Abstract
The conformational change in the selectivity filter of KcsA as a function of ambient potassium concentration is studied with solid state NMR. This highly conserved region of the protein is known to chelate potassium ions selectively. We report solid-state NMR chemical shift fingerprints of two distinct conformations of the selectivity filter; significant changes are observed in the chemical shifts of key residues in the filter as the buffer potassium ion concentration is changed from 50 mM to 1 µM. Potassium ion titration studies reveal that the site-specific Kd for K+ binding at the key pore residue Val 76, is on the order of ∼7 µM and that relatively high sample hydration is necessary to observe the low K+ conformer. Simultaneous detection of both conformers at low ambient potassium concentration suggests that the high K+ and low K+ states are in slow exchange on the NMR timescale (kex < 500 s−1). The slow rate and tight binding for evacuating both inner sites simultaneously, differ from prior observations in detergents in solution, but agree well with measurements by electrophysiology, and appear to result from our use of a hydrated bilayer environment. These characteristics rule out participation of the low K+ state on the timescale of ion transmission, which has been assumed to involve interchange of states where one of the inner binding sites is always occupied. On the other hand, these kinetic and thermodynamic characteristics of evacuation of the inner sites certainly could be compatible with participation in a control mechanism at low ion concentration, such as C-type inactivation, a process that is coupled to activation and involves closing of the outer mouth of the channel.
Keywords: potassium channel, KcsA, solid-state NMR, conformational dynamics, selectivity filter
Introduction
Potassium channels are highly conserved intrinsic membrane proteins that selectively conduct potassium ions across cell membranes near the rate of free diffusion. Although structures of some potassium channels have been known for over 10 years,1; 2 important mechanistic questions remain open for debate. One key mechanistic question is how potassium channels achieve a very fast throughput rate while remaining highly selective for potassium over sodium. Selectivity in binding could involve relatively high affinity for some intermediate of the process, which in turn could result in slow kinetics of release or escape from that state. While there is indeed evidence that potassium ions bind with a micromolar affinity3 inside potassium channels and with more than three orders of magnitude better affinity than other ions such as sodium4, this does not impede the inverse passage time of a potassium ion through the channel from being ∼10−8 s.5 This fast conduction rate is interpreted as being diffusion limited. Therefore the mechanisms by which diffusion-limited kinetics and high selectivity are simultaneously achieved have been of great interest.
KcsA is a 160 amino-acid potassium channel isolated from the soil bacterium S. lividans.6; 7 Because of its experimental convenience, high sequence homology and structural similarity to key regions of mammalian potassium channels, it has become a model system for studies of the biophysical details of ion-conduction and gating.8; 9; 10; 11; 12; 13; 14; 15 KcsA is a homotetrameric protein with two transmembrane helices per monomer and an ion-conducting pore in the center. The highly conserved selectivity filter motif for K+ channels consists of residues 74–79 (TVGYG). The earliest structures showed that in this region the backbone carbonyl oxygens specifically chelate dehydrated K+ ions inside the channel.
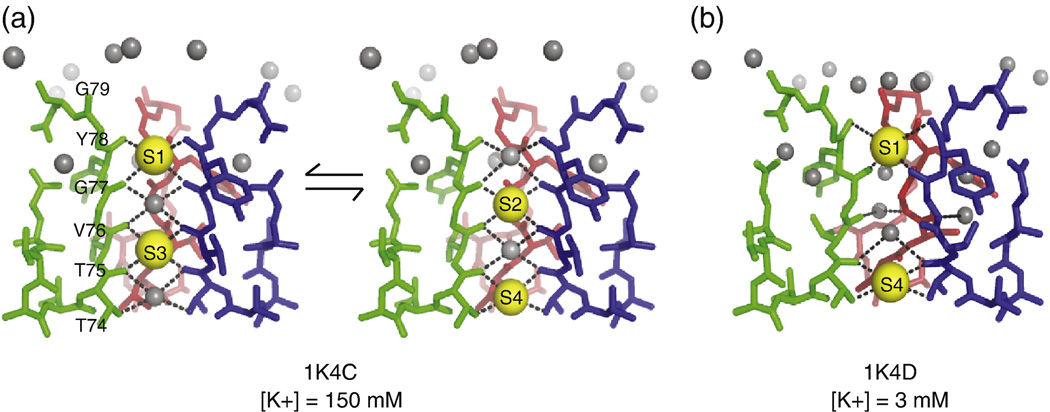
Crystal structures of KcsA2; 4 suggest that in the conductive state of the channel, four ion-binding sites are formed by carbonyl oxygens in the pore. A potassium ion fits into each of these binding-sites, thus satisfying its preference16 for an octahedral chelation environment. The ion-oxygen distance in the octahedral cages of KcsA range from 2.7 to 3.1 Å, with an average of 2.85 Å. This is close to the preferred ion-chelator distance in many biological octahedral K+ complexes like valinomycin and noactin2; 17 Sodium ions, on the other hand, prefer tetrahedral coordination environments and chelation distances of ∼2.4 Å when they interact with six fully flexible monodentate ligands.18 The ion binding sites in KcsA are thus optimized for potassium binding, in terms of both symmetry and ion-oxygen distances. These sites are typically referred to as S1-S4. S1 resides on the extracellular side and S4 on the intracellular side of the membrane. During conduction, the selectivity filter is proposed to exist in two ion configurations that may be in fast exchange: the occupancies of the sites S1-S2-S3-S4 are K+-water-K+-water and water-K+-water-K+ for the two states, which will here forth be referred to as the (1,3) and (2,4) states.16 These configurations are shown in Figure 1A.
Figure 1.
The different crystallized conformations of the selectivity filter in KcsA are shown. The selectivity filter of KcsA adopts a different conformation based on the identity and concentration of the permeant ion. The “high K+” structure, 1K4C, was crystallized at [K+] = 150 mM and has been interpreted as a mixture of two states in which potassium ions are chelated at the 1,3 and 2,4 positions. The intervening positions are occupied by water (A). The proposed “low K+” structure, 1K4D, is a single state crystallized at [K+] = 3 mM. It shows ion occupancy at sites 1 and 4. The identity of the ions is uncertain, and it has been speculated that at least one of the ions may be Na+ (B).
The crystal structures provide important support for the dominant hypothesis on the mechanism of ion selectivity: the inner ion-binding sites (S2 and S3) provide perfect chelation environments for potassium, but do not accommodate many other ions including sodium. This has been called the “snug fit” mechanism and was first proposed by Bezanilla and Armstrong.19 The Pauling ionic radii for K+ and Na+ are 1.33Å and 0.95 Å respectively5, and, as mentioned earlier, the preferred ion-oxygen distances differ by ∼ 0.4 Å. Thus, the proposed “snug-fit” mechanism has sometimes been cast as requiring the selectivity filter to retain a geometry with sufficient rigidity in the positions of the ligating carbonyls to discriminate between these ions. Ion flux measurements show that the relative free energy of selectivity ΔΔG(Na-K) is on the order of 5–6 kcal/mol.20; 21 Free energy perturbation (FEP) computations22; 23 show that the intrinsic carbonyl repulsion interactions and flexibility of the channel residues are necessary to maintain the relative difference in free energy for binding potassium vs. sodium. This led to an alternative hypothesis, which suggests that the carbonyl groups coordinating the ion in the filter are dynamic and that their intrinsic electrostatic and dynamic properties control ion selectivity.24 The crystallographic B-factors for the selectivity filter are ∼15 − 20 Å2. In principle, if the B-factors are interpreted in terms of the root-mean square fluctuation in the atom’s position5, then , indicating an RMS fluctuation of ∼ 0.8 Å in the pore. While this seems large compared to the difference in atomic radii between K+ and Na+, when compared to other parts of the protein (B-factors 30- 40 Å2, RMS ∼1–1.3 Å), the B-factors in the pore are relatively low. Since B-factors presumably report on both static and dynamic disorder at low temperature, their relation to dynamic disorder at functional temperatures and conditions is unclear. The snug fit and dynamic pore hypotheses differ in their emphasis on how much flexibility the pore has, and whether or not this is related directly to selectivity. The extent of pore-plasticity must therefore be directly measured by characterizing the dynamic behavior of the residues in the selectivity filter in an appropriate environment.
Regardless of whether dynamics in the pore contribute to the microscopic mechanism for selectivity, it is clear that the selectivity filter is a center of activity during normal channel function. Characterizing its structural plasticity and dynamic behavior is crucial to understand the transport-cycle of K+ through the channel. NMR is a natural technique to study protein dynamics. It serves as a powerful complement to x-ray crystallography, which cannot discriminate between static and dynamic disorder. Solid-state NMR gives us the unique opportunity to study channel dynamics in a native bilayer environment. Methods to study dynamics in the solid-state have recently been applied to protein systems and several studies25; 26; 27; 28; 29 report on protein dynamics at different timescales in the solid state.
Proof of the inherent plasticity of the channel emerged in a series of studies targeting structure as a function of the concentration of the permeant ions.4; 30 When the ambient potassium concentration is reduced from 200 mM to 3 mM, the crystal structures suggest that an ion binds at the outer sites, S1 and S4, but not at the inner sites S2 and S3. This structure can be called the (1,4) structure.4 In the absence of any ions in S2 and S3, the selectivity filter rearranges the key chelators V76 and G77. In the (1,4) structure the S2 and S3 sites do not appear to be capable of chelating a potassium ion, and instead the V76 carbonyl forms a water mediated hydrogen bond with G77 on the neighboring subunit.2 The G77 Cα is twisted inward and a new hydrogen bond network consisting of a belt of buried water molecules surrounds the selectivity filter around S2 and S3. This structure is assumed to be non-conductive (although studies of it are carried out at relatively high potassium concentrations of 3–5 mM) and it has sometimes described as collapsed or pinched. This low K+ conformation of the filter is shown in Fig. 1B.
We have chosen to initially focus on this dramatic transition between the low-K+ and high-K+ states in a bilayer environment. Previous characterizations11; 15 of this transition involved membrane environments that might be assumed to affect dynamics and ion affinity. We present a structural and qualitative dynamic characterization of the two limiting states of the KcsA selectivity filter as a function of permeant ion identity and concentration based on solid-state NMR spectroscopy, and illustrate ways in which the bilayer environment is very important for the detailed kinetics.
Results
Two distinct states of KcsA as a function of potassium concentration
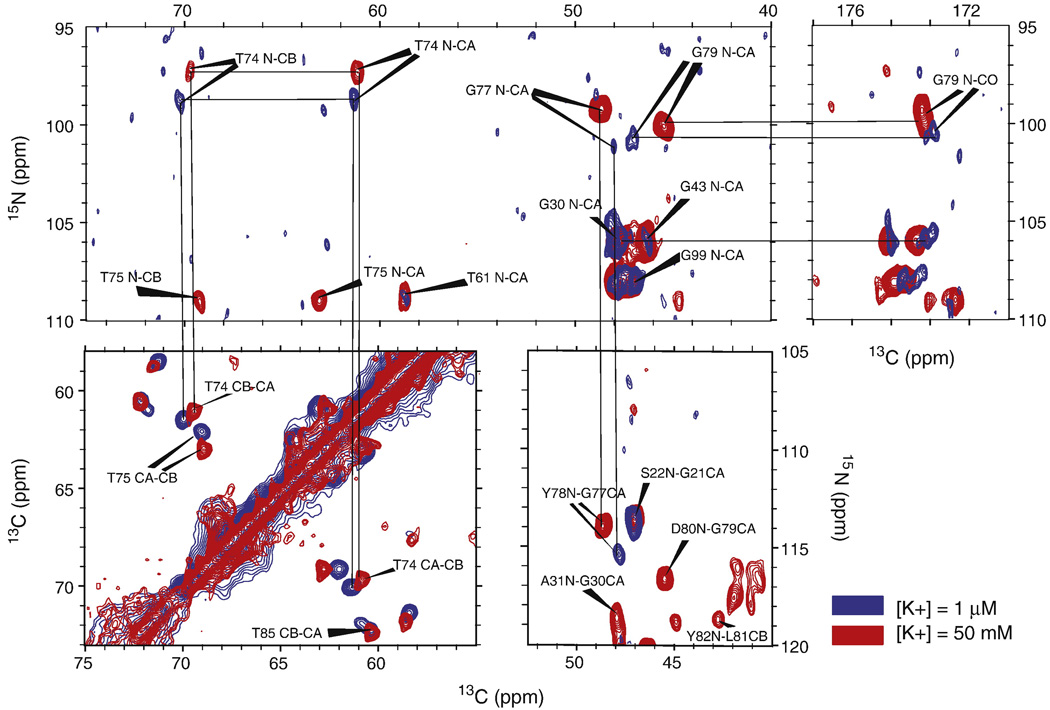
The quintessential NMR fingerprint of a protein’s conformational state is the collection of chemical shifts of its residues. Both the isotropic shift and the chemical shift anisotropy (CSA) of an atom are extremely sensitive to the local electronic environment. Changes in the electronic environment induced by changes in structure generally have strong effects on isotropic chemical shifts. We report backbone (N, Cα, C’) and sidechain (Cβ, Cγ) chemical shifts of several marker peaks at pH =7.5 and two different permeant ion conditions: (1) [K+] = 50 mM and (2)) [Na+] = 50 mM, [K+] = 1 µM. Elemental analysis revealed that the 50 mM Na+ buffer contained 1 µM K+, presumably due to minor contaminants in the reagents. Spectra and analysis of these shifts are shown in figure 2 and figure 3. Figure 2 displays homonuclear (13C-13C) and heteronuclear (13C-15N) spectra of KcsA at [K+] = 50 mM and 1 µM that illustrate significant differences in the chemical shift at several sites in the selectivity filter. The observed changes in chemical shift are reproducible to within 0.2 ppm. The chemical shifts of V76 and G77, which are in the middle of the selectivity filter and chelate sites S2 and S3, show some of the strongest dependence on potassium ion concentration. V76 Cβ is shifted by 1.8 ppm and the C’ is shifted by ∼ 3 ppm between the high and low K+ states. These differences are much larger than the typical linewidth of our peaks (∼ 0.3–0.5 ppm in the 13C dimension) and suggest that there is a potassium dependent structural rearrangement that occurs specifically in the selectivity filter. The backbone chemical shifts of residues at the edges of the filter, T74 and G79, also exhibit significant changes.
Figure 2.
Homonuclear 13C-13C and heteronuclear 15N-13C solid-state NMR spectra at two different K+ conditions are shown. Chemical shift changes in the selectivity filter at sites T74 (N,Cα,Cβ), T75 (N,Cα,Cβ), G77 (N, Cα), Y78 (N) and G79 (N,Cα,Cβ,C’) between the 50 mM K+ (red) and 50 mM Na+/1 µM K+ (blue) states are very clear in all spectra. Similar spectra for V76 (Cα,Cβ,C’) are shown in the supplement in Fig S4. The spectra shown are 15N-13C 2Ds in the T74 and G 77/79 N-Cα and N-CO regions (Top), a 13C-13C 2D in the T74 Cβ-Cα region and a 15N-13C 2D in the G77 N-CO region (Bottom). These data support the existence of at least two distinct [K+] dependent states of the selectivity filter.
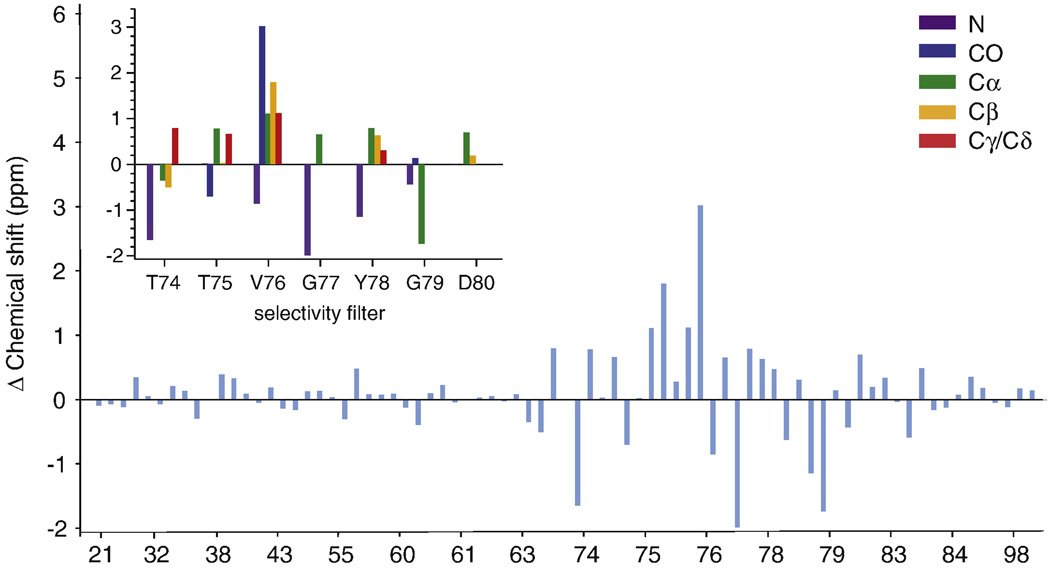
Figure 3.
The difference in the chemical shifts between the 50 mM [K+] and 1 µM [K+] states is plotted for all assigned residues resolved on a CC2D (blue). Unless otherwise specified, [K+] refers to the ion concentration in the buffer solution bathing the sample. On changing [K+] from 50 mM to 1 µM, significant changes (> 0.2 ppm) are seen in the chemical shifts at several sites in the selectivity filter (T74-D80) but not in other assigned regions of the protein. The inset shows chemical shift changes for different types of nuclei in the filter. N: purple CO: blue, Cα: green, Cβ: yellow, Cγ/Cδ: red. Spectra for these sites are shown in Fig.2. The observed chemical shift changes suggest changes in the structure and ion occupancy of the filter as a function of [K+].
Chemical shift analysis using SHIFTX and SPARTA
We used two different chemical shift prediction programs – SHIFTX31 and SPARTA32 to compare the observed experimental chemical shifts at 50 mM and 1 µM potassium, to the shifts predicted from various reported crystal structures. The high K+ shifts were compared to three different high potassium crystal structures: 1K4C, 1BL8 and 3EFF, and the low K+ chemical shifts to two different low K+ crystal structures: 1K4D and 2ITC. Our analysis indicates that within the error of the prediction tools, our high K+ shifts are most consistent with the structure reported in 1K4C and the low K+ shifts are consistent with both the 1K4D and 2ITC structures. The root mean square deviations (RMSDs) between the experimental shifts and the predicted shifts for the two conformers are presented in Table 1. The RMSDs that we report are in the 1–1.5 ppm range at the Cα and Cβ sites, which is consistent with RMSDs reported for proteins using these tools.32; 33 Previous studies have shown that the predictive accuracy of SPARTA and SHIFTX is much higher for the Cα and Cβ sites compared to C’ and N. While these tools are able to distinguish between different protein folds fairly accurately, they lack the predictive sensitivity to subtle changes in structure, which are often important for biology. Thus, it is important to underline that our high and low K+ shifts are consistent with the predictions from 1K4C and 1K4D within the error of these tools. The chemical shift databases used in these programs rely on peptide conformation and do not explicitly include electrostatic effects due to ion binding. We therefore cannot distinguish a sodium-bound state from a potassium-bound state using these tools.
Table 1.
Experimentally measured chemical shifts of residues in the selectivity filter are compared with shifts predicted from deposited high K+ crystal structures 1K4C, 1BL8, 3EFF and low K+ structures 1K4D and 2ITC using the SHIFTX and SPARTA prediction programs. The RMSDs we see are well within the range reported for other proteins studied by ssNMR33.The analysis suggests that within the error of the available prediction tools, the [K+] dependent states we detect in the bilayer are consistent with deposited crystal structures 1K4C at 50 mM K+ and with 1K4D and 2ITC at 1 µM K+.
| RMSD (Experimental Shifts vs Predicted Shifts) ppm |
||||||
|---|---|---|---|---|---|---|
| SHIFTX | SPARTA | |||||
| High K+ structures | 1K4C | 1BL8 | 3EFF | 1K4C | 1BL8 | 3EFF |
| N | 4.55 | 6.99 | 4.95 | 5.93 | 7.23 | 6.83 |
| CA | 1.18 | 2.02 | 1.79 | 0.97 | 1.84 | 1.47 |
| CB | 1.21 | 1.69 | 1.68 | 1.37 | 1.68 | 1.92 |
| CO | 1.43 | 2.11 | 1.88 | 1.74 | 2.10 | 1.69 |
| Low K+ structure (1K4D) | ||||||
| 1K4D | 2ITC | 1K4D | 2ITC | |||
| N | 3.84 | 4.09 | 5.42 | 5.52 | ||
| CA | 1.33 | 1.41 | 1.06 | 1.17 | ||
| CB | 1.39 | 1.29 | 1.74 | 1.51 | ||
| CO | 1.53 | 1.56 | 1.63 | 1.49 | ||
It is also important to note that there are many differences in the way samples are prepared for X-ray crystallography and for solid-state NMR. These include, (1) the presence of a lipid bilayer in the solid-state NMR samples, which is substituted by detergent in protein crystals, (2) the temperature at which data are collected and (3) often there are other differences in pH and buffer content that help to form crystals and may be important effectors of functional state. All reported x-ray structures of KcsA to date have been measured at cryogenic temperatures, whereas solid-state NMR data are often acquired at a sample temperature of ∼ 0 − 10° C (and were in this study). Thus, while these predictive tools are not the yet ideal to probe subtle changes in structure, they present a useful starting point to analyze chemical shift data.
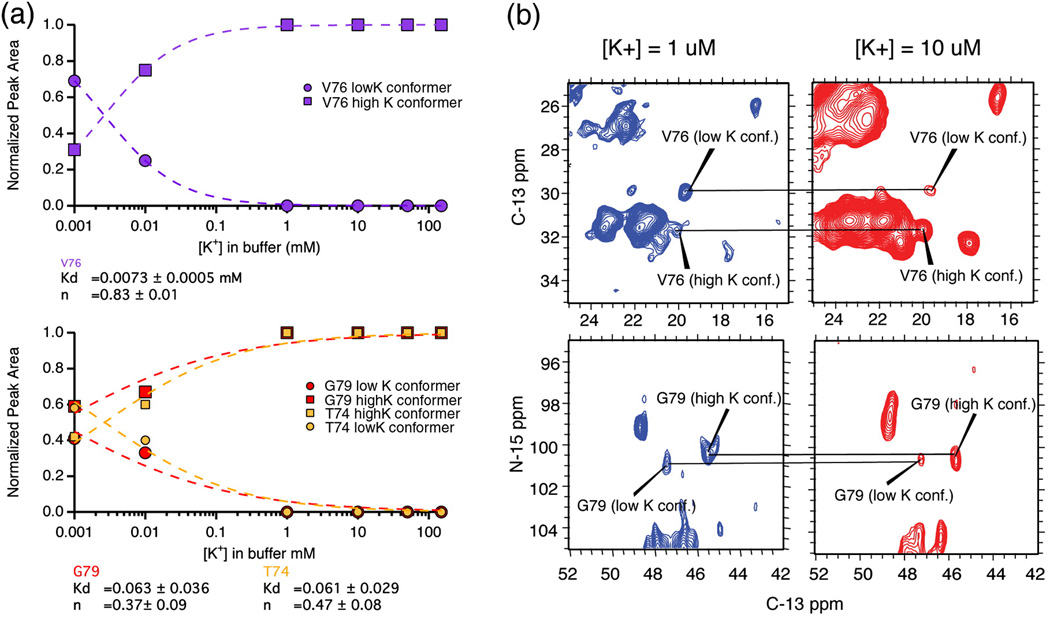
Potassium titration studies suggest strong binding (Kd ∼ 7 µM) for K+ at the V76 site, and slow exchange (< 500 s−1) between the two conformers
Since we observed two states of the filter at different K+ concentrations, we titrated the K+ between 1 µM and 50 mM to identify the ion affinity associated with the structural transition. We measured the chemical shifts of selectivity filter peaks resolved in a series of homonuclear 13C-13C correlation spectra, where the sample was prepared with ambient potassium ion concentration of 1 µM, 10 µM, 1 mM, 10 mM and 150 mM. Figure 4 plots the differences in chemical shifts of six key marker peaks in the selectivity filter as a function of potassium ion concentration in the buffer. The data show that the transition between the two states occurs at a potassium concentration between 1 and 10 µM. Both conformations of the filter were observed in the spectra at [K+] = 1 µM and [K+] = 10 µM. In order to quantify the affinity, we analyzed the relative population of both conformers as a function of potassium. Figure 5B shows homonuclear and heteronuclear spectra of two markers, V76 and G79, at potassium concentrations of 1 µM and 10 µM. At [K+] = 1 µM, the dominant conformation (the low K+ state) appears to be 2-fold more populated than the minor conformation as assessed using peaks associated with V76. At [K+] = 10 µM, the low K+ state becomes the minor conformation and is 4-fold less populated than the major conformation. This implies that the Kd for K+ binding at V76 in the pore is on the order of a few µM. Figure 5A shows the populations of these two states as reported by the NMR signal strength at three different sites: V76, T74 and G79. T74 and G79 are both at the periphery of the filter, while V76 is right in the center. Fitting the populations of the conformers using the Hill equation yielded a Kd of 7.3 µM and a Hill coefficient of 0.83 at the V76 site. When the Hill coefficient was forced to 1, the fit yielded a Kd of 2 µM. Normalization of the signal intensity led to redundancy in the fit for the low K+ populations. The normalization procedure is described in the methods section. The standard formulation of the Hill equation did not fit our data at T74 and G79 well, possibly because these outer sites can bind to potassium, sodium and water while the inner sites bind only to potassium and water. It is also possible that the degree of cooperativity is different between inner and outer sites or that the outer site has a different transition to an unoccupied state. In any case, the conformational transitions at G79 and T74 follow a different pattern compared to those at V76, suggesting that while V76 reports on structure and ion binding inside the filter, the peripheral sites likely report on some other process.
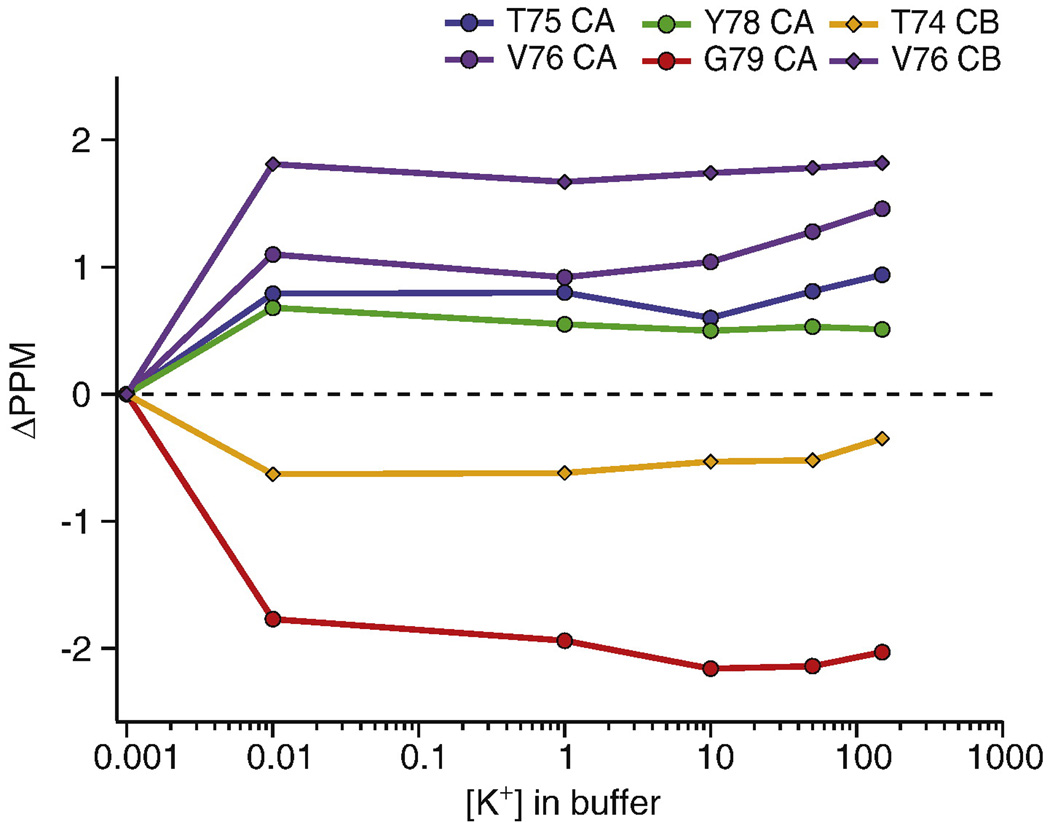
Figure 4.
[K+] titration studies at pH=7.5 show that the transition between the high K+ and the low K+ states of the filter occurs between [K+] = 1 µM and 10 µM. The plot shows the change in the chemical shifts of Cα and Cβ atoms in the filter as a function of the [K+] in the bathing solution. The chemical shifts of marker peaks of the major conformer at [K+] = 10 µM, 1mM, 10mM, 50mM and 150mM are identical within experimental error (0.2 ppm) and significantly differ from those at [K+] = 1 µM. These data show that K+ binds very strongly inside the filter with a Kd on the order of a few µM.
Figure 5.
Panel A plots the normalized peak area of the major and minor conformations of three marker peaks, V76 CB-CG, T74 CA-CB and G79 N-CA, as a function of [K+]. The curves were fit to the Hill equation as described in the methods. This yielded a Kd for K+ binding is 7.3 µM at V76. Solid-state NMR spectra of the same two peaks are shown in the right panel (B). Both C-C (top) and N–C (bottom) correlation spectra reveal a [K+] dependent population distribution between two states. The simultaneous detection of two conformations (a low K+ conformation at ω1 ∼ 30 ppm and the high K+ conformation at ω1 = 32 ppm) suggests that when 1 µM < [K+] < 10 µM, the two states are in slow exchange on the NMR timescale. Based on the chemical shift difference between the two resonances, the upper limit for this rate of exchange is 500 s−1.
Simultaneous detection of two different conformations is the hallmark of slow-exchange on the NMR timescale. When conformational exchange between two discrete states occurs at a rate slower than the difference in the Larmor precession frequencies of nuclei in those two states, both frequencies are detected. The spectra in figure 5A show that we see slow exchange at both the V76 and the G79 sites when 1–10 µM of K+ is present in the buffer solution. The chemical shift difference between the two states ranges from 0.4 ppm at the T74 Cα to 1.8 ppm at the V76 Cβ site. Simulations of the lineshape at both these sites using SPINEVOLUTION34 suggest that the upper limit to the rate of exchange, is 500 s−1. At exchange rates higher than 500 s−1, the simulation suggests that significant line broadening would occur due to exchange. Since there is no observable line broadening at these sites in our spectra, the exchange between the high K+ and the low K+ state is probably slower than 500 s−1 in the bilayer.
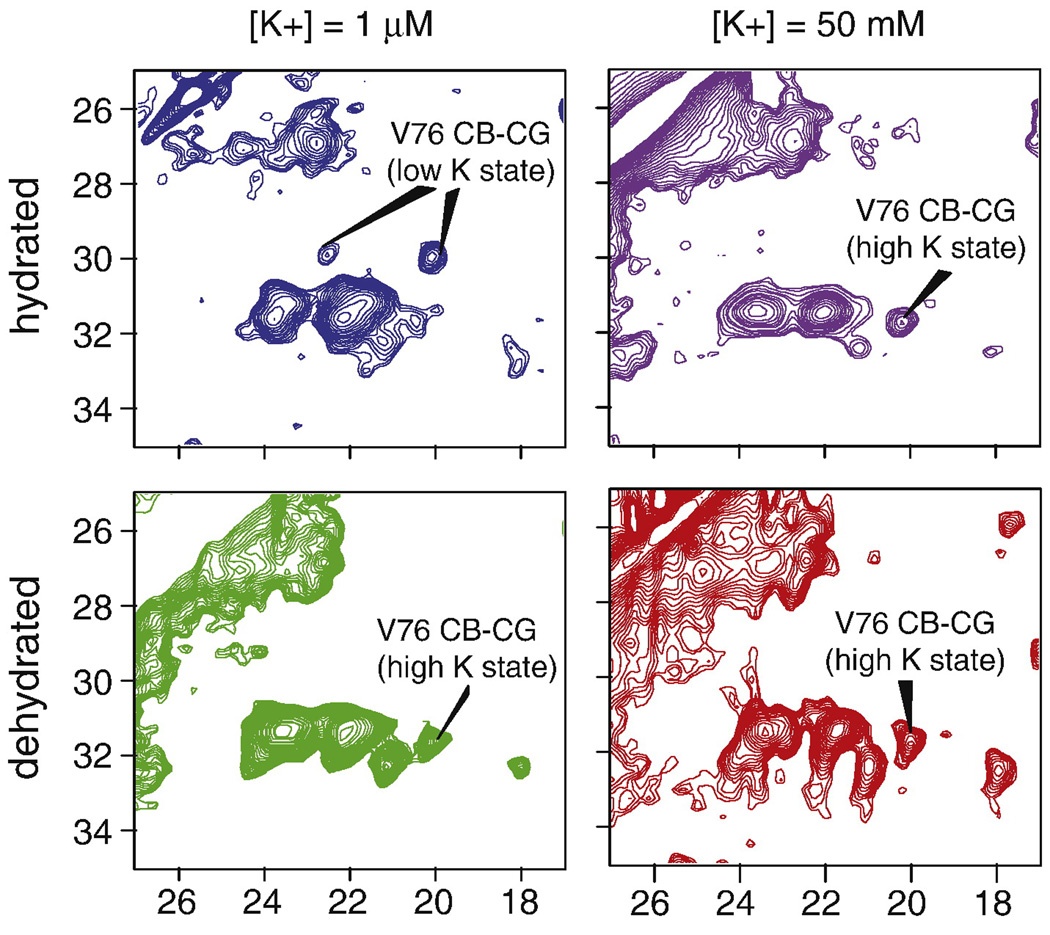
Hydration dependence of the low K+ state
We observed that the population of the collapsed state of the filter was dependent on the hydration level of the sample. This was an important observation because typically the hydration level for protein samples in SSNMR is not closely monitored, and is complicated by the fact that much of the bulk water of the sample collects in the center of the rotor during magic angle spinning. During our experiments, we observed that if the bulk buffer was removed from a low K+ KcsA sample after spinning, all of the characteristic resonance peaks corresponding to the low K+ state disappeared, indicating that it is important for the channel to be in equilibrium with surrounding excess water. When buffer was re-added, the low K+ shifts reappeared. This phenomena was reproducibly observed with two different samples [K+] = 1 µM and [K+] = 10 µM, and on two different NMR instruments. Samples prepared at higher concentrations of potassium did not show the same hydration dependence. Figure 6 shows the effect of hydration on our spectra: The blue and green panels show spectra of the same low-K+ sample ([Na+] = 50 mM, [K+] = 1 µM) first measured in a fully hydrated state and then measured in a dehydrated state. The purple and red panels show spectra of a high K+ sample ([K+] = 50 mM) in a hydrated and dehydrated environment. The signature chemical shifts of the low K+ state are visible only when the sample is fully hydrated. Both X-ray diffraction2 and MD studies35 have suggested that the low K+ state is stabilized by a ring of water-mediated hydrogen bonds around the filter, so it is not surprising that water may be important to stabilize this state. The slow exchange between the high K+ and low K+ states mentioned above is therefore also an exchange between K+ in the pore and the bound water. This process is reversible i.e. a dehydrated low K+ sample, that does not show the signature low K+ NMR shifts can be rehydrated to recover the low K+ shifts.
Figure 6.
13C-13C 2D correlation spectra of KcsA are shown as a function of potassium concentration and hydration level. The low K+ state of the selectivity filter is stabilized at pH=7.5 by the hydration level of the sample. This is illustrated by the appearance of V76 low K+ marker peaks at (ω1, ω2) = (29 ppm, 19.8 ppm) and (29 ppm, 22 ppm) when the sample is bathed in 1 µM [K+] and measured while fully hydrated (blue). When the 1 µM [K+] sample is dehydrated it reconverts into a state that resembles the 50 mM state, as seen by the disappearance of crosspeaks at ω1=29 ppm in the green panel. Samples bathed in 50 mM K+ do not show such hydration dependence. There is no significant change seen in the hydrated (purple) and dehydrated (red) spectra of KcsA at 50 mM K+. The data suggest that the 1 µM K+ state requires a critical amount of water in order to stabilize its structure.
Discussion
Our data reported here show that significant potassium ion concentration dependent changes in the chemical shifts of the selectivity filter residues can be seen at (constant) neutral pH. The low K+ state of the channel is only observed when the ambient potassium ion concentration is less than 10 µM, and observation of this conformer requires that the water-content of the sample be controlled. KcsA has been studied previously using solid-state NMR by the group of Marc Baldus.36; 37; 38 A recent series of solid-state NMR studies of a KcsA-Kv1.3 chimera in soy asolectin bilayers report differences in the chemical shifts of residues in the selectivity filter and the TM2 hinge region when the pH is dropped from 7.5 to 4 and the ambient potassium ion concentration is low,33; 39 but do not report any differences in the channel in the absence of a pH drop. Our results are different from theirs, and significant because we illustrate that ion dependent differences can also be seen at constant neutral pH. We highlight the importance of maintaining the hydration level of the sample in order to observe these functional states. In the presence of 1 µM potassium ion concentration and full hydration, we resolve both conformers of the channel. To the best of our knowledge, this is the first experimental result to report that the bulk water content of solid-state NMR samples can affect the physiological state of a protein. In the specific case of KcsA, the hydration dependence has a precedent in the literature: the crystal structure of the low K+ state (1K4D) suggests that a water-mediated hydrogen bond stabilizes the pore when K+ is limited. A recent liquid state NMR study40 echoes a similar observation based on water-protein NOEs to Val 76, and suggests that inactivation of the channel is mediated by replacing a potassium by a water molecule in the pore. Our observation of hydration-dependent functional states of this system could be related to specific waters that are involved at the active site, or could be a more generalized effect related to overall hydration of the protein or bilayer. It could also be a more macroscopic and indirect effect, in that under low hydration conditions the kinetic and thermodynamic control of local ion concentrations might be altered. This result, together with several recent reports in the literature26; 41; 42, illustrates that the hydration level of the samples is an important variable to consider and control in biophysical studies.
We report a Kd for potassium binding at the V76 site in the pore that is on the order of 2–7 µM and a combined exchange rate (kf + kb) of < 500 s−1 between the K+ bound and deficient states at neutral pH and micromolar concentrations of potassium ion. Chill et al. studied KcsA in SDS micelles11 at 50°C, used the V76 15N-1H peak as a marker, and measured an exchange rate of > 104/sec between the two states at pH = 7.5. Imai et al. conducted similar studies, albeit in DDM micelles at 45°C and measured V76 Cγ1/2 chemical shift changes as a function of potassium concentration. Baker et al. studied KcsA in foscholine micelles at 37°C and pH = 4. They used the Y78 15N-1H peak as a marker and report millisecond-timescale slow exchange between the K+ bound and deficient states.15 Our results from measurements in hydrated bilayers at a sample temperature of ∼10°C are consistent with a slow exchange rate. They provide an atomistic, site-by-site reporter for Kd of ion binding, which is also relatively tight compared to previous measurements made in detergent micelles.
The Kd for potassium binding in the selectivity filter has been measured before by several different techniques. Thermodynamic measurements of ion binding using isothermal titration calorimetry (ITC) report a Kd of ∼0.43 mM for K+ binding.4 Electrophysiological experiments on the KcsA-Kv1.3 chimera report two high affinity binding sites in the pore: an inner site that has a Kd of 6.5 µM and the outer site has a Kd of 0.9 mM. These Kd were obtained by fitting experimentally measured inactivation time constants to a potassium concentration response curve.33 Two different liquid-state NMR measurements of KcsA in detergent micelles report a Kd of ∼3 mM11 and 6.1 mM40 for potassium binding at V76, and crystallographic studies report that the “collapsed” conformation of the filter is seen at K+ concentrations < 5mM.2; 4 Our Kd is significantly lower than that measured by ITC and by solution NMR, but is in very good agreement with that reported by electrophysiology. Our Kd is lower than that measured by ITC because ITC probes the overall process of multiple ions binding at multiple sites in the pore, while we report a single site-specific Kd at V76. The disagreement could also be attributed to differences in sample temperature. Our Kd is in good agreement with the electrophysiology measurements for the inner site33, and it is remarkable that these two completely different techniques converge on similar results. The data indicate that K+ binding at the S2 site is indeed strong, and therefore offer good validation for our assertion of the patency of the hydrated bilayer environments used in these studies.
Presumably due to this strong binding, it is noteworthy that we also observed that washing K+ quantitatively from a KcsA-liposome sample is very difficult. Procedures based on dialysis and centrifugation, resulted in unsurprising ion concentrations in the mother liquor, but proteoliposome pellets typically exhibited higher K+ ion concentrations, approximately consistent with stoichiometries of about 2 ions per channel (details in the supplement). In order to be consistent with the literature, we report Kd in terms of the concentration of K+ in the buffer solution with which the membranes were equilibrated rather than the local concentration in the concentrated proteoliposome pellet.
Measurements of Kd are complicated by the fact that there are multiple ion-binding processes that occur in the pore of KcsA. Each of the four ion binding sites, S1-S4, has in principle a different affinity for potassium, and the Kd at any particular site can also be modulated by the ion-occupancy of its neighbors. Our data at V76 and G79 indicate that there are at least two different kinds of changes that occur in the pore in response to ion concentration. Electrophysiology measurements that report binding affinities probe the outer site, S4, and the inner site, S1, but not S2 and S3. For reasons of spectral resolution, the marker peaks that we used to determine Kd belong to V76. V76 chelates the S2 and S3 sites, which are hypothesized to be the most selective sites for K+ binding in the pore. Unlike S1 and S4, Na+ cannot bind at the octahedral sites of S2 and S3. Thus, it is not surprising that the Kd for potassium binding, when measured via V76, is so low. Our studies demonstrate the potential for solid-state NMR to complement techniques like electrophysiology and ITC and make atom-specific measurements at important buried sites that are inaccessible via these techniques.
Our studies also demonstrate the potential for solid-state NMR to measure key dynamic processes of membrane proteins in a lipid bilayer. The upper limit for the combined exchange rate (kf + kb) for inter-conversion between the K+ bound and deficient states from our data, is 500 s−1. The exchange observed in the NMR might be due to either or both of two related molecular processes. Possibly the protein is undergoing conformational exchange between the two conformations illustrated by crystallography experiments at low and high K+. The association with the crystallography states is supported by our chemical shift measurements, which suggest that at [K+] =1 µM, the chemical shifts of the dominant conformation agree with a low K+ state (1K4D or 2ITC) and those of the minor conformation agree with a high K+ state (1K4C). Also, in one direction of this reaction, the protein is exchanging a bound K+ for a bound Na+ at the outer sites. This exchange rate, at low micromolar concentrations of K+, could be limited by the collision of K+ ions with the filter.43 When [K+]=Kd, then kon=koff and indeed both statements could be true, i.e. the off-rate of the ion being limited by the conformational process and the on-rate being collision limited. In the prior solution NMR studies ion affinities lower than those from electrophysiology are seen, and faster exchange kinetics were observed. The loosening of ion binding might occur due to use of detergent and warmer temperatures. This connected to the disagreement in our rate constants and theirs in that when the available potassium is on the order of the Kd, a looser binding constant indeed would directly lead to faster off kinetics for the ion. Furthermore, at the higher ion concentrations used in that study, ion binding is not expected to be rate limiting.
This conformational exchange of the channel stands as a testimony for the inherent plasticity of the channel. It is important to identify more clearly what physiological processes it is likely associated with. If this conformational exchange indeed has slow kinetics in the bilayer, then it would not be a candidate for involvement in transmission of ions. Indeed the high affinity of the inner sites for K+, and their inability to competitively bind Na+, suggest that there would be a large energetic and kinetic penalty to evacuate both S2 and S3 simultaneously. Conduction and reorganization of water in the pore is known to be fast under osmotic pressure even with low ambient potassium, suggesting that reorganization of water is unlikely to be the rate limiting step.44 In this regard, the secret to the channel’s achieving both selectivity and fast conductivity is that it probably never needs to evacuate both the selective tight binding sites during the “knock-on” mechanism, but rather it exchanges the nearly iso-energetic S2 and S3 occupied states with great facility. The transition to the low K+, sodium-bound, “collapsed” or (1,4) state of the filter would then be an off pathway that is not sampled during conduction. Indeed it has sometimes been alluded to as a prototype for the C-type inactivated state of the channel.35 Channel inactivation occurs on the timescale of seconds, and the selectivity filter is proposed to house a voltage sensitive inactivation gate.13 The C-type inactivation rate constant has been measured by electrophysiology, albeit under conditions that are somewhat different from ours. These measurements report that the inactivation time constant is on the order of 2 s (rate ∼ 0.5/s) under a symmetric potassium concentration of 200 mM, pH = 3 and membrane voltage = −100 mV. The time constant reduces to ∼ 500 ms (rate ∼ 2/s) when the potassium concentration is 5 mM and we expect that since our samples are at potassium concentrations much lower than 5 mM K+, the inactivation rate is likely even faster than 2/s. Based on the discussion above it is plausible that the slow exchange rate we measure reports on conformational exchange that is related to channel inactivation, which, under our sample conditions, occurs at a rate between 2 and 500/s.
Other NMR experiments have attempted to probe the C-type inactivation using increased acidity and by blocking the channel with porphyrin.37 The chemical shift differences that we see in the selectivity filter are consistent with theirs, although they also observe changes in other regions of the protein like the TM2 gate and the pH gate because of their sample conditions. Here, we show that it is possible to site specifically measure the Kd for K+ binding sites inside the pore and to probe conformational dynamics of the channel in a native bilayer using solid-state NMR. Our results are foundational for ensuing studies of channel dynamics for which these conformations can be considered limiting structures.
Conclusions
In this paper, we show the backbone and sidechain NMR assignments of filter residues of full-length KcsA reconstituted into DOPE/DOPS bilayers at [K+] concentrations ranging from 1 µM to 150 mM at pH = 7.5. We show that at least two distinct states of the selectivity filter are observed at a neutral pH of 7.5, one observed in the presence of high K+ ion concentration and the other in the presence of 1–10 µM K+, only if the bilayers were well hydrated. We demonstrate that high hydration level of protein samples can be important for mechanistic studies. We report a Kd for the transition between these two states that is on the order of 2–7 µM but which varies somewhat among the binding sites in the filter. Additionally, we show that in the presence of 1–10 µM K+, the two states exchange on the low millisecond timescale or slower.
Materials and Methods
Protein production
The KcsA gene cloned into a PQE60 vector was over-expressed in E. Coli M15 cells. After transformation, the cells were grown in an LB medium containing 100 mg/L ampicillin and 25mg/L kanamycin at 37 °C shaking at 250 rpm. When an OD600 of 0.8–1 was reached, the cells were pelleted and redissolved an M9 minimal medium45 containing 100 mg/L ampicillin and 25mg/L kanamycin and enriched with 0.5 g/L 15NH4Cl and 3 g/L U-[13C]-glucose. After 30 mins of growing in M9, the cells were induced with 1 mM IPTG and then harvested after 4 hours. The cells were lysed by 3 passages through a French Press. After lysis decyl-B-maltopyranoside (DM, Anatrace #D322) was added to a concentration of 10% w/w to extract membrane bound proteins. Unlysed cells were pelleted by centrifugation and the supernatant was loaded onto Ni-CAM resin column (Sigma #N3158) pre-equilibrated with 50 mM Tris, 150 mM KCl, 10mM DM, pH=7.5. The column was washed with the loading buffer including 50mM imidazole to get rid of impurities. KcsA was eluted using a high imidazole buffer: 50 mM Tris, 150 mM KCl, 10 mM DM, 500 mM imidazole, pH=7.5. The excess imidazole was removed by buffer replacement using an Amicon concentrator with a 10 K membrane. Purified KcsA was characterized on a 4–20% Tris-Tricine SDS gradient gel and by UV absorption. The KcsA tetramer is stable in SDS gels at room temperature, and is detected as a clean band at ∼70 kDa. The published extinction coefficient46 of 33570 M−1cm−1 was used to determine the protein concentration by UV absorption at 280 nm. Generally, 6–8 mg of purified U-[13C]-[15N]-KcsA was obtained per liter of M9 media. The protein was stored at a concentration of 0.5–1 mg/ml in a Tris/KCl buffer (50 mM Tris, 50 mM KCl, 10 mM DM, pH=7.5) at 4° C until needed and was stable for up to 1 year. In order to prepare the [K+] depleted sample, we tried to store the protein in a 50mM Tris/150mM NaCl buffer at pH 7.5, but this lead to severe protein aggregation that has been reported in the literature.47
NMR sample preparation
All of the data presented in this paper were collected on samples of U-[13C]-[15N]-KcsA reconstituted into 9:1 DOPE/DOPS liposomes. DOPE (18:1 PE, 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine, C41H78NO8P, MW 743.547, Avanti # 850725) has a zwitterionic headgroup and DOPS (18:1 PS, 1,2-Dioleoyl-sn-Glycero-3-[Phospho-L-Serine], C42H77NO10PNa, MW 809.518, Avanti # 840035) has an anionic headgroup. Anionic lipids are known to be important for KcsA function.48 DOPE/DOPS mixtures were prepared with a lipid: lipid weight ratio of 9:1. The chloroform was evaporated under nitrogen and the lipids were resuspended in 50 mM KCl, 50 mM Tris, 0.01 mM azide, 10 mM DM, pH=7.5 to a total lipid concentration of 10 mg/ml. The lipid solution was then added to the desired amount of KcsA in DM such that the protein:lipid weight ratio was 1:1. The mixture was then diluted 10X with the relevant dialysis buffer to reduce the detergent concentration and then dialyzed against a 50 mM Tris/KCl buffer at pH=7.5 with varying amounts of KCl and NaCl. A dialysis membrane with a MW cut-off of 12,000–14,000 kDa was used for the dialysis. To prepare the 1 µM K+ state, the NaCl was replaced with ultrapure NaCl. The ionic strength of the dialysis buffer was kept constant across all samples and the K+ content was determined via atomic absorption spectroscopy. The dialysis buffer was replaced 3 times over the course of 4–5 days to remove the detergent and the protein-liposome precipitate was usually seen within the first day. The precipitate was then collected by centrifugation and left in a 90–95% hydration chamber in order to remove excess bulk water. The mass of the samples was monitored and they typically lost a few milligrams of water over several days. The samples were then center-packed into Bruker 4 mm or 3.2 mm rotors. Typically samples contained 5–8 mg of KcsA and 5–8 mg of lipid and had a total volume of 25–30 µl.
Elemental Analysis
In order to confirm the K+ concentration of our samples, we sent the samples to Robertson Microlit Labs in New Jersey, to conduct elemental analysis by atomic absorption spectroscopy. Controls included deionized water, 3 mM KCl solution and 3 mM NaCl solution (for internal calibration). Samples included 20 µl of the 50 mM [K+] and the 1 µM [K+] dialysis buffers.
Spectroscopy
All data in this paper were collected on two spectrometers: A Bruker Avance DRX-750 spectrometer equipped with a 4 mm-HXY probe spinning at 14 kHz and a Bruker Avance-II 900 spectrometer equipped with a 3.2 mm HXY E-free probe spinning at 20 kHz. Typical 90° pulse lengths for 1H were ∼2.5µs on the WB-HXY probes and ∼3 µs on the E-free probe. 13C and 15N 90° pulse lengths were ∼ 5 µs. Marker peaks were identified at 50 mM [K+] and sequentially assigned using heteronuclear 2D and 3D 15N-13C-13C DCP-DARR spectra (NcaCX/NcoCX and NCACX/NCOCX) and 2D 13C-13C homonuclear correlation spectra with a DARR mixing (15ms and 150ms). Typically the 3/2–5/2 DCP49; 50 condition was used for N-CA and N-CO transfers at 14 kHz spinning. At 20 KHz spinning, the 1/2–3/2 condition was used for both N-CA and N-CO transfers. DCP contact times of 6–8 ms were found to be optimal. ∼100 kHz of CW decoupling was used during DCP and 80–100 kHz of SPINAL6451 decoupling was used during acquisition. In order to assign filter residues in the 1 µM [K+] state, heteronuclear 2D NcaCX and NcoCX spectra and 2D 13C-13C homonuclear correlation spectra with a DARR52 mixing (15 ms) and DREAM53 mixing (4.5 ms) were collected for that sample. 2D 13C-13C homonuclear correlation spectra with a DARR mixing (15 ms–25 ms) were collected for samples equilibrated with buffers at [K+] = 1 µM, 10 µM, 1 mM, 10 mM, 50 mM and 150 mM to track the chemical shifts of resolved marker peaks. Typical recycle delays of 3 s were used. A summary of spectra and images of the assignment are provided in the supplementary information. All spectra were collected at a set-temperature of 240 K. Temperature calibrations on our spectrometers have shown that the heating due to MAS is on the order of ∼ 15–18°C for a spinning speed of 14 KHz on a 750 MHz instrument. However, for conductive samples such as ours, the sample heating due to high power RF radiation is close to 20–30 °C during 80–100 KHz H-decoupling for 10–15 ms. This leads us to estimate that although the sample set-temp is 240 K, the actual sample temperature during data acquisition is closer to ∼280 K.
Data Analysis and Simulations
All 13C chemical shifts were referenced externally to the adamantane line at 40.48 ppm. 15N shifts were referenced externally to the a-glycine amine line at 32.06 ppm. Data were processed using TopSpin (Bruker Biospin), NMRPipe54 and SPARKY.55 Typically a Lorentz-to-Gauss apodization was used in both dimensions while processing the data with net matched line broadening of 50–100 Hz as necessary. Chemical shifts were recorded and compared to the shifts predicted by the database programs SHIFTX2 and SPARTA.32 The relative populations of the two conformers used in Figure 5 were obtained by integrating the peaks in SPARKY, and normalizing the integrated peak-area for each conformer to the sum of the integrated areas for both the conformers. This gave us a fractional population of the two states ranging from 0 to 1. The normalized populations were then fit to the Hill Equation below, where θ is the normalized population and n is the Hill coefficient.
In order to get an upper limit on the exchange rate between the two conformers, a simple case of two exchanging 13C nuclei was simulated using SPINEVOLUTION34. The addition of chemical shift anisotropy (CSA) and R2 relaxation did not affect the conclusion that significant line-broadening would be noticeable if the exchange rate was >500 s−1
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Crina Nimigean and members of her lab for helpful meetings and discussions, Dr. Rod Mackinnon for the KcsA plasmid, Dr. Ansgar Siemer and Dr. Yisong Tao for their helpful feedback and Dr. Boris Itin at the NYSBC for all his help during data acquisition. This work is supported by grants from the National Institute of Health.
Abbreviations
- NMR
Nuclear Magnetic Resonance
- FEP
Free Energy Perturbation
- CSA
Chemical Shift Anisotropy
- RMSD
Root Mean Square Difference
- TM2
Transmembrane 2
- DDM
Dodecyl-B-Maltopyranoside
- DM
Decyl-B-Maltopyranoside
- ITC
Isothermal Titration Calorimetry
- OD
Optical Density
- SDS
Sodium Dodecyl Sulfate
- DOPE
1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine
- DOPS
1,2-Dioleoyl-sn-Glycero-3-Phospho-L-Serine
- DCP
Double Cross Polarization
- DARR
Dipole Assisted Rotary Resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doyle D, Cabral J-M, MacKinnon R. The structure of the potassium channel: molecular basis for K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y, Cabral J-M, MacKinnon R. Chemistry of ion coordination and hydration revealed by K+ channel-Fab complex at 2.0A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 3.Ader C, Schneider R, Hornig S, Velisettty P, Vardanyan V, Giller K, Ohmert I, Becker S, Pongs O, Baldus M. Coupling of activation and inactivation gate in a K+ channel: potassium and ligand sensitivity. EMBO J. 2009:1–10. doi: 10.1038/emboj.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lockless S, Zhou M, MacKinnon R. Structural and Thermodynamic Properties of Selective Ion Binding in a K+ channel. PLOS Biology. 2007;5:1079–1088. doi: 10.1371/journal.pbio.0050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux B. Ion Conduction and Selecivity in K+ channels. Annu. Rev. Biophys. Biomol. Struct. 2005;34:153–171. doi: 10.1146/annurev.biophys.34.040204.144655. [DOI] [PubMed] [Google Scholar]
- 6.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional Reconstitution of a Prokaryotic K+ Channel. J. Gen. Phys. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heginbotham L, LeMasurier M, Kolmakova-Partensky L, Miller C. Single Streptomyces lividans K+ Channels: Functinal Asymmetries and Sidedness of Proton Activation. J. Gen. Phys. 114:551–559. doi: 10.1085/jgp.114.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson A, Posson D, Parsa P, Nimigean C. Molecular mechanism of pH sensing in KcsA potassium channels. PNAS. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrapani S, Cordero-Morales J, Perozo E. A Quantitative Description of KcsA Gating II: Single Channel Currents. J. Gen. Phys. 2007;130:479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrapani S, Cordero-Morales J, Perozo E. A Quantitative Description of KcsA Gating 1: Macroscopic currents. J. Gen. Phys. 2007;130:465–478. doi: 10.1085/jgp.200709843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chill J, Louis J, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:685–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chill J, Louis J, Delaglio F, Bax A. Local and Global structure of the monomeric subunit of the potassium channel KcsA probed by NMR. Biochem. Biophys. Acta. 2007;1768:3260–3270. doi: 10.1016/j.bbamem.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordero-Morales J, Cuello L, Perozo E. Voltage dependent gating at the KcsA selectivity filter. Nat Struct. Mol. Bio. 2006;13:319–323. doi: 10.1038/nsmb1070. [DOI] [PubMed] [Google Scholar]
- 14.Cordero-Morales J, Cuello L, Yanxiang Z, Jogini V, Cortes D, Roux B, Perozo E. Molecular determinants of gating at the potassium channel selectivity filter. Nat Struct. Mol. Bio. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 15.Baker K, Tzitzlonis C, Kwiakowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct. Mol. Bio. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabral J-M, Zhou Y, MacKinnon R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 17.Alam A, Jiang Y. Structural Analysis of Ion Selectivity in the NaK channel. Nat Struct. Mol. Bio. 2009;16:35–41. doi: 10.1038/nsmb.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma S, Dubravko S, Rempe SB. K+/Na+ selectivity in K channels and Valinomycin: Over-coordination Versus Cavity-size constraints. J. Mol. Bio. 2008;376:13–22. doi: 10.1016/j.jmb.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bezanilla F, Armstrong CM. Negative conductance caused by entri of sodium and cesium ions into the potassium channels of squid axons. J. Gen. Phys. 1972;60:588–608. doi: 10.1085/jgp.60.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nimigean C, Miller C. Na+ block and permeation in K+ channel of known structure. J. Gen. Phys. 2002;120:323–325. doi: 10.1085/jgp.20028614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neyton J, Miller C. Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high conductance Ca2+ activated K+ channel. J. Gen. Phys. 1988;92:569–596. doi: 10.1085/jgp.92.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernache S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 23.Aqvist J, Luzhkov V. Ion permeation mechanism of the potassium channel. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- 24.Nosokov SY, Bernache S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 25.Krushelnitsky A, deAzevedo E, Linser R, Reif B, Saalwachter K, Reichert D. Direct Observation of Millisecond to Second motions in Proteins by Dipolar CODEX NMR Spectroscopy. J. Am. Chem Soc. 2009;131:12097–12099. doi: 10.1021/ja9038888. [DOI] [PubMed] [Google Scholar]
- 26.Krushelnitsky A, Zinkevich T, Mukhametshina N, Tarasova N, Gogolev Y, Gnezdilov O, Fedotov V, Belton P, Reichert D. 13C and 15N NMR Study of the Hydration Response of T4 Lysozyme and αB-Crystallin Internal Dynamics. J. Phys. Chem. B. 2009;113:10022–10034. doi: 10.1021/jp900337x. [DOI] [PubMed] [Google Scholar]
- 27.McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annu. Rev. Biophys. 2009 doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 28.Lorieau J, McDermott A. Conformational flexibility of a microcrystalline globular protein: Order parameters by solid-state NMR. J. Am. Chem Soc. 2006;128:11505. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]
- 29.Lorieau JL, Day LA, McDermott AE. Conformational dynamics of an intact virus: Order parameters for the coat protein of Pf1 bacteriophage. PNAS. 2008;105:10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson AN, Kim I, T.D. P, Iverson TM, Allen TW, Nimigean CM. Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore. Nat Struct. Mol. Bio. 2009;16:1317–1324. doi: 10.1038/nsmb.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J. Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 33.Seidal K, Etzkorn M, Schneider R, Ader C, Baldus M. Comparative analysis of NMR chemical shift predictions for proteins in the solid phase. Solid State NMR. 2009;35:235–242. doi: 10.1016/j.ssnmr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Veshtort M, Griffin R. SPINEVOLUTION: A powerful tool for the simulation of solid and liquid state NMR experiments. J. Magn. Reson. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Domene C, Furini S. Dynamics, energetics, and selectivity of the low-K+ KcsA channel structure. J. Mol. Bio. 2009;389:637–645. doi: 10.1016/j.jmb.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 36.Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, Becker S, Baldus M. Solid-state NMR Spectroscopy Applied to a chimeric potassium channel in lipid bilayers. J. Am. Chem Soc. 2008;130:7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- 37.Ader C, Schneider R, Hornig S, Velisettty P, Wilson E, Lange A, Giller K, Ohmert I, Martin-Eauclair M, Trauner D, Becker S, Pongs O, Baldus M. A structural link between inactivation and block of a K+ channel. Nat Struct. Mol. Bio. 2008;15:605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 38.Baldus M. Toxin-induced conformational changes in a pottasium channel revealed by solid state NMR. Nature. 2006;440:659–662. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 39.Lange A, Giller K, Pongs O, Becker S, Baldus M. Two-dimensional solid-state NMR applied to a chimeric potassium channel. J. Recep. Sig. Trans. 2006;26:215–240. doi: 10.1080/10799890600932188. [DOI] [PubMed] [Google Scholar]
- 40.Imai S, Masanori O, Takeuchi K, Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc. Nat. Acad. Sci. 2010 doi: 10.1073/pnas.0911270107. PNAS early edition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schenkl S, Portuondo E, Zgrablic G, Chergui M, Suske W, Dolder M, Landau EM, Haacke S. Compositional heterogeneity reflects partial dehydration in three-dimensional crystals of bacteriorhodopsin. J. Mol. Bio. 2003;329:711–719. doi: 10.1016/s0022-2836(03)00508-4. [DOI] [PubMed] [Google Scholar]
- 42.Lusceac S, Vogel MR, Herbers C. 2H and 13C NMR studies on the temperature-dependent water and protein dynamics in hydrated elastin, myoglobin and collagen. Biochem. Biophys. Acta. 2009;1804:41–48. doi: 10.1016/j.bbapap.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Cantor CR, Schimmel PR. Bimolecular Rate Constant for a Diffusion-Controlled Reaction. 1 edit. W.H. Freeman; 1980. Biophysical Chemistry. [Google Scholar]
- 44.Saparov SM, Pohl P. Beyond the diffusion limit: Water flow through the empty bacterial potassium channel. Proc. Nat. Acad. Sci. 2004;101:4805–4809. doi: 10.1073/pnas.0308309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 3rd Revised edition. Cold Spring Harbor Laboratory Press, U.S; 2000. edition. [Google Scholar]
- 46.Gill SC, von Hippel PH. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989;189:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 47.Raja M, Vales E. Effects of sodium chloride on membrane fusion and on the formation of aggregates of potassium channel KcsA in Escherichia coli membrane. Biophys. Chem. 2009;142:46–54. doi: 10.1016/j.bpc.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Marius P, Zagnoni M, Sandison M, East M, Morgan H, Lee AG. Binding of Anionic Lipids to at least Three Nonannular Siters on the Potassium channel KcsA is required for channel opening. Biophys. J. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaefer J, McKay RA, Stejskal EO. Double-cross-polarization NMR of solids. J. Magn. Reson. 1979;34:443–447. [Google Scholar]
- 50.Baldus M, Geurts DG, Hediger S, Meier BH. Efficient15N-13C Polarization Transfer by Adiabatic-Passage Hartmann-Hahn Cross Polarization. J. Magn. Reson. 1996;118:140–144. [Google Scholar]
- 51.Fung BM, Khitrin AK, Ermolaev K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 52.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 53.Verel R, Ernst M, Meier BH. Adiabatic Dipolar Recoupling in Solid-State NMR: The DREAM Scheme. J. Magn. Reson. 2001;150:81–99. doi: 10.1006/jmre.2001.2310. [DOI] [PubMed] [Google Scholar]
- 54.Delaglio F, Grzesiek G, Vuister W, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 55.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.