Abstract
We are much better at taking cells apart than putting them together. Reconstitution of biological processes from component molecules has been a powerful but difficult approach to studying functional organization in biology. Recently, the convergence of biochemical and cell biological advances with new experimental and computational tools is providing the opportunity to reconstitute increasingly complex processes. We predict that this bottom-up strategy will uncover basic processes that guide cellular assembly, advancing both basic and applied sciences.
Biology presents many strange and wonderful examples of what is possible in the physical world. We might not fully understand how axons are guided during development or how stem cells divide asymmetrically, but most scientists would accept that there is a physical explanation at the cellular level — given the right molecules and boundary conditions, the known laws of physics govern the interactions that drive biological behaviour. Molecules that are crucial for numerous cellular functions, such as motility and division, have been identified. Furthermore, the importance of boundary conditions, such as membranes, and their fluidity, lateral organization and mechanical compliance, is becoming clear1,2. However, an important question remains: how are the molecules and the boundary conditions linked to generate the remarkably coordinated and robust cellular behaviour that we observe, which often involves the creation of structures with length scales of many micrometres over timescales of minutes to hours?
The concepts of self-organizing systems and cooperative behaviour were first explored in the context of chemical systems in the 1920s, and in the past ten years they have been productively applied to the study of self-assembly in cell biology3. Reconstitutions of cytoskeletal filaments and their interactions with motors and other binding proteins are prime examples of how dynamic behaviour observed inside a cell can be recreated and studied outside the cell4. Advancement towards recreating more complex cell-like behaviour (referred to here as cellular reconstitution), such as cell division and cell motility, will involve confining reactions to small volumes that are bound by a bilayer membrane and incorporating signalling pathways that are controlled by external stimuli. Experiments on living cells, which continue to be crucial for testing theories of how molecules interact to give rise to physiological functions, cannot by themselves satisfy Feynman’s idea of understanding: “That which I cannot create I do not understand”5. Reconstitution studies using cell-free extracts or purified proteins provide a complementary approach to live-cell studies; taken together, these two approaches can lead to an understanding of biological processes that advances both basic and applied sciences.
This kind of reductionist biology — perhaps more appropriately called constructionist biology — offers three important insights that complement the investigation of living cells. First, conducting in vitro reconstitution experiments can confirm and refine molecular models of processes outside the complicating environment of cells. Second, unexpected behaviour can arise in simplified reconstitutions constrained only by known physical laws and specific biochemical interactions, and explaining such behaviour might yield new insight into biological organization. Third, reconstitutions are often amenable to mathematical modelling that considers space, time and a manageable number of other variables to describe the components involved. The level of detailed understanding available from reconstitution studies can help reveal how evolved biological systems work and provide insight into how new biological functions could be engineered.
A growing number of spatially organized processes have now been reconstituted and characterized using purified components and cell-free extracts (see next section; FIG. 1). The recent surge in cellular reconstitutions reported in the literature indicates an increasingly productive convergence of biochemical knowledge, experimental tools and theoretical understanding, which is needed to build cellular structures from scratch. In this Opinion article, we trace the roots of today’s cellular reconstitution efforts back to the early days of biochemical reconstitution and discuss technical advances that enable the current work, along with the remaining limitations. We suggest that reconstituting cellular-scale structures and behaviour from component parts, an approach that is poised to become more accessible, informative, and useful than ever before, is a crucial step towards answering some of the most fundamental questions of cell biology.
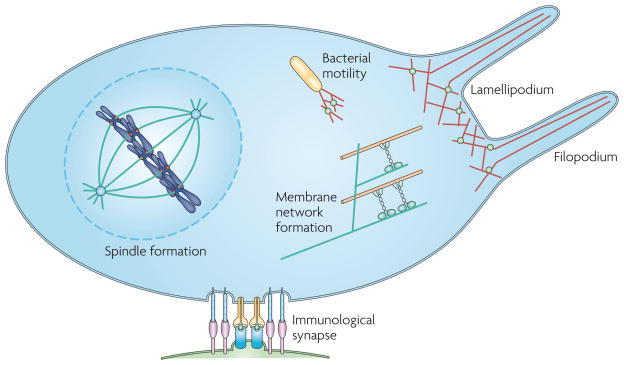
Figure 1. A cartoon of progress towards reconstitution of cellular processes.
Although rebuilding a cell is a long way off, several micrometre-scale structures involved in various cellular processes have been successfully reconstituted using either purified proteins or cell-free extracts. Spindle formation has been reconstituted using Xenopus laevis cell extract and DNA-coated microspheres. Actin-based motility of the bacterium Listeria monocytogenes has been reconstituted using purified proteins. Growing actin networks involved in lamellipodial protrusions have been shown to displace membranes, and parallel actin filaments growing against membranes have been shown to generate filopodium-like protrusions. An immunological synapse has been reconstituted using a planar lipid bilayer and live T cells. Tubular membrane networks, like those found in cells, have also been reconstituted using purified kinesin molecular motors, microtubules and giant vesicles. In each of these cases, a fairly simple set of components was found to generate a remarkably complex structure that is helping us to understand the assembly, organization and behaviour of cellular processes.
Early days of reconstitution
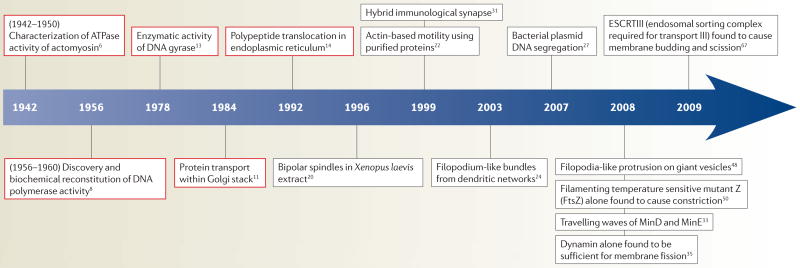
When biochemistry emerged as its own field, questions regarding the spatial organization of cellular structures were secondary to more basic questions about their molecular makeup, stoichiometric composition and functional significance (TIMELINE). In the 1940s, actomyosin threads from muscle were reconstituted and biochemically characterized by Szent-Gyoergyi, and later by Straub (see REF. 6 for an overview), leading the way for the understanding of how muscles contract. Not long after, the discovery of the structure of DNA by Watson and Crick in 1953 (REF. 7) led to a flurry of investigation on the synthesis of DNA, including Arthur Kornberg’s enzymatic study of DNA replication8. Simply mixing the four deoxynucleotide components of DNA and Escherichia coli extract proved to have little success, as unwanted nuclease activities were present in the extracts. With the purification of what is now known as DNA polymerase I, Kornberg and colleagues successfully reconstituted the synthesis of DNA9. The conceptually simple three-component enzymatic reaction, requiring the four deoxynucleotides, single-stranded template DNA and purified DNA polymerase, has paved the way for further studies of DNA replication.
Timeline. Examples of biochemical and cellular reconstitution.
Biochemical reconstitution on a molecular-length scale (red) and cellular reconstitution on a micrometre scale (black).
Biochemical reconstitution has been embraced by several fields of modern cell biology beyond the cytoskeleton and DNA replication. The study of membrane trafficking began with the identification and isolation of yeast secretion (Sec) mutants that blocked membrane traffic10. Later, using a reconstitution approach with purified proteins and semi-intact cells, coat protein I (COPI) and coat protein II (COPII) were identified as proteins involved in protein transport11,12. Biochemical reconstitution has also led to a better understanding of the protein translocation process in the endoplasmic reticulum (ER) and of how other DNA enzymes function13,14. More recently, oscillation of the circadian clock was reconstituted15.
In these examples, biochemical reconstitution is aimed at identifying specific molecular reactions that operate at molecular-length scales. How cells assemble, organize and use micrometre-scale structures is emerging as a central question in cell biology, one that is now becoming amenable to reconstitution.
Towards cellular reconstitution
Biological activities and reactions are often confined to specific locations in the cell, such as on the plasma membrane, in various organelles and bound to biopolymers, such as the cytoskeleton and chromosomes. Reconstitution of spatially organized processes often requires the specific physical or biochemical boundary conditions that localize reactions or restrict diffusion to be established on the cellular scale, an aspect that was not the focus of early biochemical reconstitutions.
In some cases, the system of biomolecules naturally includes its own boundary conditions. Cytoskeletal polymers reconstituted with binding proteins can spatially organize on the micrometre scale in the absence of physical confinement, with the polymer itself serving as a scaffold for self-assembly. Notable examples include the formation of organized structures, such as asters and vortices that arise from mixtures of microtubules and motor proteins16, as well as the spontaneous transition from a dendritic actin network to bundled actin filaments17. The large-scale organization exhibited by the microtubule system is reviewed elsewhere18.
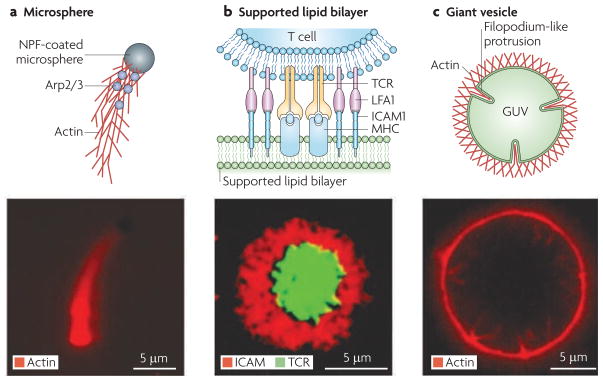
In other cases of reconstitution, the boundary conditions must be added explicitly. Using a new collection of experimental approaches, researchers are ‘seeding’ the spatial organization of cellular reconstitutions by restricting biochemical activities to surfaces, such as a micrometre-sized polystyrene bead or a planar fluid lipid bilayer (FIG. 2). Confining reactions to two dimensions is not only physiologically appropriate in many cases, but it also enables the use of advanced light microscopy techniques, which result in vivid images of spatially complex processes.
Figure 2. Strategies for reconstituting cellular processes.
Once the specific molecular components that are involved in a cellular process are identified, they must be organized by appropriate boundary conditions to generate functional behaviour. Three primary strategies have been used to define the boundary conditions that initiate spatially organized processes. a | The surface of microspheres can be functionalized with proteins such as actin nucleation-promoting factors (NPFs), that spatially organize actin assembly in purified protein solutions or cell extracts by a process that requires the actin-related protein 2/3 (Arp2/3) complex. The lower panel is an epi-fluorescence microscopy image showing an NPF-coated microsphere propelled by a growing actin network. b | supported lipid bilayers ensure that molecular components of the system being studied can freely diffuse. They have been used to reconstitute the immunological synapse. T cell receptor (TCR) and leukocyte function-associated antigen 1 (LFA1) on the T cell bind to intracellular adhesion molecule 1 (ICAM1) and major histocompatibility complex (MHC) peptide reconstituted on the supported lipid bilayer, to form the immunological synapse. The lower panel is an epi-fluorescence microscopy image showing TCR at the centre and ICAM1 at the periphery of the immunological synapse. c | Giant unilamellar vesicles (GUVs) represent deformable substrates, and they have been used to reconstitute filopodium-like protrusions. The lower panel is an epi-fluorescence microscopy image showing actin networks on a GUV spontaneously undergoing transitions to form inward protrusions that consist of long unbranched actin filaments. Part b is reproduced, with permission, from REF. 62 © (2005) American Association for the Advancement of Science. Part c is reproduced, with permission, from REF. 48 © (2008) Macmillan Publishers Ltd. All rights reserved.
In this Opinion article, we focus on cellular reconstitution in which boundary conditions are added to guide organization. A brief survey of such reconstitutions, summarized in TABLE 1, is organized according to the experimental platform used to reconstitute the cellular-scale process.
Table 1.
Summary of recent progress in cellular reconstitution
| Reconstituted process or structure | Components used | References |
|---|---|---|
| Microspheres | ||
| Cell motility | Listeria monocytogenes and NPF-coated microspheres immersed in Xenopus laevis extracts | 63,64 |
| NPF-coated microspheres, actin, profilin, cofilin, Arp2/3 complex and capping protein | 22,23 | |
| NPF-coated microspheres, actin, Arp2/3 complex and fascin | 25,24 | |
| Mitotic spindle | DNA-coated microspheres and X. laevis cytoplasmic extracts | 20,65 |
| Plasmid segregation | ParC coated microspheres, ParR and ParM | 27 |
| Supported bilayers | ||
| Immunological synapse | Supported bilayer containing MHC peptide and ICAM1, and T cells | 31,62 |
| Neuronal synapse | Supported bilayer containing neuroligin 1, and neuronal cells | 32 |
| Bacterial cell division | MinD and MinE | 33 |
| Vesicle fission | Dynamin 1 and GTP | 35 |
| Cell motility | Lipid-coated beads, N-WASP, actin, profilin, cofilin, Arp2/3 complex and capping protein | 34 |
| Giant vesicles | ||
| Bacterial cell division | Membrane-targeted FtsZ | 50 |
| Membrane tube networks | Kinesin-bound GUV and microtubule tracks | 40,66 |
| Actin–membrane interaction | PtdIns(4,5)P2-containing vesicles, actin, N-WASP and Arp2/3 complex | 46 |
| Filopodia formation | PtdIns(4,5)P2-containing vesicles, actin, N-WASP and Arp2/3 complex | 48 |
| Membrane scission | vps20, snf7 and vps24 | 67 |
Arp2/3, actin-related protein 2/3; GUV, giant unilamellar vesicle; ICAM1, intercellular adhesion molecule 1; MHC, major histocompatibility complex; NPF, nucleation-promotion factor; N-WASP, neuronal-Wiscott–Aldrich syndrome protein (also known as WASL); PtdIns(4,5)P2, phosphatidylinositol-4,5-bisphosphate; snf7, sucrose non-fermenting protein 7; Vps, vacuole protein sorting protein.
Cytoskeleton assembly on microspheres
Cellular shape change and intracellular motion are driven primarily by the assembly and disassembly of cytoskeletal polymers19. Polystyrene microspheres are a useful substrate to reconstitute many of the processes that use force generation of cytoskeletal polymers. Polystyrene microspheres come in many different sizes, and proteins or DNA can be physically adsorbed or covalently conjugated onto their surface (FIG. 2a). Furthermore, individual beads can be manipulated using optical traps, magnetic tweezers and micropipettes. However, despite the ease of manipulation, proteins that are directly conjugated to the microspheres may adopt non-physiological conformations and they will have restricted lateral movements.
The first notable use of microspheres in reconstitution was for mitotic spindle formation driven by microtubule dynamics in a cytoplasmic extract from Xenopus laevis. Mitotic spindles were found to assemble around DNA-coated microspheres acting as artificial chromosomes20. The study showed that centrosomes (the microtubule organizing centre) and kinetochores (the region of the chromosome where spindle fibres attach) were not required for spindle formation. More importantly, it showed that bipolarity is an intrinsic property of mitotic spindles because microtubules and motor proteins assemble around DNA in mitotic extracts in the absence of external polarity cues.
Actin filaments produce forces that drive membrane protrusions during processes such as cell motility. Reconstitution of crawling motility began with studies of the pathogenic bacterium Listeria monocytogenes. L. monocytogenes expresses an actin nucleation-promoting factor (NPF), ActA, which is required for hijacking the cellular machinery of the infected cell to propel its own movement21. In an impressive feat, the motility of dead L. monocytogenes was reconstituted using NPF-coated microspheres and a minimal set of proteins (actin and three actin filament regulators: the actin-related protein 2/3 (Arp2/3) complex, actin depolymerizing factor and capping protein)22. Using a similar reconstituted system on NPF-coated microspheres, a controversy was recently resolved regarding the role of capping protein, a protein that limits actin filament growth by binding to the filament’s barbed end, and yet increases the velocity of actin-based motility. Careful analysis of actin growth on microspheres revealed that actin filament capping leads to an increase in filament nucleation and branching23. Again using NPF-coated microspheres, bundles of actin filaments that resemble filopodia (long and thin actin-based protrusions) were reconstituted by introducing the actin-bundling protein fascin24. In addition, these filopodial bundles were shown to be capable of supporting L. monocytogenes motility25, which shows that architecturally different actin networks can support cellular motility.
Unlike for eukaryotes, prokaryotic cellular functions were not attributed to cytoskeletal polymers until recent years26, but they are now thought to be important in a large number of processes. Segregation of DNA-coated polystyrene microspheres driven by polymerization of the prokaryotic actin homologue partitioning system M (ParM) has been reconstituted using microspheres and the three-component system of the R1 plasmid, which consists of parC, parR and parM in an operon27. The parR gene encodes a protein that binds cooperatively to parC to form a complex. In the absence of the ParR–parC complex, the ParM polymer is dynamically unstable28, but it is stabilized when it binds to the ParR–parC complex. Insertional elongation of ParM and force generation were observed as the ParR–parC-conjugated microspheres were pushed apart by the growing polymer. Through these reconstitution experiments it was shown that the R1 plasmid contained all the components necessary for dynamically searching for and then stably separating plasmid copies.
Reconstitution on supported lipid bilayers
To reconstitute membrane-based phenomena, in which diffusion in two dimensions is crucial, a lipid bilayer can be used as a substrate (FIG. 2b). Planar supported lipid bilayers can be made by fusing small liposomes, tiny vesicles made of lipids, onto clean glass coverslips. These bilayers have been used to study lipid diffusion and have been exploited for biomedical applications29. Functional membrane proteins can be incorporated in the supported lipid bilayer using liposomes with inserted proteins30. Planar supported lipid bilayers are particularly suitable for investigating processes that occur at the plasma membrane, such as the formation of the immunological synapse, which occurs at the interface between an antigen presenting cell (APC) and a lymphocyte.
The immunological synapse is a complex supramolecular structure that is involved in the communication between an APC and a T cell. The process of T cell activation was reconstituted by replacing the APC with a planar supported lipid bilayer that contained fluorescently labelled major histocompatibility complex (MHC) peptide and intercellular adhesion molecule 1 (ICAM1)31. These molecules, which are expressed by the APC, could diffuse freely on the supported lipid bilayer and associate with cellular receptors on the T cell. The dynamic association and rearrangement of T cell antigen receptor (TCR) and leukocyte function-associated antigen 1 (LFA1) on the T cell with MHC peptide and ICAM1 in the supported lipid bilayer forms the immunological synapse. This consists of a central supramolecular cluster (c-SMAC), in which TCR predominates, and a peripheral supramolecular cluster (p-SMAC), in which adhesion molecules are localized. This reconstitution indicated that the formation of the immunological synapse provides a mechanism for sustained signalling in engaged T cells. Hybrid neuronal synapses, consisting of neuronal cells and a supported lipid bilayer, were similarly reconstituted32. These studies show that spatially organized and biologically relevant structures can be formed between a living cell and a supported lipid bilayer, providing a controlled environment in which to study cell–cell communication.
Recently, an elegant study reported the self-organization of MinD and MinE (proteins that are responsible for locating the contractile ring during bacterial cytokinesis) into travelling waves on supported lipid bilayers33. Supported lipid bilayers can also be formed on glass microspheres instead of on planar glass. Using supported lipid bilayers on glass microspheres and purified proteins involved in actin-based motility, the mechanism behind attachment of the NPF neural Wiskott–Aldrich syndrome protein (N-WASP; also known as WASL) to actin networks was elucidated34. Similarly, a reconstituted system was recently developed using lipid-coated glass microspheres to study dynamin-mediated membrane fission, and it was found that dynamin and GTP are sufficient for this process35.
Reconstitution using giant lipid vesicles
An alternative to the supported lipid bilayer is the giant lipid vesicle (FIG. 2c). Giant vesicles (10–100 μm in diameter) can be composed of a single lipid bilayer (unilamellar) or multiple bilayers (multilamellar), and they support the diffusion of molecules in-plane, as well as morphological deformations out of plane, such as the formation of protrusions or invaginations. Although small vesicles have been used in many biochemical reconstitutions in which a membrane substrate is required36,37, they are not amenable to investigations of spatial organization by light microscopy. By contrast, giant vesicles are suitable for real-time imaging to directly observe the dynamics and organization of biomolecule–membrane interactions. This approach is limited, however, by the challenge of making large numbers of giant vesicles with a defined size and composition.
Decades ago, thin-section electron microscopy revealed a labyrinth of membrane networks in cells38, and reconstitution studies are now helping us to understand how they might be formed. Tubular membrane networks reminiscent of the Golgi and ER were reconstituted using a truncated kinesin motor from Drosophila melanogaster that was bound to giant unilamellar vesicles (GUVs) on one end and to surface immobilized microtubules on the other end39,40. Processive kinesin molecules were attached to GUVs through a biotin–streptavidin interaction and ‘walked’ along stabilized microtubules to generate tubular networks. Motor-driven membrane tube networks have also been used to dissect the mechanism of dynamin-mediated membrane fission39.
The deformability of lipid bilayer vesicles makes them an ideal tool for elucidating the biophysical effects of force generation by actin41,42 and other cytoskeletal polymers. For example, membrane deformations that are dependent on the polymerization of microtubules and actin filaments encapsulated in lipid vesicles have been demonstrated experimentally43,44. Actin shells that are reminiscent of the actin cortex have also been reconstituted on the inner side of GUVs45.
The effect of both membrane fluidity and deformability on the assembly of cytoskeletal networks can be studied directly with GUVs. When phase-separated GUVs that contained phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) were mixed with purified proteins that are involved in dendritic actin network assembly, the actin networks grew external to the GUVs and only on membrane domains that were enriched with PtdIns(4,5)P2 (REF. 46). It was found that the interaction between actin networks and the membrane acted to stabilize the PtdIns(4,5)P2-containing membrane domains against temperature-induced mixing with other lipids. The potential influence of an actin network on the spatial organization of membrane components supports the requirement of an intact cytoskeleton in signalling systems47.
More recently, dynamic filopodia-like membrane deformations were reconstituted on GUVs48. It was previously thought that filopodia emerge from branched dendritic actin networks by a process that involves bundling of actin filaments by accessory proteins. Furthermore, the length of cellular filopodia was thought to be limited by membrane resistance force, with filopodia expected to buckle if they grew past the theoretical Euler buckling length — where a structural element can no longer withstand compressive stress. The reconstitution of filopodium-like structures in the absence of both the tip complex (a molecular assembly that promotes actin filament elongation) and the crosslinking protein fascin led to two physical insights: the membrane alone can facilitate the transition of a branched actin network to a parallel array of actin filaments, and the force generated by membrane tension will not buckle filopodia, even those beyond the Euler buckling length49. The reconstitution of complex actin-based structures through interactions with a lipid bilayer suggests that in early cellular life, liposome-encapsulating force-generating polymers could have exhibited rich morphological behaviours that are reminiscent of current living systems.
Last year, the use of multilamellar vesicles to study the bacterial tubulin homologue, filamenting temperature sensitive mutant Z (FtsZ), revealed that membrane-anchored FtsZ alone can self-assemble into ring-like structures known as Z rings, which serve as scaffolds for the bacterial division apparatus50. The reconstituted Z rings generated a constriction force that was dependent on GTP hydrolysis. However, because constriction of the multilamellar vesicles by the Z rings was incomplete, it remains possible that additional force-generating activity might be required to complete the process.
Tools for cellular reconstitution
Continued progress in cellular reconstitution will depend on the creation of new tools. Recently, techniques that enable efficient encapsulation of components inside cell-sized vesicles have been developed, making it possible to begin controlling both internal and external biochemical conditions for studies involving transmembrane signalling and transport (BOX 1). Micro-fabrication and patterning tools will also continue to advance, enabling the control of molecular location for activity gradients or three-dimensional confinement51,52.
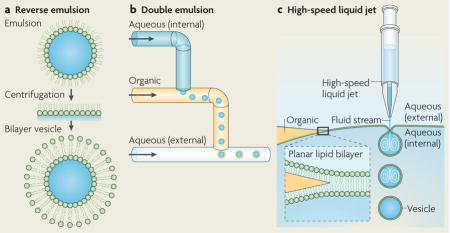
Box 1. Advances in vesicle encapsulation.
Encapsulation of biomolecules in cell-like vesicles represents an important step in cellular reconstitution, as biological systems naturally operate in confined volumes rather than in test tubes. There have been several reports in recent years that describe new methods for vesicle encapsulation and three are described here. A technique called reverse emulsion was used to make asymmetric lipid vesicles that encapsulate cytoplasmic extract (see the figure, part a)55,56. In this technique, an emulsion of lipid-stabilized aqueous droplets in oil is centrifuged through a planar interface with an aqueous solution, adding a second monolayer of lipids to create bilayer vesicles as they pass into the aqueous phase.
Microfluidic channels have been used to generate monodisperse emulsions and vesicles with high fidelity and high throughput. For example, monodisperse double emulsion (water–oil–water) structures can be made using a fabricated microfluidic device that uses a continuous flow to deposit aqueous droplets into an organic phase and then deposit the emulsion with an oil shell back into an aqueous flow (see the figure, part b)57,58. Double emulsions can become vesicles through solvent evaporation, in which lipid molecules dissolved in the organic phase form a lipid bilayer on removal of the oil59.
Recently, a microfluidic jetting technique analogous to blowing soap bubbles was demonstrated. Both double emulsions and unilamellar lipid bilayer vesicles have been formed by shooting a high-speed pulsed liquid jet against a solvent-supported planar lipid bilayer (see the figure, part c)60,61. These techniques hold great promise in cellular reconstitution for encapsulating biomolecules in lipid vesicles.
Central to the study of spatial organization in cells is the widespread use of light microscopy. Traditional biochemical reconstitution techniques typically report the average behaviour of the system and yield little information on spatial organization. By contrast, coordinated cellular processes that are dependent on the collective motion of many molecules often require microscopy to follow their dynamic evolution in real time. Commercial wide-field and confocal light microscopes have become mainstream tools in biology laboratories, and researchers continue to push light microscopy to higher spatial and temporal resolutions. Several techniques that break the diffraction-limited spatial resolution of conventional light microscopes have been demonstrated, and their uses, as well as current developments in fluorescent probe technology, have recently been reviewed53.
Computational tools have been instrumental in guiding experimental approaches to cellular reconstitution as well as in understanding the complex behaviour they create. Mathematical modelling and computer simulations will continue to be crucial for explaining the observations from in vitro reconstitution experiments16,18,45,54, and we expect their importance to grow as both experimental and computational techniques improve.
Learning by building
Cell biology has come a long way since Szent-Gyoergyi and Straub’s biochemical reconstitution of muscle actomyosin and Arthur Kornberg’s biochemical reconstitution of DNA synthesis more than half a century ago. Although the knowledge of an enzyme’s binding affinity and catalytic efficiency gives us sufficient information to predict its turnover capability, we do not yet have the power to predict how a mixture of macromolecules will assemble into spatially complex structures, even with a thorough characterization of the individual parts. Cellular reconstitution offers a way to learn by building.
Advances in imaging technology, combined with clever experimental techniques for controlling boundary conditions, have led to successful reconstitution of several self-organizing biological processes, with more on the way. However, because cellular reconstitution includes only a subset of components used by cells, the approach is not guaranteed to yield the precise mechanistic details of a biological process. So how can we know whether a reconstitution is behaving ‘biologically’? Perhaps one day we will need the cellular equivalent of a Turing test — the approach devised by Alan Turing in 1950 to test a computing machine’s ability to demonstrate intelligence — to tell the difference, asking the question: does the reconstitution behave like a normal cell in every way? Until then, we will need to use modelling to bridge the gap between small assemblies and cells, and we will need to use cellular reconstitution to complement top-down approaches, all while deliberately peeling away the levels of complexity and regulation that are at work in live cells.
The road map for cellular reconstitution will probably include experiments that are motivated by basic science as well as those that are aimed at creating biological systems that exhibit customized functional behaviour — a kind of bottom-up synthetic biology. It is conceivable that one day our ability to reconstitute basic modules of cellular life will lead us to create a growing and dividing system that can be computationally designed to crawl, swim or fight disease. Now that would be intelligent design.
Acknowledgments
We apologize to those colleagues whose original and important work could not be cited owing to space limitations. We thank D. Richmond, J. Stachowiak and the rest of the Fletcher laboratory, as well as T. Pucadyil, for helpful discussion. A.P.L. is supported by the Natural Sciences and Engineering Research Council of Canada. D.A.F. is funded by National Institutes of Health R01 grants and a Nanomedicine Development Centre grant.
Footnotes
DATABASES
UniProtKB: http://www.uniprot.org
FtsZ | ICAM1 | MinD | MinE | N-WASP
FURTHER INFORMATION
Daniel A. Fletcher’s homepage: http://fletchlab.berkeley.edu/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Allen P. Liu, Department of Cell Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, MB-6, La Jolla, California 92037, USA
Daniel A. Fletcher, Biophysics Group and Bioengineering Department, University of California, Berkeley, California 94702, USA and the Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA
References
- 1.Groves JT. Bending mechanics and molecular organization in biological membranes. Annu Rev Phys Chem. 2007;58:697–717. doi: 10.1146/annurev.physchem.56.092503.141216. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Saez AJ, Chiantia S, Schwille P. Effect of line tension on the lateral organization of lipid membranes. J Biol Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 3.Karsenti E. Self-organization in cell biology: a brief history. Nature Rev Mol Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 4.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawking SW. The Universe in a Nutshell. Bantam; New York: 2001. [Google Scholar]
- 6.Kritikou E. To see them contract for the first time. Nature Rev Mol Cell Biol. 2008;9:S6. [Google Scholar]
- 7.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg A. Biologic synthesis of deoxyribonucleic acid. Science. 1960;131:1503–1508. doi: 10.1126/science.131.3412.1503. [DOI] [PubMed] [Google Scholar]
- 9.Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J Biol Chem. 1958;233:163–170. [PubMed] [Google Scholar]
- 10.Schekman R. Protein localization and membrane traffic in yeast. Annu Rev Cell Biol. 1985;1:115–143. doi: 10.1146/annurev.cb.01.110185.000555. [DOI] [PubMed] [Google Scholar]
- 11.Orci L, Glick BS, Rothman JE. A new type of coated vesicular carrier that appears not to contain clathrin: its possible role in protein transport within the Golgi stack. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- 12.Barlowe C, et al. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 13.Higgins NP, Peebles CL, Sugino A, Cozzarelli NR. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci USA. 1978;75:1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 16.Surrey T, Nedelec F, Leibler S, Karsenti E. Physical properties determining self-organization of motors and microtubules. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- 17.Haviv L, et al. Reconstitution of the transition from lamellipodium to filopodium in a membrane-free system. Proc Natl Acad Sci USA. 2006;103:4906–4911. doi: 10.1073/pnas.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karsenti E, Nedelec F, Surrey T. Modelling microtubule patterns. Nature Cell Biol. 2006;8:1204–1211. doi: 10.1038/ncb1498. [DOI] [PubMed] [Google Scholar]
- 19.Pollard TD. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422:741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 20.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 21.Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 23.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignjevic D, et al. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brieher WM, Coughlin M, Mitchison TJ. Fascin-mediated propulsion of Listeria monocytogenes independent of frequent nucleation by the Arp2/3 complex. J Cell Biol. 2004;165:233–242. doi: 10.1083/jcb.200311040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiol Mol Biol Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 29.Sackmann E, Tanaka M. Supported membranes on soft polymer cushions: fabrication, characterization and applications. Trends Biotechnol. 2000;18:58–64. doi: 10.1016/s0167-7799(99)01412-2. [DOI] [PubMed] [Google Scholar]
- 30.Groves JT, Wulfing C, Boxer SG. Electrical manipulation of glycan-phosphatidyl inositol-tethered proteins in planar supported bilayers. Biophys J. 1996;71:2716–2723. doi: 10.1016/S0006-3495(96)79462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 32.Pautot S, Lee H, Isacoff EY, Groves JT. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nature Chem Biol. 2005;1:283–289. doi: 10.1038/nchembio737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 34.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takei K, et al. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- 37.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 38.Palade GE. The organization of living matter. Proc Natl Acad Sci USA. 1964;52:613–634. doi: 10.1073/pnas.52.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 40.Koster G, VanDuijn M, Hofs B, Dogterom M. Membrane tube formation from giant vesicles by dynamic association of motor proteins. Proc Natl Acad Sci USA. 2003;100:15583–15588. doi: 10.1073/pnas.2531786100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giardini PA, Fletcher DA, Theriot JA. Compression forces generated by actin comet tails on lipid vesicles. Proc Natl Acad Sci USA. 2003;100:6493–6498. doi: 10.1073/pnas.1031670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Upadhyaya A, van Oudenaarden A. Biomimetic systems for studying actin-based motility. Curr Biol. 2003;13:R734–R744. doi: 10.1016/j.cub.2003.08.051. [DOI] [PubMed] [Google Scholar]
- 43.Cortese JD, Schwab B, Frieden C, Elson EL. Actin polymerization induces a shape change in actin-containing vesicles. Proc Natl Acad Sci USA. 1989;86:5773–5777. doi: 10.1073/pnas.86.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fygenson DK, Marko JF, Libchaber A. Mechanics of microtubule-based membrane extension. Phys Rev Lett. 1997;79:4497. [Google Scholar]
- 45.Pontani LL, et al. Reconstitution of an actin cortex inside a liposome. Biophys J. 2009;96:192–198. doi: 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holowka D, Sheets ED, Baird B. Interactions between FcεRI and lipid raft components are regulated by the actin cytoskeleton. J Cell Sci. 2000;113:1009–1019. doi: 10.1242/jcs.113.6.1009. [DOI] [PubMed] [Google Scholar]
- 48.Liu AP, et al. Membrane-induced bundling of actin filaments. Nature Phys. 2008;4:789–793. doi: 10.1038/nphys1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pronk S, Geissler PL, Fletcher DA. Limits of filopodium stability. Phys Rev Lett. 2008;100:258102. doi: 10.1103/PhysRevLett.100.258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doh J, Irvine DJ. Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proc Natl Acad Sci USA. 2006;103:5700–5705. doi: 10.1073/pnas.0509404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cosentino Lagomarsino M, et al. Microtubule organization in three-dimensional confined geometries: evaluating the role of elasticity through a combined in vitro and modeling approach. Biophys J. 2007;92:1046–1057. doi: 10.1529/biophysj.105.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nature Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 54.Alberts JB, Odell GM. In silico reconstitution of Listeria propulsion exhibits nano-saltation. PLoS Biol. 2004;2:e412. doi: 10.1371/journal.pbio.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pautot S, Frisken BJ, Weitz DA. Engineering asymmetric vesicles. Proc Natl Acad Sci USA. 2003;100:10718–10721. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okushima S, Nisisako T, Torii T, Higuchi T. Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir. 2004;20:9905–9908. doi: 10.1021/la0480336. [DOI] [PubMed] [Google Scholar]
- 58.Utada AS, et al. Monodisperse double emulsions generated from a microcapillary device. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 59.Shum HC, Kim JW, Weitz DA. Microfluidic fabrication of monodisperse biocompatible and biodegradable polymersomes with controlled permeability. J Am Chem Soc. 2008;130:9543–9549. doi: 10.1021/ja802157y. [DOI] [PubMed] [Google Scholar]
- 60.Funakoshi K, Suzuki H, Takeuchi S. Formation of giant lipid vesicle-like compartments from a planar lipid membrane by a pulsed jet flow. J Am Chem Soc. 2007;129:12608–12609. doi: 10.1021/ja074029f. [DOI] [PubMed] [Google Scholar]
- 61.Stachowiak JC, et al. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc Natl Acad Sci USA. 2008;105:4697–4702. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 63.Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 64.Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci USA. 1999;96:4908–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaetz J, Gueroui Z, Libchaber A, Kapoor TM. Examining how the spatial organization of chromatin signals influences metaphase spindle assembly. Nature Cell Biol. 2006;8:924–932. doi: 10.1038/ncb1455. [DOI] [PubMed] [Google Scholar]
- 66.Roux A, et al. A minimal system allowing tubulation with molecular motors pulling on giant liposomes. Proc Natl Acad Sci USA. 2002;99:5394–5399. doi: 10.1073/pnas.082107299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]