an asymptomatic or mildly affected parent. Furthermore, it is likely that a subset of the identified SCN1A-DS missense mutations only causes a modest alteration in channel function, and additional inherited or de novo variants at other loci are required to produce the severe clinical presentation.
A number of studies are beginning to provide evidence for the involvement of genetic modifiers in GEFS+ and DS. We previously showed that mice with mutations in Scn8a display elevated thresholds to chemically induced seizures when compared with wild-type littermates (Martin et al., 2007). Interestingly, in the presence of an Scn8a mutation, normal seizure thresholds and life spans were restored in heterozygous Scn1a knockout mice which serve as a model of DS (Martin et al., 2007). In a screen for genetic modifiers of DS, Ohmori et al. (2008) sequenced the coding exons of SCN1B, GABRG2, and CACNB4 in 38 SCN1A-positive DS patients. The amino acid substitution R468Q in CACNB4, the voltage-gated calcium channel β4 subunit, was identified in one individual. Increased calcium currents were seen when R468Q mutant channels were examined in BHK cells. This mutation may therefore result in increased neurotransmitter release from excitatory cells, which when combined with reduced inhibition as a result of the SCN1A mutation, may cause a more severe clinical presentation. Recently, Singh et al. (2009) proposed a role for SCN9A as a genetic modifier of DS. In that study, six DS patients who harbored missense or splice site mutations in SCN1A were also found to carry SCN9A variants. This raises the possibility that alterations in SCN9A may exacerbate the impact of otherwise less deleterious SCN1A mutations on neuronal excitability.
Functional Effects of SCN1A Mutations
The biophysical effects of several SCN1A GEFS+ and DS mutations have been examined using heterologous expression systems. The locations of the mutations for which functional data are mentioned in this review are shown in Figure 1. We characterized the effects of five GEFS+ mutations (R859C, T875M, W1204R, R1648H and D1866Y) by expression in Xenopus oocytes (Spampanato et al., 2001; Spampanato et al., 2003; Spampanato et al., 2004; Barela et al., 2006), and other mutations have been characterized by expression in human embryonic kidney cells (HEK or tsA201) (Alekov et al., 2000; Alekov et al., 2001; Lossin et al., 2002; Lossin et al., 2003; Cossette et al., 2003). These mutations altered sodium channel activity in many different ways, with some of the effects predicted to either increase or decrease sodium channel activity (Table 1). For example, in oocyte expression studies, R1648H dramatically accelerated recovery from inactivation, W1204R shifted the voltage-dependence of activation and inactivation in the negative direction, R859C shifted the voltage-dependence of activation in the positive direction, and T875M enhanced slow inactivation. The alterations due to R1648H and W1204R are predicted to increase sodium channel function and neuronal excitability, whereas the alterations due to R859C and T875M should decrease channel function and neuronal excitability.
Table 1.
Functional Effects of the SCN1A Sodium Channel Mutations that Cause GEFS+
| Increased Sodium Channel Activity | |||
|---|---|---|---|
| Mutation | Channel | Cell Type | Effects |
| D188V | rNav1.2 | HEK | ↓ Use-dependence, Faster recovery from slow inactivation |
| T875M | rNav1.1 | Xenopus oocytes | ↑ Persistence |
| W1204R | rNav1.1 | Xenopus oocytes | Negative voltage-dependence |
| W1204R | hNav1.1 | tsA201 | ↑ Persistence |
| R1648H | rNav1.1 | Xenopus oocytes | ↓ Use-dependence, Faster recovery |
| R1648H | hNav1.1 | tsA201 | ↑ Persistence |
| R1648H | hNav1.4 | tsA201 | Faster recovery |
| R1648C | rNav1.1 | tsA201 | ↑ Persistence |
| D1866Y | rNav1.1 | Xenopus oocytes | ↑ Persistence |
| Decreased Sodium Channel Activity | |||
| Mutation | Channel | Cell Type | Effects |
| R859C | rNav1.1 | Xenopus oocytes | Positive Voltage-dependence, Slower recovery from slow inactivation, ↓ Current |
| T875M | rNav1.1 | Xenopus oocytes | ↑ Slow inactivation |
| T875M | hNav1.4 | tsA201 | ↑ Fast and slow inactivation |
| V1353L | hNav1.1 | tsA201 | No sodium current |
| I1656M | hNav1.1 | tsA201 | Positive voltage-dependence |
| R1657C | hNav1.1 | tsA201 | Positive voltage-dependence, ↓ Current |
| A1685V | hNav1.1 | tsA201 | No sodium current |
The observed functional effects of SCN1A mutations are also influenced by the expression system. For example, the most prominent effect of R1648H, W1204R, and T1875M in tsA201 cells was an increase in the level of persistent current, with a marked increase for R1648H and a slight increase for T875M and W1204R (Lossin et al., 2002). The increased persistent current resulting from the R1648H mutation was due to a higher probability of late reopening and a fraction of channels with prolonged open times (Vanoye et al., 2006). It was previously shown in a transgenic Scn2a mouse model that increased persistent current can lead to seizures (Kearney et al., 2001), so a reasonable hypothesis was that this is the physiological change that causes seizures in GEFS+ (Lossin et al., 2002; Cannon, 2002).
Taken together, the data from the heterologous expression systems failed to uncover a consistent mechanism of pathogenesis and suggested that SCN1A mutations causing either gain-of-function or loss-of-function in Nav1.1 can predispose the CNS to abnormal excitability. However, this conclusion assumes that the effects of the mutations in heterologous systems are a direct reflection of their effects in native neurons, which may not be the case for a number of reasons (Ragsdale, 2008). First, the specific β subunits that are present in each type of neuron (β1 or β3 and β2 or β4) could alter the properties of the sodium channels, and hence the effects of the mutations. Second, the GEFS+ mutations could alter aspects of channel processing or trafficking that may be unique to neurons, so that the physiologically relevant phenotype would not be seen in another cell type. Third, neurons express multiple sodium channel isoforms, as well as an array of other voltage- and ligand-gated ion channels. The repertoire of channels can have a dramatic effect on the properties of a mutation, as has been shown for an SCN9A mutation that causes erythermalgia, and which made sympathetic neurons hypoexcitable but sensory neurons hyperexcitable because of the presence of Nav1.8 (Rush et al., 2006). Finally, studies in heterologous systems cannot reveal whether the mutations primarily affect excitatory or inhibitory neurons. A comparable alteration in sodium channel function in excitatory or inhibitory neurons is likely to have opposite effects on network excitability. For these reasons, it is essential to determine the effects of the mutations in native neurons in which SCN1A is normally expressed. This can be done most effectively using genetic mouse models.
Mouse Models of Human SCN1A Syndromes
Two different mouse models of DS have been constructed and analyzed (Table 2). Yu et al. (2006) constructed a mouse model of DS by targeted disruption of the mouse Scn1a gene. Heterozygous mutants (Scn1a+/−) with 50% of normal Nav1.1 levels were found to exhibit spontaneous seizures, reduced thresholds to hyperthermia-induced seizures, and a high mortality rate from four weeks after birth (Yu et al., 2006; Oakley et al., 2009). Homozygous mutants, which completely lack Nav1.1, were ataxic and died 15 days after birth. Electrophysiological analysis of hippocampal pyramidal neurons from homozygous Scn1a knockout mice revealed normal levels of sodium currents, indicating that Nav1.1 does not contribute significantly to sodium currents in these neurons. In contrast, sodium currents were significantly reduced in hippocampal interneurons that are critical for GABA-mediated neuronal inhibition. In addition, GABAergic cerebellar Purkinje neurons demonstrated significantly decreased peak, persistent and resurgent sodium currents, which is a likely explanation for the severe ataxia in these mice (Kalume et al., 2007). The Scn1a knockout mice were also found to have a compensatory increase in Nav1.3 expression. This was an unexpected finding, because Nav1.3 is highly expressed in the developing embryo, but decreases to almost undetectable levels in the adult mouse. It is not clear what role the increased Nav1.3 expression plays in the pathogenesis of epilepsy in these mice.
Table 2.
Mouse Models of Human Epilepsy Caused by SCN1A Mutations
| Human Syndrome | Type of Model | Salient Features | References |
|---|---|---|---|
| DS | Knockout | Spontaneous seizures, febrile seizures, ataxia and death at P15 in homozygous mice Spontaneous seizures, febrile seizures and impaired coordination in heterozygous mice Decreased sodium current density in inhibitory interneurons and cerebellar Purkinje neurons, but not in excitatory pyramidal neurons Decreased action potential firing in inhibitory interneurons Upregulation of Nav1.3 in hippocampal interneurons |
(Yu et al., 2006; Kalume et al., 2007; Oakley et al., 2009) |
| DS | Knock-In of R1407X nonsense mutation | Spontaneous seizures, unstable gait and death by P20 in homozygous mice Spontaneous seizures in heterozygous mice Spike amplitude decrements in parvalbumin-positive interneurons from heterozygous mice Nav1.1 was clustered at the axon initial segments of parvalbumin-positive interneurons |
(Ogiwara et al., 2007) |
| GEFS+ | BAC transgene with R1648H mutation | Increased seizure susceptibility to kainic acid Decreased sodium current density in inhibitory bipolar neurons Delayed recovery from inactivation and increased use-dependent inactivation of mutant sodium channels in inhibitory bipolar neurons Hyperpolarizing shift in the voltage-dependence of inactivation of mutant sodium channels in excitatory pyramidal neurons |
(Tang et al., 2009) |
| GEFS+ | Knock-in of R1648H mutation | Spontaneous seizures and death between P16 and P26 in homozygous mice Infrequent spontaneous seizures, reduced seizure thresholds and accelerated propagation of febrile seizures in heterozygous mice Slower recovery from inactivation, greater use-dependent inactivation, reduced sodium current density and decreased action potential firing in bipolar inhibitory neurons |
(Martin et al., 2010) |
The second mouse model of DS was constructed by Ogiwara et al. (2007), who incorporated a premature stop codon (R1407X) that was found in three human patients. Both homozygous and heterozygous mice demonstrated decreased Nav1.1 expression and spontaneous seizures. Wild-type Nav1.1 channels were observed to cluster primarily at the axon initial segments of parvalbumin-positive interneurons, and trains of action potentials in these neurons from the mutant mice demonstrated decreased amplitudes late in the burst. The results from both knockout mouse models are consistent and suggest that a reduction in neuronal inhibition contributes to the severe seizures associated with DS.
To examine the effects of a GEFS+ SCN1A mutation on channel function in vivo, we generated mice carrying the R1648H mutation (Table 2), which was previously shown to decrease use-dependent inactivation in Xenopus oocytes (Spampanato et al., 2001) and increase persistent current in tsA201 cells (Lossin et al., 2002). To determine how the mutation affected sodium channel function in neurons, we first generated a bacterial artificial chromosome (BAC) transgenic mouse model (Tang et al., 2009). This model had two advantages for these studies. First, the transgene included the entire Scn1a gene, so it was likely to be regulated and expressed in the same neurons as the native channels. Second, the transgene was engineered to also include the E954Q substitution that makes the channel resistant to tetrodotoxin (TTX) and saxitoxin (STX) (Kontis & Goldin, 1993). The predominant sodium channels in the mammalian CNS are all blocked by nanomolar concentrations of TTX or STX, so the R1648H mutant channels could be studied in isolation by recording in the presence of either of these toxins.
The effects of the R1648H mutation depended on the neurons in which it was studied. In inhibitory neurons from the BAC transgenic mice, the mutation caused delayed recovery from inactivation and increased use-dependent inactivation, which is the exact opposite of the effects that were observed in oocytes (Spampanato et al., 2001). In addition, there was no detectable increase in persistent current, unlike the findings in tsA201 cells (Lossin et al., 2002). In contrast, mutant sodium channels in excitatory pyramidal neurons showed no alteration in use-dependent inactivation or recovery from inactivation, but they did have a negative shift in the voltage-dependence of inactivation. In addition, the mutation decreased the total sodium current amplitude in bipolar neurons, but it did not affect the current amplitude in pyramidal neurons, further suggesting a specific impairment of inhibitory function. The BAC transgenic mice did not exhibit spontaneous seizures, but they did have a more severe response to the proconvulsant kainic acid compared with mice expressing a control Scn1a transgene.
To more accurately model the human disorder, we also introduced the R1648H mutation into the orthologous mouse gene by constructing a knock-in mouse model of GEFS+ (Martin et al., 2010). Mice homozygous for the R1648H mutation (Scn1aRH/RH) exhibited spontaneous generalized seizures and premature death between P16 and P26, whereas heterozygous mutants (Scn1aRH/+) exhibited infrequent spontaneous generalized seizures, reduced threshold and accelerated propagation of hyperthermia-induced seizures, and decreased threshold to flurothyl-induced seizures. Inhibitory cortical interneurons from P5–P15 Scn1aRH/+ and Scn1aRH/RH mice demonstrated slower recovery from inactivation and greater use-dependent inactivation compared with wild-type cells. In contrast, excitatory cortical pyramidal neurons demonstrated slightly faster recovery from inactivation. The R1648H mutation specifically decreased the total sodium current amplitude in bipolar neurons, consistent with the results from the BAC transgenic mice (Tang et al., 2009).
The alteration in sodium channel properties in bipolar neurons from the knock-in mice had a significant effect on the excitability of those cells and would most likely result in decreased inhibition. Bipolar neurons from Scn1aRH/RH mice fired a maximum of 20 action potentials in response to a 2-s injection of 70-pA current, whereas bipolar neurons from wild-type and Scn1aRH/+ mice fired approximately 35 action potentials in response to the same stimulus. Pyramidal neurons from all three genotypes fired similar numbers of action potentials over the range of currents injected, with the exception of Scn1aRH/RH mice, which showed reduced numbers when 30–50 pA of current was injected. However, since the number of action potentials was comparable for higher levels of current (≥ 70 pA), it is unclear whether the difference at lower levels of current is physiologically significant. Reduced inhibition is consistent with the rapid, abnormal propagation of experimental febrile seizures in these mice. Most Scn1aRH/RH mutants became immobile (the typical onset of experimental febrile seizure), then immediately progressed to a tonic seizure. This progression was rarely seen in wild-type or Scn1aRH/+ mice, and when evident took several minutes to occur.
The results from both the BAC transgenic and knock-in mouse models suggest that the primary effect of the R1648H GEFS+ mutation is to cause cell-specific impairment of interneuron sodium channel activity, leading to decreased inhibition. Given the importance of inhibitory circuits in controlling the excitability of postsynaptic neurons, it seems likely that reduced firing of cortical interneurons, and possibly other GABAergic interneurons, contributes to seizure generation in the Scn1aR1648H mouse models. This model is consistent with the fact that mutations of GABAA receptor genes GABRG2, GABRD, GABRA1 and GABRB3 cause GEFS+, DS and childhood absence epilepsy by decreasing either the function or number of GABAA receptors (Kang & Macdonald, 2009).
Conclusions
Mutations in the genes encoding voltage-gated sodium channels cause a variety of epilepsy syndromes in humans, with most of the mutations occurring in SCN1A. All of these mutations are dominant, and they can result in either loss of function (most commonly observed in DS) or altered function (most commonly observed in GEFS+). Based on the analysis of mouse models of these two syndromes, decreased activity of the inhibitory circuitry is likely a major factor contributing to seizure generation in the mice, and by extension in GEFS+ and DS patients. Impairment of inhibition could be a general effect of SCN1A mutations causing epilepsy, although the analysis of mouse models with other Scn1a mutations is needed to determine whether this is a consistent mechanism.
Acknowledgments
We are grateful to Cheryl Strauss for editorial assistance. Research in the authors’ laboratories is supported by grants from the NIH (NS065187 to AE and ALG, NS046484 and NS066155 to AE, NS048336 to ALG), the McKnight Foundation (34653 to ALG), and the Epilepsy Foundation (AE).
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Footnotes
Disclosure: Andrew Escayg has received support from and has served as a paid consultant for Allergan and Alan Goldin has received support from and has served as a paid consultant for Allergan.
Contributor Information
Andrew Escayg, Email: aescayg@emory.edu.
Alan L. Goldin, Email: agoldin@uci.edu.
References
- Alekov AK, Rahman M, Mitrovic N, Lehmann-Horn F, Lerche H. Enhanced inactivation and acceleration of activation of the sodium channel associated with epilepsy in man. Eur J Neurosci. 2001;13:2171–2176. doi: 10.1046/j.0953-816x.2001.01590.x. [DOI] [PubMed] [Google Scholar]
- Alekov AK, Rahman MM, Mitrovic N, Lehmann-Horn F, Lerche H. A sodium channel mutation causing epilepsy in man exhibits defects in fast inactivation and activation in vitro. J Physiol (Lond) 2000;529:533–539. doi: 10.1111/j.1469-7793.2000.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barela AJ, Waddy SP, Lickfett JG, Hunter J, Anido A, Helmers SL, Goldin AL, Escayg A. An epilepsy mutation in the sodium channel SCN1A that decreases channel excitability. J Neurosci. 2006;26:2714–2723. doi: 10.1523/JNEUROSCI.2977-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme J-F, Baulac M, Brice A, Bruzzone R, LeGuern E. First genetic evidence of GABAA receptor dysfunction in epilepsy: a mutation in the γ2-subunit gene. Nat Genet. 2001;28:46–48. doi: 10.1038/ng0501-46. [DOI] [PubMed] [Google Scholar]
- Beckh S, Noda M, Lübbert H, Numa S. Differential regulation of three sodium channel messenger RNAs in the rat central nervous system during development. EMBO J. 1989;8:3611–3636. doi: 10.1002/j.1460-2075.1989.tb08534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JA, Cummins TR, Plumpton C, Chen YH, Hormuzdiar W, Clare JJ, Waxman SG. Upregulation of a silent sodium channel after peripheral, but not central, nerve injury in DRG neurons. J Neurophysiol. 1999;82:2776–2785. doi: 10.1152/jn.1999.82.5.2776. [DOI] [PubMed] [Google Scholar]
- Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR. Sodium channel Nav1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Natl Acad Sci USA. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SC. Sodium channel gating: no margin for error. Neuron. 2002;34:853–854. doi: 10.1016/s0896-6273(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes LRF, Deprez L, Suls A, Baets J, Smets K, Van Dyck T, Deconinck T, Jordanova A, De Jonghe P. The SCN1A variant database: a novel research and diagnostic tool. Hum Mutat. 2009;30:E904–E920. doi: 10.1002/humu.21083. [DOI] [PubMed] [Google Scholar]
- Commission on Classification and Terminology of the International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Cossette P, Loukas A, Lafrenière RG, Rochefort D, Harvey-Girard E, Ragsdale DS, Dunn RJ, Rouleau GA. Functional characterization of the D188V mutation in neuronal voltage-gated sodium channel causing generalized epilepsy with febrile seizures plus (GEFS) Epilepsy Res. 2003;53:107–117. doi: 10.1016/s0920-1211(02)00259-0. [DOI] [PubMed] [Google Scholar]
- Davidsson J, Collin A, Olsson ME, Lundgren J, Soller M. Deletion of the SCN gene cluster on 2q24.4 is associated with severe epilepsy: an array-based genotype-phenotype correlation and a comprehensive review of previously published cases. Epilepsy Res. 2008;81:69–79. doi: 10.1016/j.eplepsyres.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Depienne C, Arzimanoglou A, Trouillard O, Fedirko E, Baulac S, Saint-Martin C, Ruberg M, Dravet C, Nabbout R, Baulac M, Gourfinkel-An I, LeGuern E. Parental mosaicism can cause recurrent transmission of SCN1A mutations associated with Severe Myoclonic Epilepsy of Infancy. Hum Mutat. 2006;27:389. doi: 10.1002/humu.9419. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Feng H-J, Richards MC, Harkin LA, Hodgson BL, Scott D, Jenkins M, Petrou S, Sutherland GR, Scheffer IE, Berkovic SF, Macdonald RL, Mulley JC. GABRD encoding a protein for extra- or peri-synaptic GABAA receptors is a susceptibility locus for generalized epilepsies. Hum Molec Genet. 2004;13:1315–1319. doi: 10.1093/hmg/ddh146. [DOI] [PubMed] [Google Scholar]
- Escayg A, Heils A, MacDonald BT, Haug K, Sander T, Meisler MH. A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus and prevalence of variants in patients with epilepsy. Am J Hum Genet. 2001;68:866–873. doi: 10.1086/319524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Felts PA, Yokoyama S, Dib-Hajj S, Black JA, Waxman SG. Sodium channel α-subunit mRNAs I, II, III, NaG, Na6 and hNE (PN1): Different expression patterns in developing rat nervous system. Mol Brain Res. 1997;45:71–82. doi: 10.1016/s0169-328x(96)00241-0. [DOI] [PubMed] [Google Scholar]
- Fujiwara T. Clinical spectrum of mutations in SCN1A gene: severe myoclonic epilepsy in infancy and related epilepsies. Epilepsy Res. 2006;70:S223–S230. doi: 10.1016/j.eplepsyres.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Gennaro E, Santorelli FM, Bertini E, Buti D, Gaggero R, Gobbi G, Linial M, Granata T, Freri E, Parmeggiani A, Striano P, Veggiotti P, Cardinali S, Bricarelli FD, Minetti C, Zara F. Somatic and germline mosaicisms in Severe Myoclonic Epilepsy of Infancy. Biochem Biophys Res Commun. 2006;341:489–493. doi: 10.1016/j.bbrc.2005.12.209. [DOI] [PubMed] [Google Scholar]
- Goldin AL. Evolution of voltage-gated Na+ channels. J Exp Biol. 2002;205:575–584. doi: 10.1242/jeb.205.5.575. [DOI] [PubMed] [Google Scholar]
- Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Berwald-Netter Y, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Gong B, Rhodes KJ, Bekele-Arcuri Z, Trimmer JS. Type I and type II Na+ channel α-subunit polypeptides exhibit distinct spatial and temporal patterning, and association with auxiliary subunits in rat brain. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- Gordon D, Merrick D, Auld V, Dunn R, Goldin AL, Davidson N, Catterall WA. Tissue-specific expression of the RI and RII sodium channel subtypes. Proc Natl Acad Sci USA. 1987;84:8682–8686. doi: 10.1073/pnas.84.23.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu N, Cai J-Q, Quinn T, Zong Z-H, Zeng Y-J, Hao L-Y. Voltage-gated sodium channel Nav1.1, Nav1.3 and β1 subunit were up-regulated in the hippocampus of spontaneously epileptic rat. Brain Res Bull. 2008;75:179–187. doi: 10.1016/j.brainresbull.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Hains BC, Klein JP, Saab CY, Craner MJ, Black JA, Waxman SG. Upregulation of sodium channel Nav1.3 and functional involvement in neuronal hyperexcitability associated with central neuropathic pain after spinal cord injury. J Neurosci. 2003;23:8881–8892. doi: 10.1523/JNEUROSCI.23-26-08881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LA, Bowser DN, Dibbens LM, Singh R, Phillips F, Wallace RH, Richards MC, Williams DA, Mulley JC, Berkovic SF, Scheffer IE, Petrou S. Truncation of the GABAA-receptor γ2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2002;70:530–536. doi: 10.1086/338710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser WA, Annegers JF, Kurland LT. Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935–1984. Epilepsia. 1993;34:453–468. doi: 10.1111/j.1528-1157.1993.tb02586.x. [DOI] [PubMed] [Google Scholar]
- Heron SE, Scheffer IE, Iona X, Zuberi SM, Birch R, McMahon JM, Bruce CM, Berkovic SF, Mulley JC. De novo SCN1A mutations in Dravet syndrome and related epileptic encephalopathies are largely of paternal origin. J Med Genet. 2009;47:137–141. doi: 10.1136/jmg.2008.065912. [DOI] [PubMed] [Google Scholar]
- Holland KD, Kearney JA, Glauser TA, Buck G, Keddache M, Blankston JR, Glaaser IW, Kass RS, Meisler MH. Mutation of sodium channel SCN3A in a patient with cryptogenic pediatric partial epilepsy. Neurosci Lett. 2008;433:65–70. doi: 10.1016/j.neulet.2007.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Tian C, Li T, Yang M, Hou H, Shu Y. Distinct contributions of Nav1.6 and Nav1.2 in action potential initiation and backpropagation. Nat Neurosci. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- Isom LL, DeJongh KS, Catterall WA. Auxiliary subunits of voltage-gated ion channels. Neuron. 1994;12:1183–1194. doi: 10.1016/0896-6273(94)90436-7. [DOI] [PubMed] [Google Scholar]
- Ito M, Nagafuji H, Okazawa H, Yamakawa K, Sugawara T, Mazaki-Miyazaki E, Hirose S, Fukuma G, Mitsudome A, Wada K, Kaneko S. Autosomal dominant epilepsy with febrile seizures plus with missense mutations of the (Na+)-channel α1 subunit gene, SCN1A. Epilepsy Res. 2002;48:15–23. doi: 10.1016/s0920-1211(01)00313-8. [DOI] [PubMed] [Google Scholar]
- Kalume F, Yu FH, Westenbroek RE, Scheuer T, Catterall WA. Reduced sodium current in Purkinje neurons from Nav1,1 mutant mice: implications for ataxia in severe myoclonic epilepsy in infancy. J Neurosci. 2007;27:11065–11074. doi: 10.1523/JNEUROSCI.2162-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, Makita N, Tanaka M, Fukushima K, Fujiwara T, Inoue Y, Yamakawa K. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-Q, Macdonald RL. Making sense of nonsense GABAA receptor mutations associated with genetic epilepsies. Trends Mol Med. 2009;15:430–438. doi: 10.1016/j.molmed.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JA, Plummer NW, Smith MR, Kapur J, Cummins TR, Waxman SG, Goldin AL, Meisler MH. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102:307–317. doi: 10.1016/s0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- Kontis KJ, Goldin AL. Site-directed mutagenesis of the putative pore region of the rat IIA sodium channel. Mol Pharmacol. 1993;43:635–644. [PubMed] [Google Scholar]
- Krzemien DM, Schaller KL, Levinson SR, Caldwell JH. Immunolocalization of sodium channel isoform NaCh6 in the nervous system. J Comp Neurol. 2000;420:70–83. [PubMed] [Google Scholar]
- Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–130. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Lossin C, Rhodes TH, Desai RR, Vanoye CG, Wang D, Carniciu S, Devinsky O, George AL., Jr Epilepsy-associated dysfunction in the voltage-gated neuronal sodium channel SCN1A. J Neurosci. 2003;23:11289–11295. doi: 10.1523/JNEUROSCI.23-36-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Marini C, Mei D, Cross JH, Guerrini R. Mosaic SCN1A mutation in familial Severe Myoclonic Epilepsy of Infancy. Epilepsia. 2006;47:1737–1740. doi: 10.1111/j.1528-1167.2006.00675.x. [DOI] [PubMed] [Google Scholar]
- Marini C, Scheffer IE, Nabbout R, Mei D, Cox K, Dibbens LM, McMahon JM, Iona X, Carpintero RS, Elia M, Cilio MR, Specchio N, Giordano L, Striano P, Gennaro E, Cross JH, Kivity S, Neufeld MY, Afawi Z, Andermann E, Keene D, Dulac O, Zara F, Berkovic SF, Guerrini R, Mulley JC. SCN1A duplications and deletions detected in Dravet syndrome: implications for molecular diagnosis. Epilepsia. 2009;50:1670–1678. doi: 10.1111/j.1528-1167.2009.02013.x. [DOI] [PubMed] [Google Scholar]
- Martin MS, Dutt K, Papale LA, Dube CM, Dutton SB, de Haan G, Shankar A, Tufik S, Meisler MH, Baram TZ, Goldin AL, Escayg A. Altered function of the SCN1A voltage-gated sodium channel leads to GABAergic interneuron abnormalities. J Biol Chem. 2010;285:9823–9834. doi: 10.1074/jbc.M109.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MS, Tang B, Papale LA, Yu FH, Catterall WA, Escayg A. The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum Molec Genet. 2007;16:2892–2899. doi: 10.1093/hmg/ddm248. [DOI] [PubMed] [Google Scholar]
- Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115:2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner DJ, Catterall WA. The sodium channel from rat brain - separation and characterization of subunits. J Biol Chem. 1985;260:10597–10604. [PubMed] [Google Scholar]
- Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K, Jackson AP. β3: An additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci USA. 2000;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Mazaki E, Nishimura A, Chiyonobu T, Sawai Y, Murakami A, Nakamura K, Inoue I, Ogiwara I, Sugimoto T, Yamakawa K. SCN1A mutation mosaicism in a family with Severe Myoclonic Epilepsy in Infancy. Epilepsia. 2006;47:1732–1736. doi: 10.1111/j.1528-1167.2006.00645.x. [DOI] [PubMed] [Google Scholar]
- Mullen SA, Scheffer IE. Translational research in epilepsy genetics. Sodium channels in man to interneuronopathy in mouse. Arch Neurol. 2009;66:21–26. doi: 10.1001/archneurol.2008.559. [DOI] [PubMed] [Google Scholar]
- Mulley JC, Scheffer IE, Petrou S, Dibbens LM, Berkovic SF, Harkin LA. SCN1A mutations and epilepsy. Human Mutation. 2005;25:535–542. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- Nelson KB, Ellenberg JH. Predictors of epilepsy in children who have experienced febrile seizures. New Engl J Med. 1976;295:1029–1033. doi: 10.1056/NEJM197611042951901. [DOI] [PubMed] [Google Scholar]
- Noebels JL. The biology of epilepsy genes. Annu Rev Neurosci. 2003;26:599–625. doi: 10.1146/annurev.neuro.26.010302.081210. [DOI] [PubMed] [Google Scholar]
- Oakley JC, Kalume F, Yu FH, Scheuer T, Catterall WA. Temperature- and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc Natl Acad Sci USA. 2009;106:3994–3999. doi: 10.1073/pnas.0813330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Ito K, Sawaishi Y, Osaka H, Mazaki E, Inoue I, Montal M, Hashikawa T, Shike T, Fujiwara T, Inoue Y, Kaneda M, Yamakawa K. De novo mutations of voltage-gated sodium channel αII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–1053. doi: 10.1212/WNL.0b013e3181b9cebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, Takeuchi T, Itohara S, Yanagawa Y, Obata K, Furuichi T, Hensch TK, Yamakawa K. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori I, Ouchida M, Miki T, Mimaki N, Kiyonaka S, Nishiki T, Tomizawa K, Mori Y, Matsui H. A CACNB4 mutation shows that altered Cav2.1 function may be a genetic modifier of severe myoclonic epilepsy in infancy. Neurobiol Dis. 2008;32:349–354. doi: 10.1016/j.nbd.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Ottman R, Hirose S, Jain S, Lerche H, Lopes-Cendes I, Noebels JL, Serratosa J, Zara F, Scheffer IE. Genetic testing in the epilepsies - report of the ILAE genetics commission. Epilepsia. 2010 doi: 10.1111/j.1528-1167.2009.02429.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino GA, Claes LRF, Lopez-Santiago LF, Slat EA, Dondeti RSR, Chen C, O’Malley HA, Gray CBB, Miyazaki H, Nukina N, Oyama F, De Jonghe P, Isom LL. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale DS. How do mutant Nav1.1 sodium channels cause epilepsy? Brain Res Rev. 2008;58:149–159. doi: 10.1016/j.brainresrev.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Rush AM, Dib-Hajj SD, Liu S, Cummins TR, Black JA, Waxman SG. A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc Natl Acad Sci USA. 2006;103:8245–8250. doi: 10.1073/pnas.0602813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller KL, Caldwell JH. Developmental and regional expression of sodium channel isoform NaCh6 in the rat central nervous system. J Comp Neurol. 2000;420:84–97. [PubMed] [Google Scholar]
- Schaller KL, Krzemien DM, Yarowsky PJ, Krueger BK, Caldwell JH. A novel, abundant sodium channel expressed in neurons and glia. J Neurosci. 1995;15:3231–3242. doi: 10.1523/JNEUROSCI.15-05-03231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic SF. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain. 1997;120:479–490. doi: 10.1093/brain/120.3.479. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic SF. The genetics of human epilepsy. Trends Pharmacol Sci. 2003;24:428–433. doi: 10.1016/S0165-6147(03)00194-9. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Zhang Y-H, Jansen FE, Dibbens L. Dravet syndrome or genetic (generalized) epilepsy with febrile seizures plus? Brain Dev. 2009;31:394–400. doi: 10.1016/j.braindev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Shi X, Yasumoto S, Nakagawa E, Fukasawa T, Uchiya S, Hirose S. Missense mutation of the sodium channel gene SCN2A causes Dravet syndrome. Brain Dev. 2009;31:758–762. doi: 10.1016/j.braindev.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Singh NA, Pappas C, Dahle EJ, Claes LRF, Pruess TH, De Jonghe P, Thompson J, Dixon M, Gurnett C, Peiffer A, White HW, Filloux F, Leppert MF. A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome. PLoS Genet. 2009;5:e1000649. doi: 10.1371/journal.pgen.1000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Scheffer IE, Crossland K, Berkovic SF. Generalized epilepsy with febrile seizures plus: A common childhood-onset genetic epilepsy syndrome. Ann Neurol. 1999;45:75–81. doi: 10.1002/1531-8249(199901)45:1<75::aid-art13>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Spampanato J, Escayg A, Meisler MH, Goldin AL. Functional effects of two voltage-gated sodium channel mutations that cause generalized epilepsy with febrile seizures plus type 2. J Neurosci. 2001;21:7481–7490. doi: 10.1523/JNEUROSCI.21-19-07481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Escayg A, Meisler MH, Goldin AL. The generalized epilepsy with febrile seizures plus type 2 mutation W1204R alters voltage-dependent gating of Nav1.1 sodium channels. Neuroscience. 2003;116:37–48. doi: 10.1016/s0306-4522(02)00698-x. [DOI] [PubMed] [Google Scholar]
- Spampanato J, Kearney JA, de Haan G, McEwen DP, Escayg A, Aradi I, MacDonald BT, Levin SI, Soltesz I, Benna P, Montalenti E, Isom LL, Goldin AL, Meisler MH. A novel epilepsy mutation in the sodium channel SCN1A identifies a cytoplasmic domain for β subunit interaction. J Neurosci. 2004;24:10022–10034. doi: 10.1523/JNEUROSCI.2034-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinlein OK. Channelopathies can cause epilepsy in man. Eur J Pain. 2002;6 (Suppl A):27–34. doi: 10.1053/eujp.2001.0319. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Tsurubuchi Y, Agarwala KL, Ito M, Fukuma G, Mazaki-Miyazaki E, Nagafuji H, Noda M, Imoto K, Wada K, Mitsudome A, Kaneko S, Montal M, Nagata K, Hirose S, Yamakawa K. A missense mutation of the Na+ channel αII subunit gene Nav1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc Natl Acad Sci USA. 2001;98:6384–6389. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Claeys KG, Goossens D, Harding B, Van Luijk R, Scheers S, Deprez L, Audenaert D, Van Dyck T, Beeckmans S, Smouts I, Ceulemans B, Lagae L, Buyse G, Barisic N, Misson J-P, Wauters J, Del-Favero J, De Jonghe P, Claes LRF. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Human Mutation. 2006;27:914–920. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- Tang B, Dutt K, Papale L, Rusconi R, Shankar A, Hunter J, Tufik S, Yu FH, Catterall WA, Mantegazza M, Goldin AL, Escayg A. A BAC transgenic mouse model reveals neuron subtype-specific effects of a Generalized Epilepsy with Febrile Seizures Plus (GEFS+) mutation. Neurobiol Dis. 2009;35:91–102. doi: 10.1016/j.nbd.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS, Rhodes KJ. Localization of voltage-gated ion channels in mammalian brain. Annu Rev Physiol. 2004;66:477–519. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- Tzoumaka E, Tischler AC, Sangameswaran L, Eglen RM, Hunter JC, Novakovic SD. Differential distribution of the tetrodotoxin-sensitive rPN4/NaCh6/Scn8a sodium channel in the nervous system. J Neurosci Res. 2000;60:37–44. doi: 10.1002/(SICI)1097-4547(20000401)60:1<37::AID-JNR4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Vanoye CG, Lossin C, Rhodes TH, George AL., Jr Single-channel properties of human Nav1.1 and mechanism of channel dysfunction in SCN1A-associated epilepsy. J Gen Physiol. 2006;127:1–14. doi: 10.1085/jgp.200509373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG, Williams DA, Sutherland GR, Mulley JC, Scheffer IE, Berkovic SF. Mutant GABAA receptor γ2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet. 2001a;28:49–52. doi: 10.1038/ng0501-49. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Scheffer IE, Barnett S, Richards M, Dibbens L, Desai RR, Lerman-Sagie T, Lev D, Mazarib A, Brand N, Ben-Zeev B, Goikhman I, Singh R, Kremmidiotis G, Gardner A, Sutherland GR, George AL, Jr, Mulley JC, Berkovic SF. Neuronal sodium-channel α1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2001b;68:859–865. doi: 10.1086/319516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Scheffer IE, Parasivam G, Barnett S, Wallace GB, Sutherland GR, Berkovic SF, Mulley JC. Generalized epilepsy with febrile seizures plus: mutation of the sodium channel subunit SCN1B. Neurology. 2002;58:1426–1429. doi: 10.1212/wnl.58.9.1426. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- Wang J, Kurahashi H, Ishii A, Kojima T, Ohfu M, Inoue T, Ogawa A, Yasumoto S, Oguni H, Kure S, Fujii T, Ito M, Okuno T, Shirasaka Y, Natsume J, Hasegawa A, Konagaya A, Kaneko S, Hirose S. Microchromosomal deletions involving SCN1A and adjacent genes in severe myoclonic epilepsy in infancy. Epilepsia. 2008;49:1528–1534. doi: 10.1111/j.1528-1167.2008.01609.x. [DOI] [PubMed] [Google Scholar]
- Westenbroek RE, Merrick DK, Catterall WA. Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- Whitaker W, Faull R, Waldvogel H, Plumpton C, Burbidge S, Emson P, Clare J. Localization of the type VI voltage-gated sodium channel protein in human CNS. NeuroReport. 1999;10:3703–3709. doi: 10.1097/00001756-199911260-00044. [DOI] [PubMed] [Google Scholar]
- Whitaker WRJ, Faull RLM, Waldvogel HJ, Plumpton CJ, Emson PC, Clare JJ. Comparative distribution of voltage-gated sodium channel proteins in human brain. Mol Brain Res. 2001;88:37–53. doi: 10.1016/s0169-328x(00)00289-8. [DOI] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- Yu FH, Westenbroek RE, Silos-Santiago I, McCormick KA, Lawson D, Ge P, Ferriera H, Lilly J, Distefano PS, Catterall WA, Scheuer T, Curtis R. Sodium channel β4, a new disulfide-linked auxiliary subunit with similarity to β2. J Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]