Abstract
Irritable bowel syndrome is a frequent gastrointestinal disorder of unknown etiology. The serotonin transporter regulates the intensity and duration of serotonin signaling in the gut and is, therefore, an attractive candidate gene for irritable bowel syndrome. Previous studies investigating the 5-HTTLPR and Stin2 VNTR polymorphisms of the serotonin transporter have proved inconclusive. In this exploratory study we therefore expanded the search for a possible association of the serotonin transporter with irritable bowel syndrome to include not only the 5-HTTLPR and Stin2 VNTR length polymorphisms, but also the functional single nucleotide polymorphism rs25531. We genotyped 186 patients with irritable bowel syndrome and 50 healthy control subjects raging in age from 18 to 70 years. Carriers of the rare G allele of rs25531 had approximately threefold increased odds of irritable bowel syndrome compared with healthy controls (OR 3.3, 95% CI 1.1–9.6). Our findings suggest that further investigation of the possible role of the serotonin transporter in the etiology of IBS is warranted.
Keywords: Irritable bowel syndrome, Functional gastrointestinal disorders, Serotonin transporter, Serotonin, Genetic polymorphism
Introduction
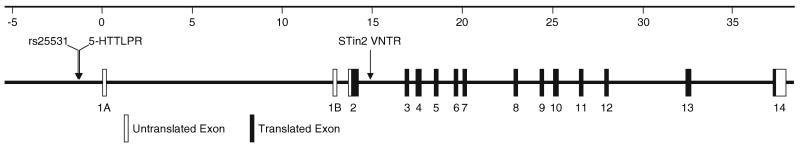
Irritable bowel syndrome (IBS) is a gastrointestinal (GI) disorder affecting around 10% of the population in Western countries [1, 2]. The disease is characterized by abdominal discomfort or pain associated with defecation or altered bowel habits, with no structural or biochemical explanation of the symptoms [1]. Evidence from twin and familial aggregation studies suggests that genetic factors contribute to the etiology of IBS [3, 4]. Because the serotonin system has been widely implicated in the pathophysiology of IBS [5–7], a number of studies have investigated possible association of serotonin transporter (SERT) polymorphisms with IBS, with conflicting results. Localized on the pre-synaptic membrane of serotonergic neurons, SERT acts as the major regulator of serotonergic neurotransmission throughout the body by controlling intensity and duration of serotonergic signaling via re-uptake of the neurotransmitter into the synapse [8]. The SERT gene is located on chromosome 17q11.1–17q12 and organized into 14 exons spanning approximately 38 kb (Fig. 1) [9, 10]. Its most frequently studied variant, 5-HTTLPR, located in the promoter region, is subdivided into a short (s) and long (l) allele, based on the presence or absence of a 43 bp insertion/deletion polymorphism [11, 12]. Evidence for an association of 5-HTTLPR with IBS is inconclusive, because some studies found an association between IBS and the 5-HTTLPR s/s genotype [13, 14] whereas others did not [15–20]. A second SERT polymorphism, STin2 VNTR, located in intron 2 and consisting of a variable number (usually 9, 10, or 12) of nearly identical 17-bp segments, has been found to be associated with IBS in only one study, with the 10/12 genotype more frequent in patients than in controls [21]. Most authors, however, have found no association between STin2 VNTR and IBS [13, 15, 18, 20].
Fig. 1.
Structure of the SERT gene. The scale at the top gives the distance in kb from the transcription start site. Exons 1A and 1B are alternately spliced. The positions of rs25531, 5-HTTLPR, and STin2 VNTR are indicated by arrows
The objective of this exploratory study was to contribute to our understanding of the role of SERT polymorphisms in IBS by not only genotyping the 5-HTTLPR and STin2 VNTR polymorphisms but also a single nucleotide polymorphism, rs25531, which is located immediately upstream of 5-HTTLPR and has not previously been investigated in IBS [12]. Because of their close proximity, rs25531 and 5-HTTLPR are strongly linked, yet they have opposing effects on SERT expression. The 5-HTTLPR l-allele has been associated with higher SERT expression and function than the s-allele [11, 22–25]. In contrast, the comparatively rare rs25531 G allele, occurring most frequently in combination with 5-HTTLPR – l, lowers SERT transcription compared with the more frequent A-allele [26]. As previous association studies indicated possible association of IBS with low-expression SERT genotypes [13, 14], we hypothesized that the low-expressing variant of rs25531 would be more frequent in patients with IBS than in controls.
Methods
Study Participants
This study was reviewed and approved by the University of Washington human participants institutional review board prior to participant recruitment. Participants were recruited through community advertisement. Subjects with IBS were part of an intervention study at the University of Washington investigating the effects of cognitive-behavioral therapy in IBS patients (Nursing Management of IBS: Improving Outcomes). Of 255 subjects enrolled in the parent study 188 consented separately to this supplemental genetic study. In two of these 188 cases DNA isolation or genotyping failed, resulting in a total of 186 samples from IBS patients. To be included in the study, IBS patients had to have a prior diagnosis of IBS made by a health-care provider and had to be currently experiencing IBS symptoms as described in the Rome-II criteria [27], i.e. at least 12 weeks in the preceding 12 months of abdominal discomfort or pain with two of the following three features:
relieved by defecation;
onset associated with a change in frequency of stool; or
onset associated with a change in form (appearance) of stool.
Fifty healthy age, gender, and race/ethnicity-matched volunteers were recruited by community advertisement for this genetic study only. Participants with IBS and control subjects were excluded if they had another GI disorder, previous GI surgery, other painful chronic health conditions, or a history of severe psychiatric illness. Participants lived in the Seattle area and reflect the predominantly Caucasian racial/ethnic mix of the region.
Sample Collection and Genotyping
A sample of 10 ml EDTA-anticoagulated blood was collected at a local laboratory. Samples were identified by subject number only. DNA isolation and genotyping was performed by investigators unaware of any subject information. DNA was isolated from blood using buffy coat preparations in a modification of the procedure of Miller [28], using Puregene DNA Purification Kits (Gentra Systems, MN, USA) and following the manufacturer's instructions. For genotyping of 5-HTTLPR, 0.5 μM oligonucleotide primers flanking 5-HTTLPR (forward: 5′-ATGCCAGCACCTAACCCCTAATGT-3′) and reverse: 5′-GGACCGCAAGGTGGGCGGGA-3′) were used in 10 μl PCR reactions containing 5 μl HotStar Taq Master Mix (Qiagen, CA, USA), 2.5 μl betaine (Sigma–Aldrich, MO, USA), and 100 ng genomic DNA from each subject. PCR reactions were run on an MJ Research PCT-200 DNA engine, using the following cycling conditions: 15-min incubation at 95°C, followed by 33 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min, followed by a 10-min final extension step of 72°C for 10 min. Results were size-fractionated on a 3% agarose gel which enabled easy distinction of the s allele, yielding a 376-bp fragment, the l allele, resulting in a 419 bp fragment, and the xl allele, resulting in an approximately 460-bp fragment. For genotyping of STin2 VNTR, 0.5 μM oligonucleotide primers flanking the polymorphic site (forward: 5′-GTCAGTATCACAGGCTGCGAG-3′ and reverse: 5′-TGTTCCTAGTCTTACGCCAGTG-3′) were used in PCR reactions using the same reagents and cycling conditions as described above for 5-HTTLPR. Reaction products were size-fractionated on a 5% non-denaturing polyacrylamide gel, enabling distinction of the 214-bp STin2.7, 248-bp STin2.9, 265-bp STin2.10, and 299-bp STin2.12 alleles. rs25531 was genotyped using an ABI7000 Gene Expression System. Genomic DNA (100 ng) was amplified in the presence of gene-specific primers (forward: 5′-CCCTCGCGGCATCCC-3′, reverse: 5′-ATGCTGGAAGGGCTGCA-3′) and allele-specific fluorescent probes (VIC-CTGCACCCCCAGCAT-NFQ and FAM-CTGCACCCCCGGCAT-NFQ) obtained through the Applied Biosystems Custom TaqMan SNP genotyping assay service, and following the manufacturer's instructions.
Data Analysis
Calculations for deviation from Hardy–Weinberg equilibrium were performed using χ2 tests. Comparisons between IBS subjects and controls by SERT genotype and other comparisons involving categorical variables were made using the generalization of Fisher's exact test. Continuous variables, for example age at onset of IBS, were compared between the genotype groups using two-sample t-tests. Odds ratios for IBS by SERT genotype were determined by logistic regression with and without controlling for membership in one of four race categories: White, African American, Asian, or other/multiple races.
Results
The demographics of IBS subjects and controls are shown in Table 1. Genotype distributions for the 5-HTTLPR, STin2 VNTR, and rs25531 polymorphisms were checked for deviation from Hardy–Weinberg equilibrium and no deviation was observed (IBS: 5-HTTLPR χ2 = 2.3, P = 0.51; STin2 VNTR χ2 = 2.5, P = 0.47; rs25531 χ2 = 2.9, P = 0.09. Controls: 5-HTTLPR χ2 = 2.1, P = 0.15; STin2 VNTR χ2 = 0.58, P = 0.90; rs25531 χ2 = 0.09, P = 0.77). Allele frequencies are shown in Table 2.
Table 1.
Comparison of subjects with IBS and controls by demographic variables
| IBS n (%) |
Controls n (%) |
||
|---|---|---|---|
| Gender | Male | 27 (15) | 9 (18) |
| Female | 159 (85) | 41 (82) | |
| Race | White only | 153 (82) | 40 (80) |
| African American only | 7 (4) | 1 (2) | |
| Asian only | 7 (4) | 1 (2) | |
| American Indian/Alaskan Native only | 1 (1) | 0 | |
| Native Hawaiian/Pacific Islander only | 0 | 0 | |
| More than one race | 17 (9) | 5 (10) | |
| Unknown | 1 (1) | 3 (6) | |
| Age | 44.6 ± 14.2 | 43.9 ± 14.8 |
Note: For categorical variables (gender and race) the numbers (n) and the percentages (%) of subjects with IBS (n = 186) and controls (n = 50) that met the demographic criteria listed are given. For age, mean ± SD is given. Differences between subjects with IBS and controls were not significant (Fisher's exact test or two-sample t-test)
Table 2.
Distribution of allele frequencies for 5-HTTLPR, STin2 VNTR, and rs25531
| Polymorphism | Allele | Allele frequency | |
|---|---|---|---|
| IBS | Controls | ||
| 5-HTTLPR | s | 41 | 49 |
| l | 59 | 51 | |
| xl | 0.3 | 0 | |
| STin2 VNTR | STin2.9 | 3 | 1 |
| STin2.10 | 35 | 35 | |
| STin2.12 | 62 | 64 | |
| rs25531 | A | 89 | 96 |
| G | 11 | 4 | |
Note: Shown are the allele frequencies (%) for subjects with IBS (n = 186) and controls (n = 50)
Comparing genotype distributions between subjects with and without IBS (Table 3) showed that the rs25531 A/G genotype was significantly more frequent among subjects with IBS than among controls. There was also a trend towards association of the 5-HTTLPR l/l genotype with IBS. These results remained the same whether men and women were analyzed together, as shown, or when the analysis was performed for women only (data not shown).
Table 3.
Influence of SERT genotype on IBS risk
| Polymorphism | Genotype | IBS n (%) |
Controls n (%) |
Odds ratio | 95% CI | Adjusted odds ratio | 95% CI | P-values |
|---|---|---|---|---|---|---|---|---|
| 5-HTTLPR | s/s | 35 (19%) | 9 (19%) | 1 | 1 | |||
| s/l | 81 (44%) | 29 (60%) | 0.7 | 0.3–1.7 | 0.7 | 0.3–1.6 | ||
| l/l | 69 (37%) | 10 (21%) | 1.8 | 0.7–4.8 | 1.8 | 0.6–5.0 | ||
| Fisher's exact test: | 0.07 | |||||||
| Logistic regression likelihood ratio test: | 0.06 | |||||||
| Logistic regression likelihood ratio test controlling for race: | 0.05 | |||||||
| STin2 VNTR | 10/10 | 27 (15%) | 6 (13%) | 1 | 1 | |||
| 10/12 | 75 (40%) | 22 (46%) | 0.7 | 0.3–1.9 | 0.6 | 0.2–1.8 | ||
| 12/12 | 74 (40%) | 19 (40%) | 0.8 | 0.3–2.2 | 0.7 | 0.2–2.0 | ||
| 9/12 | 8 (4%) | 1 (2%) | 1.7 | 0.2–15.8 | 1.4 | 0.1–13.9 | ||
| 9/10 | 2 (1%) | 0 | ||||||
| Fisher's exact test: | 0.94 | |||||||
| Logistic regression likelihood ratio test: | 0.78 | |||||||
| Logistic regression likelihood ratio test controlling for race: | 0.72 | |||||||
| rs25531 | A/A | 145 (78%) | 46 (92%) | 1 | 1 | |||
| A/G | 41 (22%) | 4 (8%) | 3.3 | 1.1–9.6 | 2.9 | 1.0–8.7 | ||
| Fisher's exact test: | 0.03 | |||||||
| Logistic regression likelihood ratio test: | 0.02 | |||||||
| Logistic regression likelihood ratio test controlling for race: | 0.04 | |||||||
Note: Shown are the numbers (n) and the percentages (%) of subjects with IBS (n = 186) and healthy controls (n = 50) with any given genotype. One subject carrying the l/xl genotype was excluded from the 5-HTTLPR analysis, and two controls failed genotyping for 5-HTTLPR and STin2 VNTR. Shown are odds ratios and 95% confidence intervals (95% CI). Adjusted odds ratios were controlled for membership in one of four race categories: White, African American, Asian, or other/multiple races
In addition, genotype frequencies for the STin2VNTR and rs25531 polymorphisms differed significantly by population group (Table 4). In particular, the STin2.9 allele was exclusively observed among subjects who had classified themselves as White only. The STin2.12 allele, on the other hand, was less common among White subjects (allele frequency 60%) than among individuals who had classified themselves as Black (69%), Asian (94%), or of American Indian/Alaskan Native or mixed origin (70%). Likewise, the rs25531 G-allele was much less frequent among Whites (8%) than among Blacks (19%), Asians (31%), or subjects of American Indian/Alaskan Native or mixed origin (11%). Neither the observed association of IBS with the rs25531 G-allele, nor the trend towards an association with the 5-HTTLPR l/l genotype changed if results were controlled for race, however (Table 3).
Table 4.
Distribution of SERT genotypes by population group
| Polymorphism | Genotype | White n (%) |
Non-White n (%) |
|---|---|---|---|
| 5-HTTLPR | s/s | 34 (18) | 9 (23) |
| s/l | 91 (48) | 17 (44) | |
| l/l | 66 (35) | 12 (31) | |
| l/xl | 0 | 1 (3) | |
| STin2 VNTR | 10/10 | 27 (14) | 5 (13) |
| 10/12 | 86 (45) | 10 (26) | |
| 12/12 | 67 (35) | 24 (62) | |
| 9/12 | 9 (5) | 0 | |
| 9/10 | 2 (1) | 0 | |
| rs25531 | A/A | 162 (84) | 26 (67) |
| A/G | 31 (16) | 13 (33) |
Note: Shown are comparisons between subjects describing themselves as “White” only (n = 193) versus all other and mixed population groups combined (n = 39). Subjects with unknown race/ethnicity or missing genotypes were excluded. There was no statistically significant difference in genotype distribution for 5-HTTLPR (Fisher's exact test P = 0.24) whereas for STin2VNTR (Fisher's exact test P = 0.03) and rs25531 (Fisher's exact test P = 0.02) genotype distributions differed significantly
Genotypes at the three polymorphic sites, 5-HTTLPR, STin2 VNTR, and rs25531, were not independent of each other. As shown in Table 5, in White subjects the 5-HTTLPR s allele occurred more frequently in combination with the STin2.12 allele and the rs25531 A allele than with STin2.9, STin2.10, or rs25531 G, indicating the presence of linkage disequilibrium (LD) between the three sites. There was less evidence for LD among non-White subjects. These results remained the same whether subjects with IBS and controls were analyzed together, as shown, or when the analysis was done for IBS subjects only (data not shown).
Table 5.
Associations of genotypes between 5-HTTLPR, STin2 VNTR, and rs25531
| White | Non-White | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5-HTTLPR | STin2 VNTR | 5-HTTLPR | STin2 VNTR | ||||||||
| 10/10 | 10/12 | 12/12 | 9/12 | 9/10 | 10/10 | 10/12 | 12/12 | 9/12 | 9/10 | ||
| A | |||||||||||
| s/s | 2 (7%) | 7 (8%) | 25 (37%) | 0 | 0 | s/s | 0 | 2 (20%) | 7 (29%) | 0 | 0 |
| s/l | 7 (26%) | 47 (55%) | 32 (48%) | 5 (56%) | 0 | s/l | 1 (20%) | 4 (40%) | 12 (50%) | 0 | 0 |
| l/l | 18 (67%) | 32 (37%) | 10 (15%) | 4 (44%) | 2 (100%) | l/l | 4 (80%) | 4 (40%) | 4 (17%) | 0 | 0 |
| l/xl | 0 | 0 | 0 | 0 | 0 | l/xl | 0 | 0 | 1 (4%) | 0 | 0 |
| P-value | <0.001 | P-value | 0.19 | ||||||||
| rs25531 | STin2 VNTR | rs25531 | STin2 VNTR | ||||||||
| 10/10 | 10/12 | 12/12 | 9/12 | 9/10 | 10/10 | 10/12 | 12/12 | 9/12 | 9/10 | ||
| B | |||||||||||
| A/A | 23 (85%) | 79 (92%) | 54 (81%) | 3 (33%) | 1 (50%) | A/A | 3 (60%) | 8 (80%) | 15 (63%) | 0 | 0 |
| A/G | 4 (15%) | 7 (8%) | 13 (19%) | 6 (67%) | 1 (50%) | A/G | 2 (40%) | 2 (20%) | 9 (38%) | 0 | 0 |
| P-value | 0.000 | P-value | 0.61 | ||||||||
| rs25531 | 5-HTTLPR | rs25531 | 5-HTTLPR | ||||||||
| s/s | s/l | l/l | l/xl | s/s | s/l | l/l | l/xl | ||||
| C | |||||||||||
| A/A | 34 (100%) | 76 (84%) | 50 (76%) | 0 | A/A | 9 (100%) | 11 (65%) | 6 (50%) | 0 | ||
| A/G | 0 | 15 (16%) | 16 (24%) | 0 | A/G | 0 | 6 (35%) | 6 (50%) | 1 (100%) | ||
| P-value | 0.002 | P-value | 0.02 | ||||||||
Note: All observed genotype combinations between pairs of polymorphic sites are tabulated separately for subjects describing themselves as “White” only (n = 193) versus all other and mixed population groups combined (n = 39). Subjects with unknown race/ethnicity or missing genotypes were excluded. Shown are number (n) and column percent (%) of genotype carriers and P-values of Fisher's exact test for association between genotypes within each of the two subject groups. A: 5-HTTLPR and STin2 VNTR, B: rs25531 and STin2 VNTR, C: rs25531 and 5-HTTLPR
Because of the presence of LD one might expect that the trend towards association of 5-HTTLPR l/l with IBS that we observed might be because subjects with the 5-HTTLPR l/l genotype also carry an rs25531 G allele more frequently than subjects with other 5-HTTLPR genotypes. However, when rs25531 G allele carriers were excluded from the analysis, the trend towards an association of 5-HTTLPR l/l with IBS persisted (Fisher's exact P = 0.061).
SERT genotype distributions did not differ significantly by IBS subtype (Table 6). In addition, age at onset of disease, average pain intensity, or extent to which IBS subjects reduced their daily activities due to pain were not significantly associated with SERT genotype (data not shown).
Table 6.
Distribution of SERT genotypes by IBS subtype
| Polymorphism | Genotype | Constipation-predominant total n = 42 n (%) |
Diarrhea-predominant total n = 98 n (%) |
Alternators total n = 36 n (%) |
Normal stool total n = 10 n (%) |
|---|---|---|---|---|---|
| 5-HTTLPR | s/s | 8 (19) | 18 (18) | 7 (19) | 2 (20) |
| s/l | 19 (45) | 41 (42) | 15 (42) | 6 (60) | |
| l/l | 15 (36) | 39 (40) | 13 (36) | 2 (20) | |
| l/xl | 0 | 0 | 1 (3) | 0 | |
| STin2 VNTR | 10/10 | 2 (5) | 16 (16) | 9 (25) | 0 |
| 10/12 | 20 (48) | 39 (40) | 13 (36) | 3 (30) | |
| 12/12 | 19 (45) | 35 (36) | 14 (39) | 6 (60) | |
| 9/12 | 1 (2) | 6 (6) | 0 | 1 (10) | |
| 9/10 | 0 | 2 (2) | 0 | 0 | |
| rs25531 | A/A | 36 (86) | 74 (76) | 29 (81) | 6 (60) |
| A/G | 6 (14) | 24 (24) | 7 (19) | 4 (40) |
Note: Shown are the comparisons between IBS subjects with different bowel patterns. Genotype distributions did not differ significantly by bowel pattern for any of the three polymorphisms shown (Fisher's exact test)
Discussion
Our study is the first to show that the G allele of the rs25531 polymorphism of SERT is associated with IBS. The frequency of the rs25531 G allele in our healthy control subjects agrees with previous observations giving the frequency of the rs25531 G allele in Caucasian subjects as 7% [12]. Our observations are also in agreement with earlier studies that demonstrated population differences in the genotype distribution of SERT polymorphisms, with population differences in the degree of LD between these loci [29–32]. It should be noted that the presence of LD within the SERT gene implies the possibility that the association between rs25531 and IBS we observed might not be because of the functional effects of rs25531 itself, but could also be because of the effects of another, as yet not investigated, marker in the SERT gene that is in LD with rs25531.
Our finding that the presence of the rs25531 G allele raises the odds for IBS about threefold is in keeping with prior observations that have reported an association of IBS with the lower-expressing 5-HTTLPR s/s genotype [13, 14]. The connection between IBS and low SERT expression and/or function is also supported by observations of decreased SERT expression in the colonal mucosa of IBS patients [33], and decreased SERT function in blood platelets of patients with diarrhea-predominant IBS [34, 35]. Substitution of the common rs25531 A allele with a G allele creates an additional AP2 transcription factor binding site in the SERT promoter which has previously been shown to lower SERT expression in vitro [26]. Yet it cannot necessarily be concluded that this polymorphism has the same functional effect in IBS. The transcription of most genes depends on the cellular environment, and the effect of SERT polymorphisms on gene expression has been shown to differ in different types of cell [36]. Depending on the cellular environment, AP2 transcription factors can act as transcriptional activators and repressors [26, 37]. Hence, the functional significance of our observed association of rs25531 G with IBS in the gut is still unknown. Moreover, we also found a trend towards association of the higher-expressing 5-HTTLPR l/l genotype with IBS, which cannot be solely explained on the basis of LD between these polymorphisms. In common with most prior studies of SERT in IBS, we did not observe any association of the STin2 VNTR polymorphism with the disease. Moreover, we saw no effect of SERT genotype on the predominant bowel pattern of IBS or age at onset of disease.
Studies in mice lacking SERT have shown that most SERT knock-out animals experience diarrhea, yet a smaller group has predominantly constipation. Moreover, for a single mouse lacking SERT, diarrhea and constipation alternate irregularly [38]. The presence of constipation even though increased SERT availability should predispose to diarrhea has been attributed to enteric serotonin receptor desensitization in the presence of chronically enhanced serotonergic neurotransmission in the gut. This mechanism could explain why we found the presumably lower-expressing rs25531 G-allele to be associated with all forms of IBS, not just the diarrhea-predominant type. The G-allele was more common in diarrhea-predominant subjects than in constipation-predominant or alternator subjects; because of the small subject size, however, these differences were not significant. A recent meta-analysis concluded that the 5-HTTLPR polymorphism of SERT is not associated with IBS [39]. Our results show that we cannot generalize from this and similar negative genetic association studies that SERT plays no role in IBS, because other, previously not investigated SERT polymorphisms, for example rs25531, may very well be relevant to the disease. Our finding that the rs25531 G allele raises the odds for IBS about threefold is further evidence of the role of serotonin in the pathophysiology of IBS.
The main limitation of this study is its small sample size. Not only would a larger sample size have established the association between rs25531 and IBS with greater certainly, it would also have been better able to answer the question of whether the rs25531 G-allele increases the odds for diarrhea-prominent IBS more than those for constipation-predominant or alternator subtypes. Moreover, it can be argued that our group of IBS patients represents a potentially biased sample, as patients were originally recruited for participation in a cognitive-behavioral trial of IBS and not chosen from a clinic population. However, because both subjects and controls were recruited via community advertisement and utilizing the same venues, the differences in SERT genotype distributions reported here should still be valid. Nonetheless, confirmation of the association of rs25531 with IBS in a larger sample of subjects is clearly needed, along with further studies on SERT function in IBS and the effects of genetic variants on SERT function in gut cells.
Acknowledgments
This study was supported by National Institute of Nursing Research Grants NIH NR01094, P30 NR04001, the NIH Office of Research in Women's Health, and by resources from the Veterans Affairs Puget Sound Health Care System, Seattle, Washington.
Abbreviations
- SERT or 5-HTT
Serotonin transporter
- 5-HTTLPR
Serotonin transporter length polymorphic region, i.e. a 43 bp insertion–deletion polymorphism in the promoter region
- STin2 VNTR
Serotonin transporter intron 2 variable number tandem repeat polymorphism
- rs25531
Unique identifier for a single nucleotide polymorphism in the serotonin transporter gene promoter region
- IBS
Irritable bowel syndrome
- PCR
Polymerase chain reaction
- GI
Gastrointestinal
- LD
Linkage disequilibrium
Contributor Information
Ruth Kohen, Email: ruko@u.washington.edu, Department of Psychiatry & Behavioral Sciences, University of Washington, Seattle, WA 98195-6560, USA; Geriatric Research, Education and Clinical Center, Veterans Affairs Puget Sound Health Care System, 1660 South Columbian Way, Seattle, WA 98108, USA.
Monica E. Jarrett, Department of Biobehavioral Nursing & Health Systems, University of Washington, Seattle, WA 98195-7226, USA
Kevin C. Cain, Department of Biostatistics, School of Public Health, University of Washington, Seattle, WA 98195-7230, USA
Sang-Eun Jun, Department of Biobehavioral Nursing & Health Systems, University of Washington, Seattle, WA 98195-7226, USA.
Grace P. Navaja, Geriatric Research, Education and Clinical Center, Veterans Affairs Puget Sound Health Care System, 1660 South Columbian Way, Seattle, WA 98108, USA
Sarah Symonds, Geriatric Research, Education and Clinical Center, Veterans Affairs Puget Sound Health Care System, 1660 South Columbian Way, Seattle, WA 98108, USA.
Margaret M. Heitkemper, Department of Biobehavioral Nursing & Health Systems, University of Washington, Seattle, WA 98195-7226, USA
References
- 1.Drossman DA, Camilleri M, Mayer EA, Whitehead WE. AGA technical review on irritable bowel syndrome. Gastroenterology. 2002;123(6):2108–2131. doi: 10.1053/gast.2002.37095. [DOI] [PubMed] [Google Scholar]
- 2.Cremonini F, Talley NJ. Irritable bowel syndrome: epidemiology, natural history, health care seeking and emerging risk factors. Gastroenterol Clin North Am. 2005;34(2):189–204. doi: 10.1016/j.gtc.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Park MI, Camilleri M. Genetics and genotypes in irritable bowel syndrome: implications for diagnosis and treatment. Gastroenterol Clin North Am. 2005;34(2):305–317. doi: 10.1016/j.gtc.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Saito YA, Petersen GM, Locke GR, III, Talley NJ. The genetics of irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3(11):1057–1065. doi: 10.1016/S1542-3565(05)00184-9. [DOI] [PubMed] [Google Scholar]
- 5.Crowell MD. Role of serotonin in the pathophysiology of the irritable bowel syndrome. Br J Pharmacol. 2004;141(8):1285–1293. doi: 10.1038/sj.bjp.0705762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mawe GM, Coates MD, Moses PL. Review article: intestinal serotonin signalling in irritable bowel syndrome. Aliment Pharmacol Ther. 2006;23(8):1067–1076. doi: 10.1111/j.1365-2036.2006.02858.x. [DOI] [PubMed] [Google Scholar]
- 7.Andresen V, Camilleri M. Irritable bowel syndrome: recent and novel therapeutic approaches. Drugs. 2006;66(8):1073–1088. doi: 10.2165/00003495-200666080-00004. [DOI] [PubMed] [Google Scholar]
- 8.Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4(1):13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 9.Ramamoorthy S, Bauman AL, Moore KR, et al. Antidepressant- and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc Natl Acad Sci USA. 1993;90(6):2542–2546. doi: 10.1073/pnas.90.6.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesch KP, Balling U, Gross J, et al. Organization of the human serotonin transporter gene. J Neural Transm. 1994;95(2):157–162. doi: 10.1007/BF01276434. [DOI] [PubMed] [Google Scholar]
- 11.Heils A, Teufel A, Petri S, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 12.Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11(3):224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- 13.Yeo A, Boyd P, Lumsden S, et al. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut. 2004;53(10):1452–1458. doi: 10.1136/gut.2003.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JM, Choi MG, Park JA, et al. Serotonin transporter gene polymorphism and irritable bowel syndrome. Neurogastroenterol Motil. 2006;18(11):995–1000. doi: 10.1111/j.1365-2982.2006.00829.x. [DOI] [PubMed] [Google Scholar]
- 15.Pata C, Erdal ME, Derici E, Yazar A, Kanik A, Ulu O. Serotonin transporter gene polymorphism in irritable bowel syndrome. Am J Gastroenterol. 2002;97(7):1780–1784. doi: 10.1111/j.1572-0241.2002.05841.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, Camilleri M, Carlson PJ, et al. Association of distinct alpha(2) adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53(6):829–837. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee DY, Park H, Kim WH, Lee SI, Seo YJ, Choi YC. Serotonin transporter gene polymorphism in healthy adults and patients with irritable bowel syndrome. Korean J Gastroenterol. 2004;43(1):18–22. [PubMed] [Google Scholar]
- 18.Li YY, Nie YQ, Xie J, Tan HZ, Zhou YJ, Wang H. Serotonin transporter gene polymorphisms in irritable bowel syndrome and their impact on tegaserod treatment. Chung-Hua Nei Ko Tsa Chih Chin J Intern Med. 2006;45(7):552–555. [PubMed] [Google Scholar]
- 19.Saito YA, Locke GR, III, Zimmerman JM, et al. A genetic association study of 5-HTT LPR and GNbeta3 C825T polymorphisms with irritable bowel syndrome. Neurogastroenterol Motil. 2007;19(6):465–470. doi: 10.1111/j.1365-2982.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Nie Y, Xie J, et al. The association of serotonin transporter genetic polymorphisms and irritable bowel syndrome and its influence on tegaserod treatment in Chinese patients. Dig Dis Sci. 2007;52(11):2942–2949. doi: 10.1007/s10620-006-9679-y. [DOI] [PubMed] [Google Scholar]
- 21.Wang BM, Wang YM, Zhang WM, et al. Serotonin transporter gene polymorphism in irritable bowel syndrome. Chung-Hua Nei Ko Tsa Chih Chin J Intern Med. 2004;43(6):439–441. [PubMed] [Google Scholar]
- 22.Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 23.Hanna GL, Himle JA, Curtis GC, et al. Serotonin transporter and seasonal variation in blood serotonin in families with obsessive-compulsive disorder. Neuropsychopharmacology. 1998;18(2):102–111. doi: 10.1016/S0893-133X(97)00097-3. [DOI] [PubMed] [Google Scholar]
- 24.Little KY, McLaughlin DP, Zhang L, et al. Cocaine, ethanol, and genotype effects on human midbrain serotonin transporter binding sites and mRNA levels. Am J Psychiatry. 1998;155(2):207–213. doi: 10.1176/ajp.155.2.207. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg BD, Tolliver TJ, Huang SJ, Li Q, Bengel D, Murphy DL. Genetic variation in the serotonin transporter promoter region affects serotonin uptake in human blood platelets. Am J Med Genet. 1999;88(1):83–87. doi: 10.1002/(SICI)1096-8628(19990205)88:1<83∷AID-AJMG15>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 26.Hu XZ, Lipsky RH, Zhu G, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78(5):815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drossman DA. Rome-II: The Functional Gastrointestinal Disorders: Diagnosis, Pathophysiology, and Treatment: a Multinational Consensus. McLean, VA: Degnon Associates; 2000. [Google Scholar]
- 28.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelernter J, Kranzler H, Cubells JF. Serotonin transporter protein (SLC6A4) allele and haplotype frequencies and linkage disequilibria in African- and European-American and Japanese populations and in alcohol-dependent subjects. Hum Genet. 1997;101(2):243–246. doi: 10.1007/s004390050624. [DOI] [PubMed] [Google Scholar]
- 30.Gelernter J, Cubells JF, Kidd JR, Pakstis AJ, Kidd KK. Population studies of polymorphisms of the serotonin transporter protein gene. Am J Med Genet. 1999;88(1):61–66. doi: 10.1002/(SICI)1096-8628(19990205)88:1<61∷AID-AJMG11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura M, Ueno S, Sano A, Tanabe H. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 2000;5(1):32–38. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- 32.Lotrich FE, Pollock BG, Ferrell RE. Serotonin transporter promoter polymorphism in African Americans: allele frequencies and implications for treatment. Am J Pharmacogenomics. 2003;3(2):145–147. doi: 10.2165/00129785-200303020-00007. [DOI] [PubMed] [Google Scholar]
- 33.Coates MD, Mahoney CR, Linden DR, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126(7):1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Bellini M, Rappelli L, Blandizzi C, et al. Platelet serotonin transporter in patients with diarrhea-predominant irritable bowel syndrome both before and after treatment with alosetron. Am J Gastroenterol. 2003;98(12):2705–2711. doi: 10.1111/j.1572-0241.2003.08669.x. [DOI] [PubMed] [Google Scholar]
- 35.Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2006;130(1):34–43. doi: 10.1053/j.gastro.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 36.Sakai K, Nakamura M, Ueno S, et al. The silencer activity of the novel human serotonin transporter linked polymorphic regions. Neurosci Lett. 2002;327(1):13–16. doi: 10.1016/S0304-3940(02)00348-8. [DOI] [PubMed] [Google Scholar]
- 37.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6(13):246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JJ, Li Z, Pan H, et al. Maintenance of serotonin in the intestinal mucosa and ganglia of mice that lack the high-affinity serotonin transporter: Abnormal intestinal motility and the expression of cation transporters. J Neurosci. 2001;21(16):6348–6361. doi: 10.1523/JNEUROSCI.21-16-06348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26(7):979–986. doi: 10.1111/j.1365-2036.2007.03453.x. [DOI] [PubMed] [Google Scholar]