Abstract
Rationale: Several family-based studies have identified genetic linkage for lung function and airflow obstruction to chromosome 2q.
Objectives: We hypothesized that merging results of high-resolution single nucleotide polymorphism (SNP) mapping in four separate populations would lead to the identification of chronic obstructive pulmonary disease (COPD) susceptibility genes on chromosome 2q.
Methods: Within the chromosome 2q linkage region, 2,843 SNPs were genotyped in 806 COPD cases and 779 control subjects from Norway, and 2,484 SNPs were genotyped in 309 patients with severe COPD from the National Emphysema Treatment Trial and 330 community control subjects. Significant associations from the combined results across the two case-control studies were followed up in 1,839 individuals from 603 families from the International COPD Genetics Network (ICGN) and in 949 individuals from 127 families in the Boston Early-Onset COPD Study.
Measurements and Main Results: Merging the results of the two case-control analyses, 14 of the 790 overlapping SNPs had a combined P < 0.01. Two of these 14 SNPs were consistently associated with COPD in the ICGN families. The association with one SNP, located in the gene XRCC5, was replicated in the Boston Early-Onset COPD Study, with a combined P = 2.51 × 10−5 across the four studies, which remains significant when adjusted for multiple testing (P = 0.02). Genotype imputation confirmed the association with SNPs in XRCC5.
Conclusions: By combining data from COPD genetic association studies conducted in four independent patient samples, we have identified XRCC5, an ATP-dependent DNA helicase, as a potential COPD susceptibility gene.
Keywords: emphysema, genetic linkage, metaanalysis, single nucleotide polymorphism
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Chromosome 2q has been linked to lung function and airflow obstruction in family-based genetics studies. SERPINE2 has been identified as a potential COPD susceptibility gene on chromosome 2q, but there are likely to be additional COPD genes in the region.
What This Study Adds to the Field
Combining results of two case-control and two family-based COPD genetics studies, we have identified XRCC5 as an additional COPD susceptibility gene on chromosome 2q.
Genomewide linkage analysis has been widely used to localize genes involved in complex diseases. Over the past few years, genomewide association studies (GWAS) have become more common in the search for genetic influences on common chronic diseases, including asthma and chronic obstructive pulmonary disease (COPD) (1–3). Both of these genomewide techniques may point to chromosomal regions of interest. Fine mapping, by genotyping a dense panel of single nucleotide polymorphisms (SNPs), may still be required to localize the specific gene or genes of interest. The method used in most GWAS to date has included initial analysis of the genomewide SNP panel, with serial replications of the most significant single SNP or the top few SNPs. However, in many common diseases, GWAS have been performed by multiple research groups in different study populations (4), often with different SNP panels, due to differences in chip manufacturers or versions. Methodologies to effectively combine these data sets will be required for additional gene identification beyond the top SNPs found in each individual study.
Our group has previously reported a genomewide linkage analysis in the Boston Early-Onset COPD Study families, which found significant linkage to a region on chromosome 2q for COPD-related traits (5, 6). A subsequent paper integrated murine and human gene expression data with human SNP genotyping to identify SERPINE2 as a COPD-susceptibility gene on chromosome 2q (7). Using conditional linkage models, SNPs in SERPINE2 were demonstrated to explain some, but not all, of the chromosome 2q linkage signal. In addition, the presumed functional variant or variants in SERPINE2 have yet to be identified. Chromosome 2q has also been linked to lung function traits in a general population sample of 264 individuals in 26 families from Utah (8) and in a Dutch study of 1,183 individuals in 200 families ascertained through a proband with asthma (9). Therefore, we hypothesized that additional COPD-susceptibility genes are located on chromosome 2q and that these genes can be identified by merging SNP-based fine mapping data from multiple COPD study populations. To test this hypothesis, we genotyped SNPs in the chromosome 2q region in four study samples—two case-control and two family-based—and tested for association with COPD and related traits. Results of this study have been reported as an abstract (10).
METHODS
Detailed methods are available in the online supplement.
Study Populations
Subject enrollment and phenotype determination in the National Emphysema Treatment Trial (NETT)-Normative Aging Study (NAS) case-control study, the Boston Early-Onset COPD Study, the Norway case-control study, and the International COPD Genetics Network (ICGN) have been described previously (11–13). NETT cases consisted of 309 non-Hispanic white patients with COPD with severe airflow obstruction from across the United States (14). Three hundred thirty control smokers were derived from the NAS, conducted by the Boston Veterans Affairs Healthcare System (15). The Boston Early-Onset COPD Study included 949 individuals in 127 extended pedigrees recruited through a proband with FEV1 less than 40% predicted at age less than 53 years. In this analysis, subjects analyzed in the Bergen, Norway case-control study were restricted to 806 white COPD cases with FEV1 less than 70% predicted and FEV1/FVC less than 0.7 and 779 control smokers with FEV1 greater than 85% predicted and FEV1/FVC greater than or equal to 0.7. We included 1,839 white individuals from 603 families in the ICGN study, recruited in the United States and Europe. ICGN probands had FEV1 less than 60% predicted with FEV1/FVC less than 90% predicted at age 45 to 65 years. Details of the Lung Health Study subjects are provided in the online supplement. Studies were approved by the relevant institutional review boards, and subjects provided written informed consent.
SNP Genotyping
In NETT-NAS, 2,484 SNPs were genotyped at Brigham and Women's Hospital using Illumina (San Diego, CA) GoldenGate custom SNP panels. We selected SNPs to ensure linkage disequilibrium coverage (16) across the chromosome 2q linkage region (5, 6) based on Phase I data from white subjects in the International HapMap project (17). In the ICGN Study, preliminary linkage analysis identified a region on chromosome 2q to be linked to quantitative computed tomography emphysema traits (18). A set of 2,843 SNPs across this linkage interval was genotyped by GlaxoSmithKline in both the ICGN and Norway studies, using Illumina GoldenGate custom panels. SNPs in the Boston Early-Onset COPD Study were genotyped using Sequenom (San Diego, CA) or TaqMan (Applied Biosystems, Foster City, CA) assays at Brigham and Women's Hospital.
Statistical Analysis
In both the NETT-NAS and Norway case-control studies, SNPs were analyzed initially using the Cochran-Armitage trend test, which corresponds to an additive genetic model without covariate adjustment, using SAS/Genetics (SAS Institute, Cary, NC). In the Boston Early-Onset COPD and ICGN studies, SNPs were tested for association with COPD status, defined as Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2 or greater (19), without covariate adjustment (20). SNPs were analyzed under additive models, using PBAT software (21). Fisher method was used to combine P values of overlapping SNPs across studies (22).
After the initial association analysis was completed, genotype imputation (23) was performed in the NETT-NAS and Norway studies in the region encompassing genes MREG (melanoregulin), PECR (peroxisomal trans-2-enoyl-CoA reductase), TMEM169 (transmembrane protein 169), and XRCC5 (X-ray repair complementing defective repair in Chinese hamster cells 5). Imputed genotypes were analyzed as above.
DNA Sequencing
The exons, intron–exon boundaries, and promoter region of XRCC5 and the neighboring gene PECR were resequenced in 23 probands from the Boston Early-Onset COPD Study and 1 Centre d'Etude du Polymorphisme Humain control subject using dye-labeled dideoxy sequencing reactions and an Applied Biosystems 3100 DNA sequencing machine. SNPs with greater than 5% minor allele frequency identified by sequencing were subsequently genotyped in the NETT-NAS and the Boston Early-Onset COPD Studies using TaqMan or Sequenom assays.
RESULTS
Genetic Association Analysis
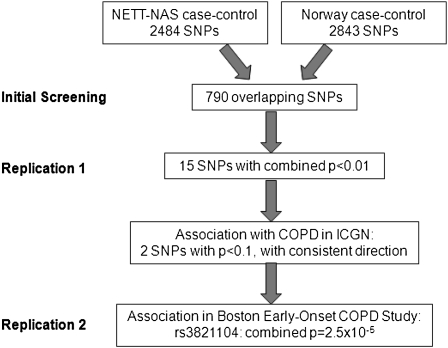
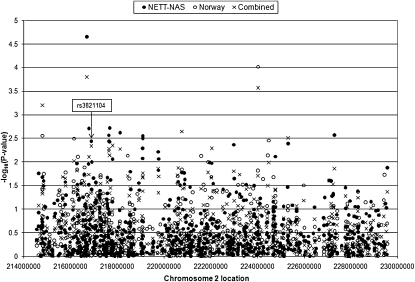
Although all subjects participating in the four studies were current or ex–cigarette smokers, they displayed a range of ages, smoking histories, and levels of lung function (Tables 1 and 2). Data from the four study samples were combined according to the scheme in Figure 1. The two case-control samples were used for screening, with the two family-based samples used for replication studies, to ensure an adequate sample size in the replication stage. Genotyping from the NETT-NAS and Norway case-control studies included 790 overlapping SNPs (Figure 2). Merging the results from these 790 SNPs, there were 15 SNPs with combined P < 0.01 (Table 3). The direction of association was consistent between the two case-control studies for 14 of the 15 SNPs. The 14 SNPs included 3 SNPs from a 200-kb region encompassing four genes—MREG (rs3770564), PECR, TMEM169 (rs828922), and XRCC5 (rs3821104)—and 5 SNPs within 170 kb of each other (rs1011502–rs2068218), located in a gene desert.
TABLE 1.
CHARACTERISTICS OF STUDY SUBJECTS IN CASE-CONTROL STUDIES
| NETT-NAS |
Norway |
|||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| Subjects, n | 309 | 330 | 806 | 779 |
| Age, yr | 67.3 ± 5.7 | 66.6 ± 8.6 | 65.7 ± 10.1 | 55.7 ± 9.8 |
| Male sex | 226 (73.1) | 330 (100) | 476 (59.1) | 384 (49.3) |
| Pack-years of smoking | 68.4 ± 32.2 | 37.8 ± 26.5 | 32.5 ± 19.2 | 18.7 ± 13.3 |
| FEV1 % predicted* | 24.5 ± 6.7 | 97.8 ± 12.8 | 45.7 ± 15.3 | 96.5 ± 8.2 |
| FEV1/FVC ratio* | 0.32 ± 0.06 | 0.80 ± 0.05 | 0.49 ± 0.13 | 0.79 ± 0.04 |
Definition of abbreviations: NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; Norway = Bergen, Norway study.
Values are presented as mean ± SD or n (%).
Because post-bronchodilator spirometry was not available in NAS control subjects, prebronchodilator values are listed in NETT-NAS. Post-bronchodilator values are listed for Norway cases and control subjects.
TABLE 2.
CHARACTERISTICS OF STUDY SUBJECTS IN FAMILY-BASED STUDIES
| Boston Early-Onset COPD |
ICGN |
|||
|---|---|---|---|---|
| Probands | Relatives | Probands | Relatives | |
| Subjects, n | 127 | 822 | 603 | 1,236 |
| Age, yr | 48.1 ± 4.7 | 46.2 ± 18.8 | 58.4 ± 5.4 | 58.4 ± 9.5 |
| Male sex | 32 (25.2) | 364 (44.3) | 360 (59.7) | 632 (51.1) |
| Pack-years of smoking | 38.9 ± 21.9 | 19.0 ± 25.0 | 51.6 ± 26.7 | 40.7 ± 24.9 |
| FEV1 % predicted, post-bronchodilator | 21.9 ± 8.4 | 87.2 ± 20.3 | 36.2 ± 13.0 | 77.4 ± 26.1 |
| FEV1/FVC ratio, post-bronchodilator | 0.31 ± 0.10 | 0.73 ± 0.13 | 0.37 ± 0.12 | 0.61 ± 0.15 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; ICGN = International COPD Genetics Network.
Values are presented as mean ± SD or n (%).
Figure 1.
Study design combining single nucleotide polymorphism (SNP) genotypes from four studies: National Emphysema Treatment Trial (NETT)-Normative Aging Study (NAS case-control study), Norway case-control study, International COPD Genetics Network (ICGN family-based study), and Boston Early-Onset COPD Study family-based study. COPD = chronic obstructive pulmonary disease.
Figure 2.
Genetic association results for 790 overlapping single nucleotide polymorphisms across the chromosome 2q region in the National Emphysema Treatment Trial (NETT)-Normative Aging Study (NAS) and Norway case-control studies. Combined P values were calculated using Fisher method. Chromosomal locations are based on National Center for Biotechnology Information build 35.
TABLE 3.
GENETIC ASSOCIATION WITH CHRONIC OBSTRUCTIVE PULMONARY DISEASE FOR THE 15 SINGLE NUCLEOTIDE POLYMORPHISMS WITH COMBINED P < 0.01 IN THE NATIONAL EMPHYSEMA TREATMENT TRIAL–NORMATIVE AGING STUDY AND NORWAY CASE-CONTROL STUDIES
| NETT-NAS |
Norway |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location* | Nearest Gene 50kb | MAF Case | MAF Control | P Value (Trend) | MAF Case | MAF Control | P Value (Trend) | Consistent Direction | Combined P Value |
| rs6756154 | 214791150 | SPAG16 | 0.39 | 0.33 | 0.021 | 0.41 | 0.36 | 0.0028 | Yes | 0.00063 |
| rs1250217 | 216144130 | FN1 | 0.48 | 0.51 | 0.39 | 0.47 | 0.52 | 0.0032 | Yes | 0.0096 |
| rs3770564 | 216696986 | MREG | 0.21 | 0.13 | 0.000022 | 0.15 | 0.15 | 0.59 | Yes | 0.00016 |
| rs828922 | 216782174 | TMEM169 | 0.34 | 0.26 | 0.0019 | 0.34 | 0.33 | 0.50 | Yes | 0.0077 |
| rs3821104 | 216883352 | XRCC5 | 0.43 | 0.35 | 0.0036 | 0.39 | 0.37 | 0.15 | Yes | 0.0046 |
| rs1011502 | 217633709 | — | 0.33 | 0.42 | 0.0027 | 0.35 | 0.37 | 0.26 | Yes | 0.0058 |
| rs1921987 | 217647419 | — | 0.33 | 0.42 | 0.0036 | 0.34 | 0.37 | 0.17 | Yes | 0.0052 |
| rs2244235 | 217658905 | — | 0.36 | 0.44 | 0.0038 | 0.38 | 0.40 | 0.24 | Yes | 0.0073 |
| rs1882421 | 217675923 | — | 0.30 | 0.22 | 0.0019 | 0.28 | 0.27 | 0.47 | Yes | 0.0072 |
| rs2068218 | 217798954 | — | 0.29 | 0.22 | 0.0087 | 0.26 | 0.24 | 0.060 | Yes | 0.0045 |
| rs1382435 | 218121509 | — | 0.36 | 0.27 | 0.0024 | 0.36 | 0.34 | 0.27 | Yes | 0.0053 |
| rs6728853 | 220757082 | — | 0.26 | 0.32 | 0.017 | 0.28 | 0.32 | 0.015 | Yes | 0.0023 |
| rs1346799 | 222044816 | — | 0.33 | 0.27 | 0.011 | 0.31 | 0.28 | 0.057 | Yes | 0.0051 |
| rs1517635 | 224028620 | — | 0.43 | 0.40 | 0.24 | 0.47 | 0.40 | 0.000096 | Yes | 0.00027 |
| rs1908252 | 225310241 | CUL3 | 0.39 | 0.47 | 0.0040 | 0.50 | 0.47 | 0.086 | No | NA |
Definition of abbreviations: MAF = minor allele frequency; NA = not applicable; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; Norway = Bergen, Norway study; SNP = single nucleotide polymorphism.
Chromosomal locations based on National Center for Biotechnology Information build 35.
The 14 SNPs with combined P < 0.01 across the two case-control studies, with consistent direction, were then examined for association with COPD (defined as GOLD stage 2 or greater) in the family-based ICGN study. Only two SNPs, rs828922 and rs3821104, had P < 0.1 and consistent direction of effect (i.e., one-sided P values) for association with COPD in the ICGN study (Table 4). Because P values are often lower in the initial study (“winner's curse”) (24), we used a less stringent P < 0.1 in the follow-up study to avoid missing relevant associations. SNP rs828922 also showed a trend for association with post-bronchodilator FEV1 (P = 0.076), but was not significantly associated with FEV1/VC ratio (P = 0.21) in the ICGN. However, rs3821104 was significantly associated with FEV1 (P = 0.021) and had a trend for association with FEV1/FVC ratio (P = 0.10) in the ICGN.
TABLE 4.
GENETIC ASSOCIATION WITH CHRONIC OBSTRUCTIVE PULMONARY DISEASE IN CASE-CONTROL STUDIES AND FAMILY-BASED STUDIES FOR TWO SINGLE NUCLEOTIDE POLYMORPHISMS
| NETT-NAS |
Norway |
ICGN |
EOCOPD |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | OR | P Value | OR | P Value | COPD risk | P Value* | COPD risk | P Value* | Combined P Value |
| rs828922 | TMEM169 | 1.44 | 0.0019 | 1.05 | 0.50 | Increased | 0.059 | Increased | 0.083 | 0.0019 |
| rs3821104 | XRCC5 | 1.41 | 0.0036 | 1.11 | 0.15 | Increased | 0.066 | Increased | 0.00066 | 2.51 × 10−5 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; EOCOPD = Boston Early-Onset COPD; ICGN = International COPD Genetics Network; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; Norway = Bergen, Norway study; OR = odds ratio; SNP = single nucleotide polymorphism.
Genetic association with COPD in the NETT-NAS and Norway case-control studies and ICGN and EOCOPD family studies for two SNPs associated with COPD at P < 0.1 with consistent directionality in the ICGN.
One-sided P values in follow-up family-based studies.
These two SNPs were then tested in the Boston Early-Onset COPD Study families. SNP rs3821104 was significantly associated with COPD, defined as GOLD 2 or greater, but rs828922 was not (Table 4). Across all four study populations, the combined P value for rs3821104 for COPD association was 2.51 × 10−5, which remained “region-wide” significant using a conservative Bonferroni correction to adjust for multiple testing of the 790 overlapping SNPs in the NETT-NAS and Norway case-control studies used in the screening phase (adjusted P = 0.020). In all four studies, both SNPs were associated with an increased risk of COPD. In the Boston Early-Onset COPD Study, both SNPs were also significantly associated with post-bronchodilator FEV1/FVC ratio, the phenotype demonstrated to be linked to chromosome 2q in that study (5, 6): rs3821104 P = 0.00058; rs828922 P = 0.043. SNP rs3821104 was also significantly associated with post-bronchodilator FEV1 (P = 0.00073), whereas rs828922 showed a trend for association (P = 0.054) with this trait. However, rs3821104 was not associated with FEV1 or FEV1/FVC ratio in subjects with established COPD in NETT and Norway.
Because the initial linkage was performed in families from the Boston Early-Onset COPD Study, who were recruited through probands with COPD at a young age, we performed an age-stratified analysis in the two case-control studies, using an a priori cutoff of 65 years old. In the Norway study, rs3821104 had a lower P value for association with COPD in subjects less than 65 years old (odds ratio [OR], 1.20; P = 0.052) than in subjects 65 years and older (OR, 1.11; P = 0.44), but rs828922 did not show stronger association in younger subjects. In NETT-NAS, the effect of rs3821104 was similar in subjects less than 65 years old (OR, 1.48; P = 0.050) and in subjects 65 and older (OR, 1.41; P = 0.022). The sample sizes in the stratified analyses in both studies were limited, especially in NETT-NAS (Norway age <65 yr: 378 cases, 626 control subjects; NETT-NAS age <65 yr: 83 cases, 128 control subjects). The effect of rs828922 was actually greater in the older subjects in NETT-NAS (age ≥65 yr: OR, 1.64; P = 0.0020; age <65 yr: OR, 1.22; P = 0.36). Age-stratified analyses were not performed in the ICGN or Boston Early-Onset COPD studies, because subjects tended to be younger than in the two case-control studies (Table 2) based on the enrollment criteria for probands in both family-based studies.
To account for cigarette smoking, the major environmental risk factor for COPD, we performed additional analyses adjusted for pack-years of smoking. In the Norway study, the effect estimates for rs828922 and rs3821104 did not change when adjusted for pack-years in all subjects. However, the association with rs3821104 in subjects less than 65 years of age was strengthened in the adjusted model (OR, 1.26; P = 0.027). In the NETT-NAS study, associations with both SNPs were attenuated in the adjusted models (rs828922: OR, 1.34; P = 0.034; rs3821104: OR, 1.27; P = 0.072). There was no evidence of SNP by pack-years interaction in either case-control sample. Because all subjects in NETT-NAS and Norway were current or former smokers, we could not test for an interaction with ever-smoking status. In the ICGN families, evidence of association for rs828922 was diminished in a model adjusted for packs (P = 0.23) but was similar for rs3821104 in the adjusted model (P = 0.071). In the Boston Early-Onset COPD study, models were additionally adjusted for ever-smoking status, to account for nonsmoking relatives. The P value for association with rs828922 was unchanged (P = 0.18), whereas the P value for rs3821104 was attenuated, although still significant (P = 0.0080). Across the four populations, rs3821104 remained significantly associated in an analysis of the more homogenous phenotype of severe COPD, defined as GOLD stage 3 or greater (combined P = 4.3 × 10−6; see Table E2 in the online supplement).
We tested whether rs3821104 was a modifier of lung function in 5,606 subjects with mild-to-moderate COPD participating in the Lung Health Study (25). Based on regression models adjusted for age, sex, and pack-years of smoking, rs3821104 was not associated with quantitative values for baseline FEV1 or FEV1/FVC ratio. There was also no association in the comparison between individuals with fast versus slow decline in FEV1 over the 5-year course of the study (minor allele frequency 0.41 in 603 fast decliners vs. 0.43 in 581 slow decliners; allele OR = 0.92).
Examining the full set of 2,484 SNPs genotyped in NETT-NAS, no SNP was significant when adjusted for multiple testing (P < 2.0 × 10−5). SNP rs3770564 was nearly significant (P = 2.2 × 10−5) in NETT-NAS (Table 3), but was not significant in Norway or ICGN. In the full set of 2,843 SNPs in Norway, no SNP was significant after multiple testing correction. The SNP with the lowest P value (rs1517635, P = 9.6 × 10−5) was not significant in NETT-NAS or ICGN (Table 3).
Genotype Imputation
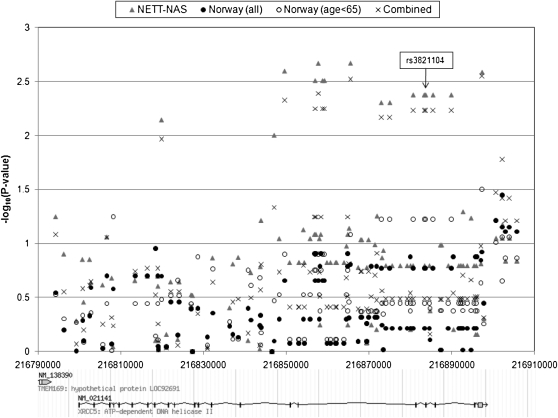
In the 285-kb interval encompassing genes MREG, PECR, TMEM169, and XRCC5, 81 SNPs had been genotyped in the NETT-NAS study and 51 SNPs had been genotyped in the Norway study. In the region, 29 SNPs overlapped between the two case-control studies. Phase II of the HapMap included 303 SNPs in the region that had been genotyped in whites (CEU). Using the HapMap genotypes as a reference, genotypes in NETT-NAS and Norway studies were imputed for the ungenotyped loci of the 303 SNPs. Forty-seven SNPs failed imputation in NETT-NAS and 87 failed in Norway, due to either minor allele frequency less than or equal to 0.001, a P value ≤ 0.001 for deviation from Hardy-Weinberg Equilibrium, or an MACH quality score less than 0.9. Nine SNPs had P < 0.1 for association with COPD in both the NETT-NAS study and in one of two Norway study analyses—an analysis in all subjects or an analysis restricted to subjects less than age 65 (Table 5); eight of the nine SNPs had a consistent direction of effect. The eight SNPs are located in or near XRCC5 (Figure 3). The eight SNPs are in the same haplotype block as rs3821104 (Figures E1 and E2); rs207936 (intronic) is in high linkage disequilibrium (LD) with rs3821104 (NETT-NAS r2 = 0.83; Norway r2 = 0.86) and the other SNPs are in complete LD with rs3821104. These eight SNPs encompass three exons and the corresponding intronic regions in XRCC5.
TABLE 5.
GENETIC ASSOCIATION WITH COPD FOR SINGLE NUCLEOTIDE POLYMORPHISMS FROM THE 285-KB INTERVAL ENCOMPASSING GENES MREG, PECR, TMEM169, AND XRCC5
| NETT-NAS |
Norway |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location* | Gene | Assay | MAF Case | MAF Control | P Value | Assay | MAF Case | MAF Control | P Value, All Subjects | P Value, Age <65 | Consistent Direction |
| rs828920 | 216780841 | TMEM169 | G | 0.092 | 0.13 | 0.023 | G | 0.10 | 0.086 | 0.070 | 0.10 | No |
| rs207936 | 216865539 | XRCC5 | I | 0.39 | 0.31 | 0.0021 | I | 0.36 | 0.33 | 0.16 | 0.082 | Yes |
| rs3770498 | 216872966 | XRCC5 | I | 0.43 | 0.35 | 0.0049 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs6704622 | 216875041 | XRCC5 | I | 0.43 | 0.35 | 0.0049 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs12470053 | 216880677 | XRCC5 | I | 0.43 | 0.35 | 0.0042 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs3821104 | 216883352 | XRCC5 | G | 0.43 | 0.35 | 0.0042 | G | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs3770492 | 216883696 | XRCC5 | I | 0.43 | 0.35 | 0.0042 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs4674066 | 216885470 | XRCC5 | G | 0.43 | 0.35 | 0.0042 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
| rs7573191 | 216889870 | XRCC5 | I | 0.43 | 0.35 | 0.0042 | I | 0.39 | 0.37 | 0.17 | 0.060 | Yes |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; G = genotyped; I = imputed; MAF = minor allele frequency; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; Norway = Bergen, Norway study; SNP = single nucleotide polymorphism.
Genetic association with COPD for SNPs from the 285-kb interval encompassing genes MREG, PECR, TMEM169, and XRCC5 with P < 0.1 in the NETT-NAS and in at least one Norway case-control analysis (all subjects or restricted to age <65 yr), including imputed genotypes for ungenotyped SNPs and subjects with missing genotypes.
Chromosomal locations based on National Center for Biotechnology Information build 35.
Figure 3.
Genetic association results for genotyped and imputed single nucleotide polymorphisms in gene XRCC5 in the National Emphysema Treatment Trial (NETT)-Normative Aging Study (NAS) and Norway case-control studies (in all subjects and limited to subjects <65 yr old). Combined P values were calculated using Fisher method. Chromosomal locations are based on National Center for Biotechnology Information build 35.
DNA Sequencing
We resequenced the 21 exons, the intron–exon boundaries, and the promoter region of XRCC5 and the three exons, the intron–exon boundaries, and the promoter region of the nearby gene PECR in 23 probands from the Boston Early-Onset COPD Study and 1 Centre d'Etude du Polymorphisme Humain control sample. Although TMEM169 is located between XRCC5 and PECR, we did not sequence this gene, which codes for hypothetical protein LOC92691. In XRCC5, we identified 21 SNPs with minor allele frequency greater than 5%. Only 2 of the 21 SNPs had not been previously identified in dbSNP build 123. The only common coding SNP identified led to a synonymous amino acid substitution Thr524Thr (rs207906). This SNP was genotyped and was not significantly associated with COPD in the NETT-NAS study, the Norway case-control study, or the ICGN families, but was significant in the Boston Early-Onset COPD Study (P = 0.0053). One other SNP identified by sequencing (rs12470053, intronic), which was not included in the NETT-NAS Illumina panel, was found to be significantly associated with COPD in NETT-NAS (Table 5). This SNP was also significant in the Boston Early-Onset COPD Study (P = 0.037).
In PECR, we identified eight SNPs with minor allele frequency greater than 5%, two of which were nonsynonymous variants, Phe297Leu and Glu149Lys. Phe297Leu (rs9288513) was significantly associated with COPD in the Norway case-control study (P = 0.029) but was not significant in the other three populations. Glu149Lys (rs1429148) showed a trend for association in Norway (P = 0.093). It was not significant in NETT-NAS or in the ICGN families and was not tested in the Boston Early-Onset COPD Study.
SERPINE2
SERPINE2 had been previously identified as a COPD susceptibility gene in the chromosome 2q linkage region, based on genotype data from the Boston Early-Onset COPD Study and the NETT-NAS case-control study (7). However, the current analysis used independent genotyping data from NETT-NAS, with a smaller set of NAS control subjects genotyped in the current study (330 vs. 441). In a subsequent paper, the association with SERPINE2 was replicated in the ICGN families and in the Norway case-control study (12). The current analysis also includes a smaller sample of both cases and control subjects from Norway, based on more stringent phenotype definitions (FEV1 <70% predicted in cases; FEV1>85% predicted in control subjects). From the 790 common SNPs across the NETT-NAS Illumina genotyping and the Norway case-control study, 15 SNPs mapped to the SERPINE2 gene (Table E1). Several SNPs had P < 0.1 in either sample, but none of the 15 SNPs was significantly associated across both case-control studies and none had a combined P < 0.01, the criterion used to identify SNPs from the combination of the two case-control screening samples (Figure 1).
DISCUSSION
We combined the data from two COPD case-control fine-mapping studies on chromosome 2q as an initial screening step and validated the findings in two family-based studies of COPD. The multistudy fine-mapping strategy identified XRCC5 as a potential COPD-susceptibility gene. Although SNP rs3821104 was not significant in all of the cohorts individually, the direction of effect was consistent in the two case-control and two family-based studies, and the combined P value remained significant even when adjusted for multiple testing. Associations with XRCC5 were more pronounced in stratified analyses of younger subjects with COPD, which corresponds to the initial linkage findings in the Boston Early-Onset COPD Study, which recruited probands with severe airflow obstruction at an early age (5, 6). Genotype imputation pointed to a 25-kb region of XRCC5 most strongly associated, although the specific functional variant or variants have yet to be identified, despite resequencing of the exonic regions of XRCC5.
The product of XRCC5 is an ATP-dependent DNA helicase involved in DNA double-strand break repair and immunoglobulin V(D)J rearrangement (26). XRCC5 is also known as Ku80 or Ku86. There are several potential mechanisms for the role of XRCC5 in the development of COPD, specifically early-onset COPD. Pulmonary emphysema has been described as an aging-related phenomenon (27, 28). For example, mice deficient in the antiaging gene klotho develop emphysema among other manifestations of early senescence (29). Ku86−/− mice also develop early aging (30), although the lungs of these mice have not been specifically examined for emphysema (P. Hasty, personal communication).
Ku80/86 has been identified as an autoantigen in systemic lupus erythematosus and other autoimmune diseases (31). Several groups have proposed an autoimmune component to COPD (32–35). These studies have identified antiendothelial cell and antielastin antibodies, but not antinuclear antibodies that are relevant in systemic lupus erythematosus and other rheumatic diseases. However, an older study did find a higher incidence of low-titer antinuclear antibody positivity in patients with chronic bronchitis compared with control subjects (36).
In mouse lung epithelial cell lines, Maeda and colleagues demonstrated that Ku80 coimmunoprecipitated with thyroid transcription factor-1 and poly(ADP-ribose) polymerase-2 (37). This complex activated the surfactant protein-B (Sftpb) promoter in mouse lung epithelial cells and in Hela cells, but not the other surfactant protein genes in the murine cell line. Our group and others have found association between variants in SFTPB and COPD in human populations (11, 38, 39).
Despite the previously published association with SERPINE2 and subsequent replication study, which used the same study populations as the current analysis (7, 12), we did not identify SERPINE2 based on the screening methods used in the current analysis. Several factors may explain this discrepancy, including differences in sample sizes, especially regarding the NETT-NAS analysis. Both of the prior papers used a family-based study (Boston Early-Onset COPD Study and ICGN, respectively) as the initial screening sample, with follow-up in the case-control samples (NETT-NAS and Norway), whereas the current study used the opposite approach of screening in the two case-control samples and serial replication in the two family-based studies. Moreover, the strongest associations for SERPINE2 SNPs in the Norway study were with quantitative spirometric traits, and not COPD status, which was the main phenotype used in the present analysis. The differences in results demonstrate that genetic association studies of carefully selected candidate genes— including multiple SNPs in the gene and additional quantitative as well as qualitative phenotypes—may provide different insights than regional fine mapping or even GWAS. In addition, a case-control study from the United Kingdom failed to demonstrate association with SERPINE2 (40). This lack of replication is common throughout complex trait genetics.
The linkage regions from the Boston Early-Onset COPD Study and the ICGN Study overlapped, but did not correspond exactly at the margins. SNP selection strategies were based on different tagging approaches in the NETT-NAS and the Norway studies. Genotyping was conducted independently, although both studies used the Illumina GoldenGate genotyping platform. Therefore, the fine mapping SNP sets are not the same in the two case-control studies used in the initial screening step. This is a limitation to our analysis. However, we were able to use the overlapping set of 790 SNPs to identify significant associations. Genotype imputation has been proposed as a method to combine data from different SNP sets, for example from different genomewide SNP chips (41). However, imputed genotypes are clearly less accurate than direct SNP genotyping (42), as evidenced by the large number of SNPs that failed imputation in our two case-control cohorts. Therefore, we elected to use SNP imputation only in a limited region as a confirmatory step, instead of imputing genotypes across the entire linkage region.
The differences in phenotype definitions across the study samples, specifically regarding subject ages and COPD severity, is a potential limitation to our study, which may have contributed to the heterogeneity of the association across the included populations. The associations with XRCC5 were more prominent in younger individuals, which is not surprising given the initial linkage in families recruited through a proband with severe, early-onset COPD. However, we did find significant evidence of association in subjects of older ages in the NETT-NAS Study. The family members in both the Boston Early-Onset COPD Study and the ICGN Study represent a broader range of ages and lung function (Table 2). Although possibly more important in severe early-onset COPD, the association with XRCC5 may be generalizable to subjects with a wider range of COPD severity. Although XRCC5 may be a COPD-susceptibility gene, we did not find evidence that XRCC5 modified lung function in subjects with established COPD in NETT, Norway, or the Lung Health Study. As with all complex traits, additional replication studies will be required to confirm the role of XRCC5 variants in COPD susceptibility. Studies in other racial groups may also be important, given the variation in minor allele frequency of SNP rs3821104 across racial groups in the HapMap project. In addition, functional studies will be required to determine the potential impact of XRCC5 variants on COPD pathophysiology.
Supplementary Material
Acknowledgments
NETT Genetics Ancillary Study Co-Investigators: Joshua Benditt, Gerard Criner, Malcolm DeCamp, Philip Diaz, Mark Ginsburg, Larry Kaiser, Marcia Katz, Mark Krasna, Neil MacIntyre, Barry Make, Rob McKenna, Fernando Martinez, Zab Mosenifar, Andrew Ries, Paul Scanlon, Frank Sciurba, and James Utz. Lung Health Study, University of Utah: Richard Kanner.
Supported by National Institutes of Health grants HL080242, HL094635, HL075478, HL084323, P01 HL083069, U01 HL065899, and P01 HL072903, and a grant from the Alpha-1 Foundation. The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119), Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), Boston, MA. The International COPD Genetics Network and the Bergen Norway COPD study are supported by GlaxoSmithKline. The Lung Health Study (LHS) is conducted and supported by the NHLBI in collaboration with the LHS Study Investigators.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200910-1586OC on May 12, 2010
Author Disclosure: C.P.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.G.P. was a full-time employee of GlaxoSmithKline until September 2009, is a full-time employee of Roche Pharmaceuticals since September 2009, and owns stock in GlaxoSmithKline. G.Z. is a full-time employee of GlaxoSmithKline. D.A.L. received $10,001–$50,000 from GlaxoSmithKline, $1,001–$5,000 from Novartis, up to $1,000 from Genzyme, and up to $1,000 from Amicus in consultancy fees; $10,001–$50,000 from GlaxoSmithKline, $5,001–$10,000 from Talecris, and $1,001–$5,000 from Thorax in advisory board fees; $1,001–$5,000 from GlaxoSmithKline, up to $1,000 from AstraZeneca, and $1,001–$5,000 from Boehringer Ingelheim in lecture fees; and more than $100,001 from GlaxoSmithKline and more than $100,001 from Merck Sharp & Dohme in industry-sponsored grants. P.B. received $1,001–$5,000 from GlaxoSmithKline and $1,001–$5,000 from AstraZeneca in lecture fees, and $10,001–$50,000 from GlaxoSmithKline in industry-sponsored grants as a principal investigator. A.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.L.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J.K. holds a patent from the Massachusetts Institute of Technology, US Patent 6703228, “Methods and products related to genotyping and DNA analysis,” receiving $0 in past 3 years. R.L. received more than $100,001 from the NIH in sponsored grants. A.A.L. received up to $1,000 from UpToDate in royalties as an author for an online reference. D.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.J.R. holds a patent from Brigham and Women's Hospital for software algorithm for CT scan analysis submitted (no financial benefit) and up to $1,000 from UpToDate in royalties for serving as an author of two chapters in online reference text. A.A. received $5,001–$10,000 from GlaxoSmithKline, $5,001–$10,000 from Almirall, and $5,001–$10,000 from Nycomed in advisory board fees; $5,001–$10,000 from GlaxoSmithKline, $5,001–$10,000 from AstraZeneca, and $5,001–$10,000 from Almirall in lecture fees; more than $100,001 from GlaxoSmithKline, $10,001–$50,000 from Pfizer, $5,001–$10,000 from Boehringer Ingelheim, and more than $100,001 from Almirall in industry-sponsored grants. P.M.A.C. received $5,001–$10,000 from Pfizer, $1,001–$5,000 from AstraZeneca, and $5,001–$10,000 from Chiesi in consultancy fees; $5,001–$10,000 from GlaxoSmithKline in advisory board fees; $1,001–$5,000 from Altana, and $10,001–$50,000 from GlaxoSmithKline in lecture fees; and $10,001–$50,000 from Nycomed, $50,001-$100,000 from GlaxoSmithKline, and $10,001-$50,000 from Boehringer Ingelheim in industry-sponsored grants. C.F.D. received $5,001–$10,000 from Pfizer and $5,001–$10,000 from Boehringer Ingelhiem in advisory board fees; $1,001–$5,000 from GlaxoSmithKline, $1,001–$5,000 from Boehringer Ingelheim, and $1,001–$5,000 from Chiesi in lecture fees; and up to $1,000 from Hodder Arnold in royalties. R.D.L. received $1,001–$5,000 from GlaxoSmithKline, $1,001–$5,000 from Actelion, $1,001–$5,000 from Pfizer, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from Nycomed, and $1,001–$5,000 from Lilly in advisory board fees; and $5,001–$10,000 from GlaxoSmithKline and $1,001–$5,000 from Actelion in lecture fees. B.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.D.P. received $1,001–$5,000 from Talecris Biotherapeutics for serving on an advisory board, more than $100,001 from GlaxoSmithKline in industry-sponsored grants for a study of CT techniques to quantify COPD phenotypes, and more than $100,001 from Merck in industry-sponsored grants for measurement of gene expression in COPD lung; and more than $100,001 from AllerGen NCE for the genetics of asthma. S.I.R. received $1,001–$5,000 from Able Associates, up to $1,000 from Adelphi Research, $10,001–$50,000 from Almirall/Prescott, $1,001–$5,000 from APT Pharma/Britnall, $1,001–$5,000 from Aradigm, $1,001–$5,000 from AstraZeneca, $5,001–$10,000 from Boehringer Ingelheim, up to $1,000 from Chiesi, up to $1,000 from Common Health, up to $1,000 from Consult Complete, $1,001–$5,000 from COPDForum, up to $1,000 from Data Monitor, up to $1,000 from Decision Resources, up to $1,000 from Defined Health, $1,001–$5,000 from Dey, up to $1,000 from Dunn Group, up to $1,000 from Eaton Associates, up to $1,000 from Equinox, up to $1,000 from Gerson, $10,001–$50,000 from GlaxoSmithKline, up to $1,000 from Infomed, up to $1,000 from KOL Connection, up to $1,000 from M. Pankove, up to $1,000 from MedaCorp, up to $1,000 from MDRx Financial, up to $1,000 from Mpex, $10,001–$50,000 from Novartis, $10,001–$50,000 from Nycomed, $1,001–$5,000 from Oriel Therapeutics, $1,001–$5,000 from Otsuka, up to $1,000 from Pennside Partners, $5,001–$10,000 from Pfizer (Varenicline), up to $1,000 from PharmaVentures, $1,001–$5,000 from Pharmaxis, up to $1,000 from Price Waterhouse, up to $1,000 from Propagate, up to $1,000 from Pulmatrix, up to $1,000 from Reckner Associates, up to $1,000 from Recruiting Resources, $1,001–$5,000 from Roche, up to $1,000 from Schlesinger Medical, up to $1,000 from Scimed, up to $1,000 from Sudler and Hennessey, $1,001–$5,000 from TargeGen, $1,001–$5,000 from Theravance, $1,001–$5,000 from UBS, $1,001–$5,000 from Uptake Medical, and $,5001–$10,000 from Vantage Point Mgmt in consultancy advisory board fees; $10,001–$50,000 from AstraZeneca, $5,001–$10,000 from Boehringer Ingelheim, $10,001–$50,000 from Creative Educational Concept, $5,001–$10,000 from the France Foundation, $1,001–$5,000 from Information TV, $1,001–$5,000 from the Network for Continuing Ed, $10,001–$50,000 from Novartis, $1,001–$5,000 from Pfizer, and $1,001–$5,000 from SOMA in lecture fees; and $50,001–$100,000 from AstraZeneca, $50,001–$100,000 from Biomarck, $50,001–$100,000 from Centocor, $50,001–$100,000 from Mpex, $50,001–$100,000 from Nabi, $50,001–$100,000 from Novartis, and $50,001–$100,000 from Otsuka in industry-sponsored grants. S.I.R. filed provisional patent applications from the University of Nebraska Medical Center covering microRNA applications for the treatment of diseases (currently marketing to various commercial entities). S.I.R. received funding from RJ Reynolds to evaluate the effect of a harm reduction product in normal smokers (1996) and in subjects with chronic bronchitis (1999) and to assess the effect of smoking cessation on lower respiratory tract inflammation (2000); participated in a Philip Morris multicenter study to assess biomarkers of smoke exposure (2002); received funding for a clinical trial from the Institute for Science and Health (2005), which receives support from the tobacco industry, to evaluate biomarkers in exhaled breath associated with smoking cessation and reduction. This study was supplemented with funding from Lorillard and RJ Reynolds. S.I.R. received a grant from the Philip Morris External Research Program (2005) to assess the impact of cigarette smoking on circulating stem cells in the mouse. S.I.R. has consulted with RJ Reynolds on the topic of harm reduction until 2007, but did not receive personal remuneration for this. There are no active tobacco-industry funded projects. All ties with tobacco industry companies and entities supported by tobacco companies were terminated in 2007. J.V.'s spouse/life partner was a full-time employee of AstraZeneca until June 2009. J.V. received $10,001–$50,000 from GlaxoSmithKline, $1,001–$5,000 from Boehringer Ingelheim, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from Nycomed, and $1,001–$5,000 from Hoffman LaRoche in consultancy fees; $5,001–$10,000 from GlaxoSmithKline, $5,001–$10,000 from Boehringer-Ingelheim, $5,001–$10,000 from AstraZeneca, $5,001–$10,000 from Nycomed, and $1,001–$5,000 from Talecris in lecture fees; and more than $100,001 from GlaxoSmithKline in industry-sponsored grants for the ECLIPSE study and HP-He substudy. E.F.M.W. received $1,001–$5,000 from Nycomed in advisory board fees; up to $1,000 from GlaxoSmithKline, up to $1,000 from AstraZeneca, and up to $1,000 from Novartis in lecture fees; and more than $100,001 from GlaxoSmithKline and more than $100,001 from AstraZeneca in industry-sponsored grants. M.B.S. received $50,001–$100,000 from Actelion, $50,001–$100,000 from Gilead, $50,001–$100,000 from InterMune, and $50,001–$100,000 from Centocor in industry-sponsored grants as a site investigator for a clinical trial. H.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.K.S. received $10,001–$50,000 from GlaxoSmithKline and $10,001–$50,000 from AstraZeneca in consultancy fees, $1,001–$5,000 from GlaxoSmithKline, $5,001–$10,000 from AstraZeneca, and $1,001–$5,000 from Bayer in lecture fees, and more than $100,001 from GlaxoSmithKline in industry-sponsored grants.
References
- 1.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–473. [DOI] [PubMed] [Google Scholar]
- 2.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 2008;358:1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topol EJ, Murray SS, Frazer KA. The genomics gold rush. JAMA 2007;298:218–221. [DOI] [PubMed] [Google Scholar]
- 5.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002;70:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 7.Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra A, Peiffer AP, Ryujin DT, Elsner T, Kanner RE, Leppert MF, Hasstedt SJ. Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med 2003;168:556–561. [DOI] [PubMed] [Google Scholar]
- 9.Postma DS, Meyers DA, Jongepier H, Howard TD, Koppelman GH, Bleecker ER. Genomewide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med 2005;172:446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh CP, Pillai SG, Zhu G, Lomas DA, Bakke P, Gulsvik A, DeMeo DL, Litonjua AA, Reilly JJ, Silverman EK, et al. Multi-study fine mapping of a COPD susceptibility locus on chromosome 2q [abstract]. Am J Respir Crit Care Med 2008;177:A907. [Google Scholar]
- 11.Hersh CP, Demeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol 2005;33:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173. [DOI] [PubMed] [Google Scholar]
- 13.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, Donner CF, Make BJ, Muller NL, Rennard SI, Vestbo J, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:500–505. [DOI] [PubMed] [Google Scholar]
- 14.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 15.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Dev 1972;3:5–17. [Google Scholar]
- 16.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004;74:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 18.Silverman EK, Lake SL, Lomas DA, Rennard SI, Pare PD, Calverley PM, Vestbo J, Wouters EF, Agusti AG, Donner CF, et al. Linkage analysis of quantitative emphysema phenotypes in chronic obstructive pulmonary disease [abstract]. Proc Am Thorac Soc 2006;3:A620. [Google Scholar]
- 19.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 2001;163:1256–1276. [DOI] [PubMed] [Google Scholar]
- 20.Poisson LM, Rybicki BA, Coon SW, Barnholtz-Sloan JS, Chase GA. Susceptibility scoring in family-based association testing. BMC Genet 2003;4:S49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher RA. Statistical methods for research workers. London: Oliver and Boyd, Ltd.; 1925.
- 23.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraft P. Curses–winner's and otherwise–in genetic epidemiology. Epidemiology 2008;19:649–651; discussion 657–658. [DOI] [PubMed] [Google Scholar]
- 25.Anthonisen NR, Connett JE, Kiley JP, Altose MD, Bailey WC, Buist AS, Conway WA Jr, Enright PL, Kanner RE, O'Hara P, et al. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. JAMA 1994;272:1497–1505. [PubMed] [Google Scholar]
- 26.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 1996;382:551–555. [DOI] [PubMed] [Google Scholar]
- 27.Tuder RM. Aging and cigarette smoke: fueling the fire. Am J Respir Crit Care Med 2006;174:490–491. [DOI] [PubMed] [Google Scholar]
- 28.Karrasch S, Holz O, Jorres RA. Aging and induced senescence as factors in the pathogenesis of lung emphysema. Respir Med 2008;102:1215–1230. [DOI] [PubMed] [Google Scholar]
- 29.Suga T, Kurabayashi M, Sando Y, Ohyama Y, Maeno T, Maeno Y, Aizawa H, Matsumura Y, Kuwaki T, Kuro OM, et al. Disruption of the klotho gene causes pulmonary emphysema in mice. Defect in maintenance of pulmonary integrity during postnatal life. Am J Respir Cell Mol Biol 2000;22:26–33. [DOI] [PubMed] [Google Scholar]
- 30.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA 1999;96:10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaneva M, Arnett FC. Antibodies against Ku protein in sera from patients with autoimmune diseases. Clin Exp Immunol 1989;76:366–372. [PMC free article] [PubMed] [Google Scholar]
- 32.Agusti A, MacNee W, Donaldson K, Cosio M. Hypothesis: does COPD have an autoimmune component? Thorax 2003;58:832–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taraseviciene-Stewart L, Burns N, Kraskauskas D, Nicolls MR, Tuder RM, Voelkel NF. Mechanisms of autoimmune emphysema. Proc Am Thorac Soc 2006;3:486–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SH, Goswami S, Grudo A, Song LZ, Bandi V, Goodnight-White S, Green L, Hacken-Bitar J, Huh J, Bakaeen F, et al. Antielastin autoimmunity in tobacco smoking-induced emphysema. Nat Med 2007;13:567–569. [DOI] [PubMed] [Google Scholar]
- 35.Cosio MG, Saetta M, Agusti A. Immunologic aspects of chronic obstructive pulmonary disease. N Engl J Med 2009;360:2445–2454. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman EA. Quantitative image analysis in predicting outcomes in NETT. American Thoracic Society International Conference. 2004. Orlando, FL.
- 37.Maeda Y, Hunter TC, Loudy DE, Dave V, Schreiber V, Whitsett JA. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J Biol Chem 2006;281:9600–9606. [DOI] [PubMed] [Google Scholar]
- 38.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J 2001;18:482–490. [DOI] [PubMed] [Google Scholar]
- 39.Seifart C, Plagens A, Brodje D, Muller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers 2002;18:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chappell S, Daly L, Morgan K, Baranes TG, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, et al. The SERPINE2 gene and chronic obstructive pulmonary disease. Am J Hum Genet 2006;79:184–186, author reply 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson CA, Pettersson FH, Barrett JC, Zhuang JJ, Ragoussis J, Cardon LR, Morris AP. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet 2008;83:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L, Wang C, Rosenberg NA. The relationship between imputation error and statistical power in genetic association studies in diverse populations. Am J Hum Genet 2009;85:692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.