Abstract
Longitudinal repeated-measures data often have been visualized with spaghetti plots for continuous outcomes. For large datasets, the use of spaghetti plots often leads to the over-plotting and consequential obscuring of trends in the data. This obscuring of trends is primarily due to the overlapping of trajectories. Here, we suggest a framework called lasagna plotting that constrains the subject-specific trajectories to prevent overlapping and utilizes gradients of color to depict the outcome. Dynamic sorting and visualization is demonstrated as an exploratory data analysis tool.
Longitudinal data are key to causal inference in epidemiologic studies. As the number of subjects, frequency of measurements, and period of active data collection grow, so does the size of the data set. As the dataset grows, so do the burden and complexity of graphically exploring and summarizing the data without obscuring salient features.
The gold standard for graphically displaying longitudinal data is the classic spaghetti plot, which plots a subject's values for the repeated outcome measure (vertical axis) versus time (horizontal axis) and connects the dots chronologically, using lines of uniform color and intensity for each subject. However, the classic spaghetti plot presents obstacles to the display of longitudinal data. Although useful for fewer subjects, trends and patterns are obscured with the larger numbers of subjects typical of modern epidemiologic studies. For example, trajectories commonly overlap in a classical spaghetti plot, as both subjects and the magnitude of the outcome measure are displayed on the vertical axis. With large datasets, the figure often succumbs to "over-plotting," in which the multiple intersecting lines have no discernible patterns. One solution has been to plot a subset of the data based on medians or deciles, but this approach fails to utilize the whole dataset.1 Furthermore, repeated-measures data containing different enrollment times, missing data, or loss to follow-up (censoring) typically are difficult to display in the classic spaghetti plot.
Exploratory plots generally are used to reveal various structures in the data: trends (how most people respond over time), outliers (whether some subjects differ from most), clusters (groups of patients responding the same, maybe due to a covariate such as treatment assignment), as well as data-quality checks of reasonable values and sensible collection patterns of multi-center repeated-measures data. Classic spaghetti plots suffering from over-plotting reduce the chances of seeing such patterns.
Spaghetti plots can be improved. Color saturation, a method of handling over-plotting, has been implemented in parallel coordinate plots, of which the spaghetti plot is a special case.2–4 In many situations a classic spaghetti plot or the more-modern parallel-coordinates plot can visualize data sufficiently. However, two common data features limit the usefulness of parallel coordinate / spaghetti plots: categorical outcomes and missing outcome values (see eFigure 16 in eAppendix; https://links.lww.com). We propose a procedure for visualization and exploration that is particularly useful for categorical outcomes and missingness.
So-called heatmaps5 provide an alternative, or at least complementary, graphical-data-exploration technique. Heatmaps enjoy frequent use in the genomics literature and other areas with high-throughput data.6–8 However, in more standard longitudinal studies, they are less popular, as evidenced by the recommendation of spaghetti plots of the raw data1,9,10 or summarized data11 in popular texts of longitudinal data analysis.
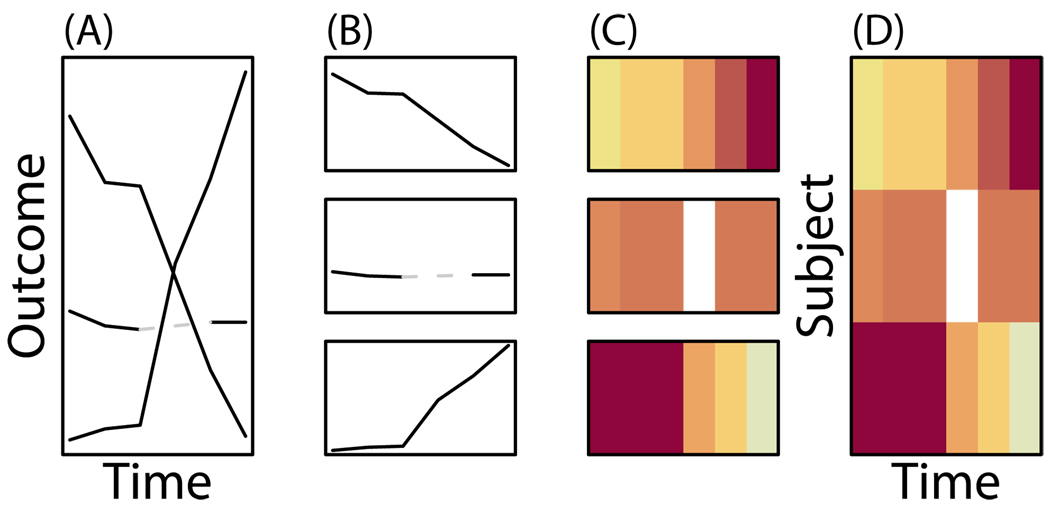
A lasagna plot is a heatmap well-suited for longitudinal data. In spaghetti plots, each subject's trajectory over time is like a noodle, that can cross other trajectories (Figure 1). In lasagna plots, each subject's trajectory over time is a horizontal layer, with the simultaneous plotting of trajectories resulting in a stacking of layers, as in lasagna.
Figure 1.
Lasagna plots as derived from spaghetti plots involve making noodles into layers. From left to right, (A) a spaghetti plot with three noodles where trajectories overlap. (B) Extracting each noodle representing repeated measures on a subject, (C) a layer is made by letting color represent the outcome. (D) Individual layers are then stacked to make a lasagna plot, with no overlapping of subject information.
The proposed lasagna plot uses color or shading to depict the magnitude of the outcome measurement and fixes the vertical dimension per subject. The lasagna plot takes advantage of color to provide a third dimension and display additional information, rather than relying upon the vertical dimension to display overlapping magnitudes of change. All information about the value of the outcome is through color (intensity), making color choice important.
Haphazard color selection can produce varying appearance in different media, induce optical illusions and after-image effects, confuse those with colorblindness, and produce misleading interpretations of data characteristics via relative aspects of color.12–15 One good principle for color selection is to have equally spaced hues with constant chroma and luminance across hues in a perceptually uniform colorspace, such as hue-chromaluminance (HCL) (as opposed to red-green-blue [RGB] or hue-saturation-value [HSV]).13 For unordered categorical outcomes, such as group membership, fixed chroma and luminance for hues equally spaced within a hue-chroma-luminance colorspace provide a flexible framework for the generation of qualitative palettes. To visualize ordered outcomes, consider the sequential-palette approach of allowing at least one and possibly all three colorspace dimensions (hue, chroma, luminance) to vary according to some function of interest based upon the ordering of the categories. If the outcomes to be visualized are ordered and have an inherent neutral value from which they diverge, say 0 for correlations, then two sequential palettes of different hues can be combined into a diverging palette to achieve color gradient symmetry about the neutral value.15 Practical and immediately accessible color selection resources include http://cran.r-project.org/web/packages/colorspace/vign as well as http://colorbrewer.org.16,17 The colors of most of the lasagna plots in this commentary were selected from the R packages colorspace or RColorBrewer.16,18
To reduce after-image effects and to afford viewers less eye strain, it is recommended to avoid fully saturated colors.16 Missing values should not be assigned a color by default but instead should be portrayed by the background color. If the background color is one of full saturation (e.g., white), then change to an off-white or directly assign missingness a color, preferably one sharing attributes of the palette in use (e.g. tantamount luminance). Epidemiologic longitudinal data sometimes have truly continuous outcomes, resulting in many unique values; this may cause some coloring procedures to categorize automatically how values are binned and assigned color. To have fullest control, it may be best to define meaningful categories of truly continuous data for visualization, such as deciles to be visualized with a palette of 10 colors and an off-white background for missing values.
There are several advantages of the lasagna plot: (1) group-, cohort-, and subject-level data are preserved regardless of the number of subjects or time points; (2) dynamic sorting of data can be used to ascertain group-level behavior over time; (3) intermittent missing data can be easily handled and clearly displayed; and (4) the distribution of onset and ending times can be easily displayed.
The Basics
Longitudinal data often are classified by two attributes: the state-space X and the time-space T, with each space independently being discrete or continuous. Truly continuous time-space is usually discretized to the level of sampling repeated measures in epidemiologic studies, so we focus on discrete-time data examples. In the case of truly continuous time-space or an instance where the discordance of common time-points of measurements among subjects in a discretized time-space, spaghetti plots may be more feasible than lasagna plots. With a reasonably large set of mutually shared time-points in a discretized time-space, lasagna plots work well for continuous and discrete-state-spaces, allowing for the usual considerations of heatmap coloring for continuous outcomes or discrete outcomes of high dimensions.
Conceptually, the process of lasagna plotting can be constructed from a spaghetti plot (Figure 1). However, a matrix construction of lasagna plots may be more practical. Epidemiologic longitudinal data following many subjects over time can be represented by a history matrix H. H is an m · n matrix where the m rows are the number of subjects in the group and n, the number of columns, is representative of the maximum number of intervals of recording of all groups comparatively. Therefore, the element hij (where i = 1, 2, …, m and j = 1, 2, …, n) is a number that corresponds to the state/outcome measurement of the ith subject during the jth interval. The matrix H is a stack of rows, with each row depicting a subject's path over time. Each row of H contains subject-specific data of repeated measurements. Each column contains cross-sectional data at the group level across subjects.
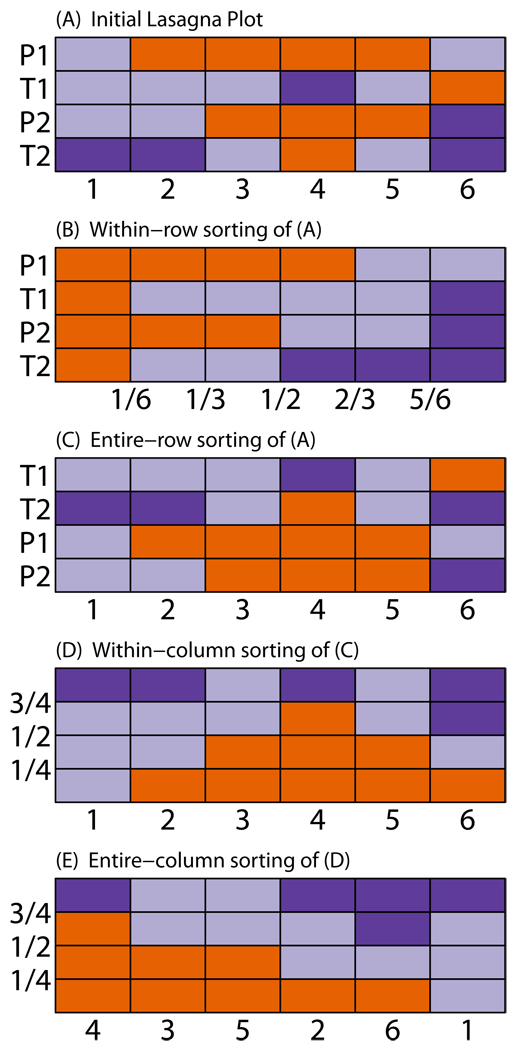
Transforming the matrix containing numbers into a graphical visualization via "painting by number" gives a snapshot of the data. A lasagna plot provides a simple image, with color representing the outcome measure. Each row contains a subject's clustered measurements, with each measurement taking place over the column variable, time (or location). Recognizing that the underpinning of the image is a matrix, we dynamically broaden the scope of information that can be obtained via sorting. Given H, five sortings are possible: within-row, entire-row, within-column, entire-column, and cluster.
Dynamic Sorting
Sortings are a way to rearrange data to reveal patterns. The proposed sortings can be done on the original H matrix or on a resultant sorting.
Within-row: sorts values of each subject in ascending order. This type of sorting preserves subject-specific information but loses the temporal ordering (Figure 2B).
Entire-row: sorts layers of subject in ascending order of a characteristic, which can be internal or external. An internal characteristic would be a feature in the lasagna plot, such as, the subject-specific mean values of the outcome. An external characteristic could be baseline age of subjects. Entire-row sorting organizes cohorts for analysis of cohort effects. This type of sorting preserves subject-specific information and temporal ordering (Figure 2C).
Within-column: sorts values across subjects in ascending order within each epoch. This sorting loses subject-specific information but can reveal group-level temporal patterns. This type of sorting is often called "vertical sorting" (Figure 2D).
Entire-column: rearranges the columns of a lasagna plot in ascending order, based on a characteristic that can be internal or external. An internal characteristic is one apparent from the information displayed in the graph, such as mean outcome value across subjects at each measurement time. An external characteristic could be measurements of a second outcome variable. This type of sorting could be useful when the external variable is seasonal, for example. Subject-specific information is preserved but temporal ordering is lost. In Figure 2E, an entire-column sort on the internal characteristic of percentage of group in state orange is applied to a vertically sorted H matrix (Figure 2D).
Cluster: A specific type of entire-row sorting in which the characteristic is internal and a clustering algorithm such as hierarchical clustering or K-means is used so that subjects that have similiar trajectories/layers are grouped together into clusters. Discrete outcomes can be ordered by strata or comparable clustering algorithms, such as correspondence analysis.
Figure 2.
An initial lasagna plot and four dynamic sorting examples. Let P denote a subject receiving placebo, T denote a subject receiving treatment. (A) The order of subjects with respect to placebo-treatment status is initially unsorted over 6 visits. (B) Within-row sorting shows subject-level composition of measurements. The horizontal axis changes to a proportion metric of the orange state. (C) Entire-row sorting on placebo-treatment status groups the cohorts. (D) Within-column sorting reveals group-level temporal patterns. The orange state has a symmetric distribution about visit 4. (E) Entire-column sorting orders the time axis. The visits are ordered according to proportion of group in the orange state.
A series of lasagna plots with sequential and cumulative sorting may allow data to be more clearly depicted than in spaghetti plots, which are static, and in the case of over-plotting obscure trends, outliers, clusters, and data-quality checks. Subject-specific trends inherently are easier to see in a lasagna plot, due to layers not overlapping as noodles do in a spaghetti plot. With dynamic sorting, trends of different cohorts (entire-row sorting on a classifying variable) and of the entire study population (within-column sorting) are possible. Outliers can be identified with various color spectra corresponding to threshold definitions as well as dynamic (entire-row) sorting. Clusters can be viewed by entire-row sorting on external variables or discovered by using cluster sorting. Data-quality checks are facilitated by the fact that lasagna plots show each datum, unobscured by other data. This unobscured depiction allows clear ascertainment of data collection patterns in a variety of time scales, allowing the time variable to change from the subject-specfic visit count to calendar month of visit, for example.
Motivating Example
The details of the study design are described in the eAppendix 1 (http://links.lww.com).
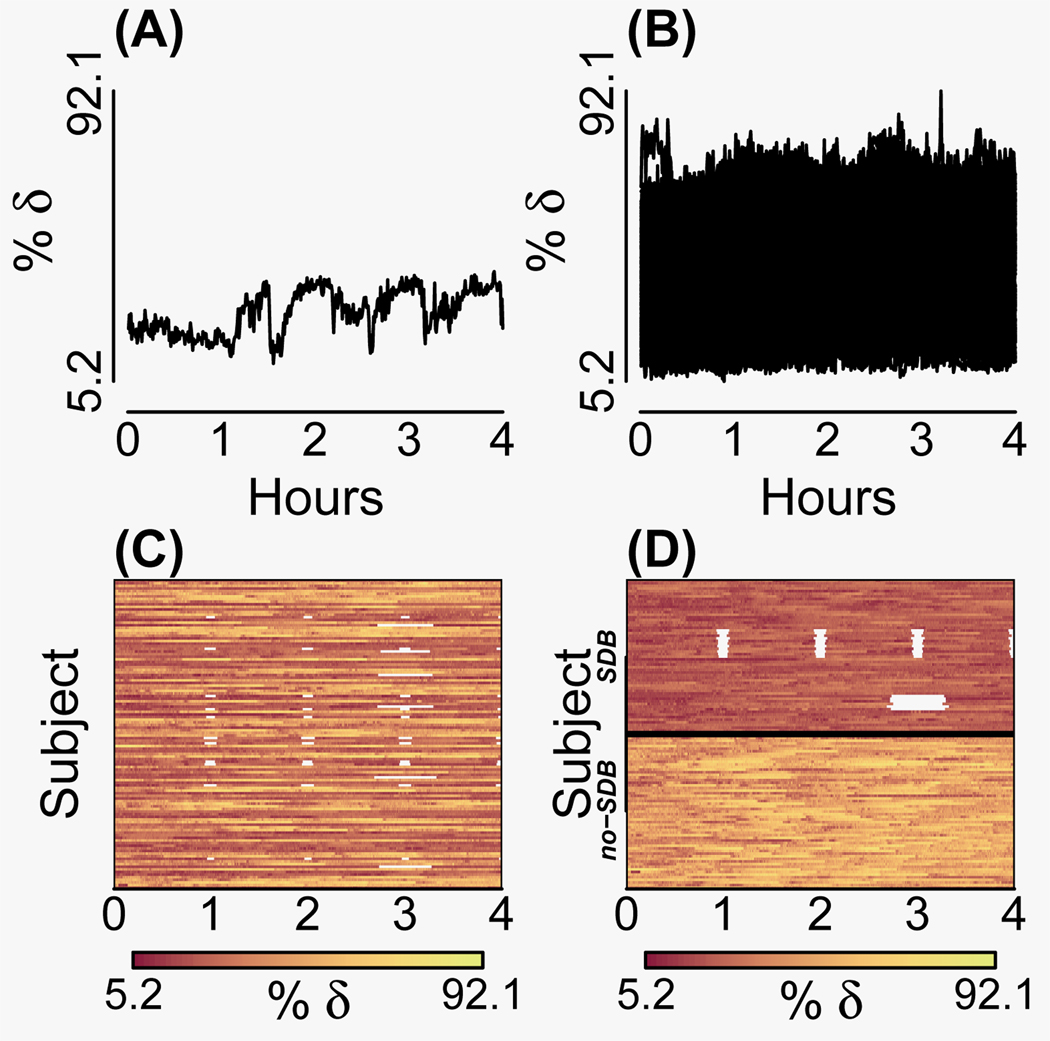
The Sleep Heart Health Study is a multicenter study on sleep-disordered breathing and cardiovascular outcomes.19 Subjects were recruited from ongoing cohort studies on respiratory and cardiovascular disease. Several biosignals for each of 6,414 subjects were collected in-home during sleep. The biosignal displayed in Figure 3 is the δ-power in the electroencephalogram (EEG) during sleep. The δ-power is a discrete-time contiuous-outcome process thought to have an important positive association with cognition and is a marker for homeostatic sleep drive.20 We explore the data by the external characteristics of sleep-disordered-breathing disease status and date of recording, looking for distinguishing patterns within subsamples of each disease group. For every 30 seconds during sleep, percent δ-power was calculated for 59 subjects with sleep-disordered breathing and 59 without. In this introductory illustrative example, we look at only the first four hours of data for each subject, so that everyone has a common onset and stopping point. We also assume that the same device was used to record sleep across different subjects and thus no two subjects had sleep recorded on the same date.
Figure 3.
Two spaghetti plots and two lasagna plots of percent delta EEG power ("% δ") recorded every thirty seconds for the first four hours of sleep. (A) Spaghetti plot for 1 subject and 479 measurements. (B) Spaghetti plot for 118 subjects and 479 measurements each. (C) Lasagna plot for the 118 (unsorted) subjects with color representing outcome. (D) Lasagna plot of the 118 subjects entire-row sorted according to disease status (sleep-disordered breathing [SDB]) and date of recording within disease status, top-down. This dynamic sort reveals the diseased overall have less % δEEG power and for some a pattern of intermittent missingness that occurs hourly over the night.
To showcase the capability of displaying intermittent missing data of the lasagna plot, a pattern of missingness is artificially applied. Via dynamic sorting, the pattern of missingness will be revealed, illustrating how patterns can be uncovered with this exploratory data analysis technique of sorting and visualizing. To showcase the process of entire-row sorting, the outcome values between disease groups were artificially made more disparate.
Figure 3 displays 2 spaghetti plots and 2 lasagna plots. Figure 3A suggests that, while a classic spaghetti plot for one subject is informative, such a plot for 118 subjects is less so (Figure 3B). The overlapping of multiple trajectories obscures trends for individual subjects, and intermittent missing data is lost. In the lasagna plot for 118 subjects, the subjects (rows) appear in random order, but the intermittent missing data (off-white) is clearly conveyed (Figure 3C). After an entire-row sort on disease status and date of EEG recording within disease status, the intermittent missing data is not only conveyed, but the sort allows the exploration of possible trends (Figure 3D). After the sort, the darker red region indicates that the diseased have less percent δEEG power during sleep. It is apparent that only the diseased have missing data, and that the recorder successfully recorded the first (going top-down) 19 subjects with sleep-disordered breathing, then malfunctioned for the next 11 recording dates in a way where it dropped measurements approximately every hour of sleep from onset. The recorder was righted and operated with full functionality for the next 14 subjects, only to malfunction again by dropping measurements about three hours from sleep onset for the next six subjects. The issue was addressed, and the recorder successfully recorded the rest of the subjects. Figures 3B and 3D contain the same outcome information, but lasagna plots more effectively depict the data. In addition to showing the extra information, such as intermittment missing data and cohort effects, lasagna plots can show informative censoring and practice effects (eAppendix 1, http://links.lww.com).
Lasagna plots complement spaghetti plots in many aspects for graphically exploring epidemiologic longitudinal data. Coding for lasagna plots is easily done in R (eAppendix 1, http://links.lww.com).21
Supplementary Material
Acknowledgments
Funding: Supported by NINDS R01NS060910, NIBIB K25EB003491, NIA R01AG10785, NIH HL086862, NIH HL089467 and NIH HL075078.
We acknowledge 2 anonymous reviewers for a very insightful review process and suggested references; Roger Peng, Alan Cohen, John Tillinghast, Nick Reich, Rafa Irizarry, Marie Thoma, and Kasper Hansen for their assistance and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SDC Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
Note to CADMUS: Please allow mention of eFigure 16 out of sequence (on p.2)— and, in general, allow the numbering of the SDC figures as the author presents them.
References
- 1.Diggle PJ, Heagerty PJ, Liang KY, Zeger SL. The Analysis of Longitudinal Data. 2nd ed. Oxford, England: Oxford University Press; 2002. Available at: http://www.amazon.com/Analysis-Longitudinal-Data-Peter-Diggle/dp/0198524846/ref_si3_rdr_bb_product. [Google Scholar]
- 2.Wegman EJ. Hyperdimensional data analysis using parallel coordinates. J Am Stat. 1990;85:664–675. [Google Scholar]
- 3.Wegman EJ, Luo Q. High dimensional clustering using parallel coordinates and the grand tour. In: Klar R, Opitz O, editors. Classification and Knowledge Organization. Berlin: Springer Verlag; 1997. pp. 93–101. [Google Scholar]
- 4.Wegman EJ. Data mining and visualization: some strategies. Bull Int Statist Inst. 1999;52:223–226. [Google Scholar]
- 5.Wilkinson L, Friendly M. The history of the cluster heat map. Am Stat. 2009;63:179–184. [Google Scholar]
- 6.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Met. 2007;4:911–913. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumgartner R, Somorjai R. Graphical display of fMRI data: visualizing multidimensional space. Magn Resonan Imag. 2001;19:283–286. doi: 10.1016/s0730-725x(01)00296-x. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar D. Lattice: Multivariate Data Visualization With R. New York: Springer Verlag; 2008. [Google Scholar]
- 9.Hedeker D, Gibbons RD. Longitudinal Data Analysis. Hoboken, NJ: Wiley-Interscience; 2006. [Google Scholar]
- 10.Fitzmaurice GM, Davidian M, Verbeke G, Molenberghs G. Longitudinal Data Analysis: A Handbook of Modern Statistical Methods. Boca Raton, FL: Chapman & Hall/CRC; 2008. [Google Scholar]
- 11.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. Hoboken, NJ: Wiley Interscience; 2004. [Google Scholar]
- 12.Poynton C. Frequently asked questions about color. 1999 Available at: http://www.inforamp.net/poynton/PDFs/ColorFAQ.Pdf.
- 13.Ihaka R. In: Hornik K, Leisch F, Zeileis A, editors. Colour for presentation graphics; Proceedings of the 3rd International Workshop on Distributed Statistical Computing; March 20–22, 2003; Vienna, Austria: Citeseer; 2003. [Google Scholar]
- 14.Lumley T. Color coding and color blindness in statistical graphics. ASA Stat Comput Stat Graphics Newslett. 2006;17:4–7. [Google Scholar]
- 15.Zeileis A, Hornik K, Murrell P. Escaping RGBland: Selecting colors for statistical graphics. Comput Stat Data Anal. 2009;53:3259–3270. [Google Scholar]
- 16.Ihaka R, Murrell P, Hornik K, Zeileis A. Colorspace: Color Space Manipulation. R package version; 2008. Available at: http://CRAN.R-project.org/package_colorspace. [Google Scholar]
- 17.Harrower M, Brewer CA. Colorbrewer.org: an online tool for selecting colour schemes for maps. Cartogr J. 2003;40:27–37. [Google Scholar]
- 18.Neuwirth E. Rcolorbrewer: Colorbrewer Palettes. R package version 1.0 –2; 2007. [Google Scholar]
- 19.Quan SF, Howard BV, Iber C, et al. The sleep heart health study: design, rationale, and methods. Sleep. 1997;20:1077–1085. [PubMed] [Google Scholar]
- 20.Marshall L, Helgad H, Matthias M, Born J. Boosting slow oscillations during sleep potentiates memory. Nature. 2006;444:610–613. doi: 10.1038/nature05278. [DOI] [PubMed] [Google Scholar]
- 21.R Development Core Team. R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. ISBN 3-900051-07-0. Available at: http://www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.