Abstract
Methylation of histone H3 by Set1 and Set2 is required for deacetylation of nucleosomes in coding regions by histone deacetylase complexes (HDACs) Set3C and Rpd3C(S), respectively. We report that Set3C and Rpd3C(S) are co-transcriptionally recruited in the absence of Set1 and Set2, but in a manner stimulated by Pol II CTD kinase Cdk7/Kin28. Consistently, Rpd3C(S) and Set3C interact with Ser5-phosphorylated Pol II and histones in extracts, but only the histone interactions require H3 methylation. Moreover, reconstituted Rpd3C(S) binds specifically to Ser5-phosphorylated CTD peptides in vitro. Hence, whereas interaction with methylated H3 residues is required for Rpd3C(S) and Set3C deacetylation activities, their co-transcriptional recruitment is stimulated by the phosphorylated CTD. We further demonstrate that Rpd3, Hos2, and Hda1 have overlapping functions in deacetylating histones and suppressing co-transcriptional histone eviction. A strong correlation between increased acetylation and lower histone occupancy in HDA mutants implies that histone acetylation is a key determinant of nucleosome eviction.
Keywords: Rpd3, Hos2, Hda1, Kin28, Cdk7, RNA Polymerase, CTD, transcription elongation, Set1, Set2, histone methylation, Gcn4
INTRODUCTION
The nucleosome forms a barrier to transcription elongation, and acetylation of histone tails is believed to stimulate passage of RNA Polymerase II (Pol II) by altering DNA-nucleosome contacts (Protacio et al., 2000) or recruiting chromatin remodeling complexes that function within transcription units (Carey et al., 2006). Histone eviction in coding sequences is correlated with transcription rate, and the H3-H4 chaperone Asf1 and SWI/SNF have been implicated in nucleosome disassembly during Pol II passage in yeast (Schwabish and Struhl, 2006, 2007; Williams and Tyler, 2007). We showed previously that the histone lysine acetyltransferase (KAT) complex SAGA is recruited to coding sequences and that its KAT subunit Gcn5 mediates increased H3 acetylation throughout the transcription unit. Inactivating Gcn5 diminished nucleosome eviction from the GAL1 ORF and decreased Pol II processivity (Govind et al., 2007). Involvement of SAGA and Gcn5 in stimulating elongation has been uncovered by other laboratories as well (Morillo-Huesca et al., 2006; Pascual-Garcia et al., 2008; Wyce et al., 2007; Zapater et al., 2007). Association of SAGA with the coding region is enhanced by phosphorylation of the Pol II CTD by CDK7/Kin28 in TFIIH (Govind et al., 2007). In addition, SAGA interacts with elongating Pol II phosphorylated on Ser5 of the CTD heptad repeats (Govind et al., 2007; Pascual-Garcia et al., 2008). The KAT complex NuA4 is also targeted to transcribed coding sequences in a manner stimulated by Kin28, and the NuA4 and SAGA complexes make additive contributions to nucleosome disassembly and transcription elongation (Ginsburg et al., 2009).
Histone deacetylases (HDAs) are also co-transcriptionally recruited to coding sequences. One of two complexes containing the HDA Rpd3, known as Rpd3C(S), deacetylates H3 and H4 in the coding sequences of many yeast genes dependent on dimethylation of Lys36 of H3 (H3-K36me2) by Set2. The chromodomain and PHD finger in the Eaf3 and Rco1 subunits of Rpd3C(S) bind to histones harboring H3-K36me2 to mediate tight association of Rpd3C(S) with chromatin and attendant deacetylation of H3 and H4 in vivo. This activity of Rpd3C(S) reduces the efficiency of transcription elongation, but also suppresses cryptic promoters in the coding sequences of certain genes (Carrozza et al., 2005; Chu et al., 2007; Joshi and Struhl, 2005; Keogh et al., 2005; Li et al., 2007a; Li et al., 2009). Recent evidence suggests that Rpd3(S) is targeted to the GAL1 promoter by methylated H3 deposited by transcription initiating from an upstream cryptic promoter (Houseley et al., 2008), although methylated H3-K4 produced by Set1, rather than H3-K36me2, appears to be responsible for recruiting Rpd3(S) in this setting (Pinskaya et al., 2009).
The Set3 complex (Set3C), containing the HDAs Hos2 and Hst1, also deacetylates nucleosomes in transcribed coding regions. Set3C is recruited by H3-K4me2, generated by Set1, via the PHD finger in Set3. This leads to H3/H4 deacetylation at the 5’ ends of coding sequences at various genes and increased association of Pol II (Kim and Buratowski, 2009), consistent with the positive role for Hos2 detected previously at the GAL and RNR3 genes (Sharma et al., 2007; Wang et al., 2002).
We showed previously that Hos2 and Rpd3 are recruited to ARG1 coding sequences during its activation by Gcn4. Interestingly, transcriptional activation by Gcn4 leads to increased H3 acetylation in the UAS but, paradoxically, to reduced H3 acetylation at the 3’ end of the ARG1 ORF. Eliminating Hos2, Hos3, and Rpd3 simultaneously, but not individually, evoked a large increase in acetylation per nucleosome in the 3’ ORF, leading us to conclude that multiple HDAs act redundantly to reverse the H3 acetylation catalyzed by SAGA in transcribed ARG1 sequences (Govind et al., 2007). In this study, we set out to determine whether Rpd3 associates with the ARG1 ORF in the context of Rpd3C(S), and whether its recruitment to ARG1 depends on H3-K36 methylation by Set2. We also explored whether recruitment of Hos2 at ARG1 requires H3-K4 methylation by Set1, according to the previously described mechanism (Kim and Buratowski, 2009), and investigated whether these HDAs collaborate to suppress co-transcriptional nucleosome eviction in the ARG1 ORF.
RESULTS
Set2-independent co-transcriptional recruitment of Rpd3C(S)
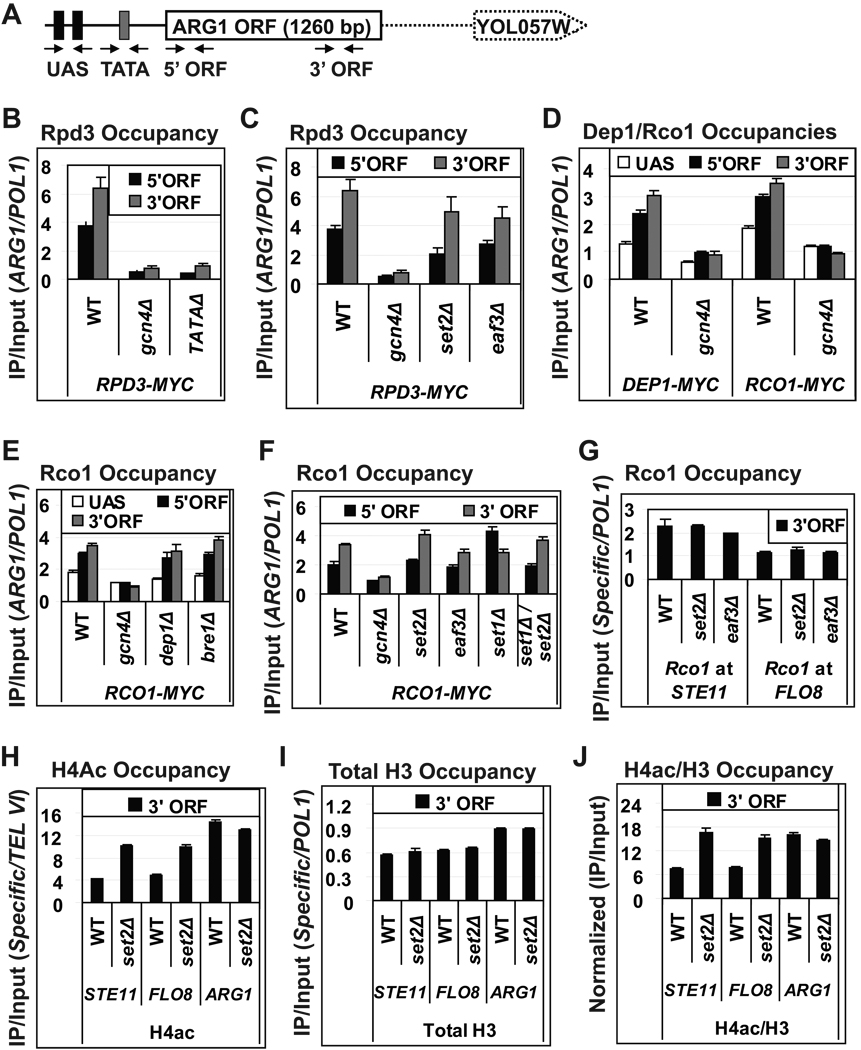
In agreement with our previous results, transcriptional induction of ARG1 by starvation for isoleucine and valine increases the occupancy of Rpd3 in the ORF, particularly near the 3’ end (Fig. 1A–B). Rpd3 occupancies were close to background levels in a gcn4Δ mutant (Fig. 1B), indicating a strict dependence on activation by Gcn4 for high-level Rpd3 association with ARG1 in vivo. To confirm that Rpd3 is recruited co-transcriptionally, we impaired PIC assembly by deleting the TATA element at ARG1 (TATAΔ), which impairs PIC assembly and reduces Pol II occupancy in the ORF (Fig. S1A) (Qiu et al., 2005). The TATAΔ mutation essentially eliminates Rpd3 occupancy in the ORF (Fig. 1B), establishing that Rpd3 association with ARG1 coding sequences requires transcription. We showed previously that the occupancies of Gcn4, TBP, and Mediator were confined to the UAS/promoter of induced ARG1 (Govind et al., 2007).
Fig. 1. Co-transcriptional recruitment of Rpd3C(S) is independent of H3-K36 methylation and Eaf3.
(A) Location of primers for ChIP analysis of ARG1. (B–F) Occupancies of Rpd3-myc, Dep1-myc and Rco1-myc at ARG1 were measured by ChIP in the indicated strains cultured in SC medium and treated with sulfometuron methyl (SM) for 30 min to induce Gcn4. Cross-linked chromatin was immunoprecipitated with anti-myc antibodies and DNA from immunoprecipitated (IP) and input samples was subjected to PCR in the presence of [33P]-dATP to amplify radiolabeled fragments of ARG1 or the POL1 ORF, analyzed as a control, which were resolved by PAGE and quantified by phosphorimaging. Ratios of ARG1 to POL1 signals in IP samples were normalized to the ratios for Input samples to yield occupancy values. (G) Occupancies of Rco1-myc at the 3’ ORF regions of STE11 and FLO8 measured by ChIP in the indicated strains. (H–I) Occupancies of acetylated H4 (H4Ac) and total H3 at the 3’ ORFs of STE11, FLO8 and ARG1 were measured by ChIP in cells grown in YEPD, using primers for these genes and non-transcribed sequences at the right-arm telomere of VI (TEL VI) (H) or POL1 (I). (J) H4Ac occupancies from (H) normalized for total H3 (I). All error bars represent the standard error of mean (SEM). For Rpb3 occupancies in TATAΔ and set2Δ see Fig. S1A.
Histone deacetylation by Rpd3C(S) requires interaction of the Eaf3 chromodomain (CD) with methylated H3-K36, such that a set2Δ or eaf3Δ mutation leads to increased histone acetylation in coding sequences (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). Surprisingly, however, both set2Δ and eaf3 Δ mutations produced relatively small decreases in Rpd3 occupancy at ARG1 compared to that given by gcn4Δ (Fig. 1C). These findings could indicate that Rpd3 is recruited to ARG1 primarily in the context of Rpd3C(L), which does not contain Eaf3 (Carrozza et al., 2005; Keogh et al., 2005), and we did observe Gcn4-dependent recruitment of an Rpd3C(L) subunit (Dep1) absent in Rpd3C(S) (Fig. 1D). However, we saw comparable Gcn4-dependent recruitment of a subunit specific to Rpd3C(S), Rco1 (Fig. 1D), indicating that Gcn4 recruits both Rpd3C(S) and Rpd3C(L) to ARG1. Rco1 occupancy was unaffected by eliminating Dep1 and by the bre1Δ mutation (Fig. 1E), which impairs H3-K4 methylation by Set1 (Hwang et al., 2003; Wood et al., 2003). Surprisingly, however, Rco1 occupancy was also unaffected by deleting SET2, even in the absence of SET1, and only slightly reduced by the eaf3 Δ mutation compared to the strong reduction given by gcn4Δ (Fig. 1F). These findings indicate that interaction of the Eaf3 CD with H3-K36me2/3 is not essential for co-transcriptional recruitment of Rpd3C(S) to the ARG1 ORF.
It was shown that deleting SET2 increases histone acetylation at the constitutively transcribed genes STE11 and FLO8, owing to impaired Rpd3C(S) function (Carrozza et al., 2005). However, similar to our findings at ARG1, we observed little reduction in Rco1 occupancy at either of these genes on deletion of SET2 or EAF3 (Fig. 1G). In contrast to the lack of any effect on Rco1 recruitment, H4 acetylation at the 3’ ends of both STE11 and FLO8 was elevated in set2Δ cells (Fig. 1H). Eliminating SET2 had no effect on occupancy of total H3 (Fig. 1I), so that the level of H4 acetylation per nucleosome (H4ac/H3 occupancy ratio) was increased at both STE11 and FLO8 in set2Δ cells (Fig. 1J). Thus, while Set2 is crucial for deacetylation of ORF nucleosomes at STE11 and FLO8, as observed previously (Carrozza et al., 2005), it is dispensable for Rpd3C(S) recruitment at these genes. In contrast to STE11 and FLO8, set2Δ does not increase H4 acetylation at ARG1 (Fig. 1H–J), consistent with our previous findings on an rpd3Δ mutant. This is due to the fact that multiple HDACs function redundantly to regulate the acetylation level at ARG1 (Govind et al., 2007).
Co-transcriptional recruitment of Rpd3C(S) to ARG1 is stimulated by Pol II CTD kinase Kin28
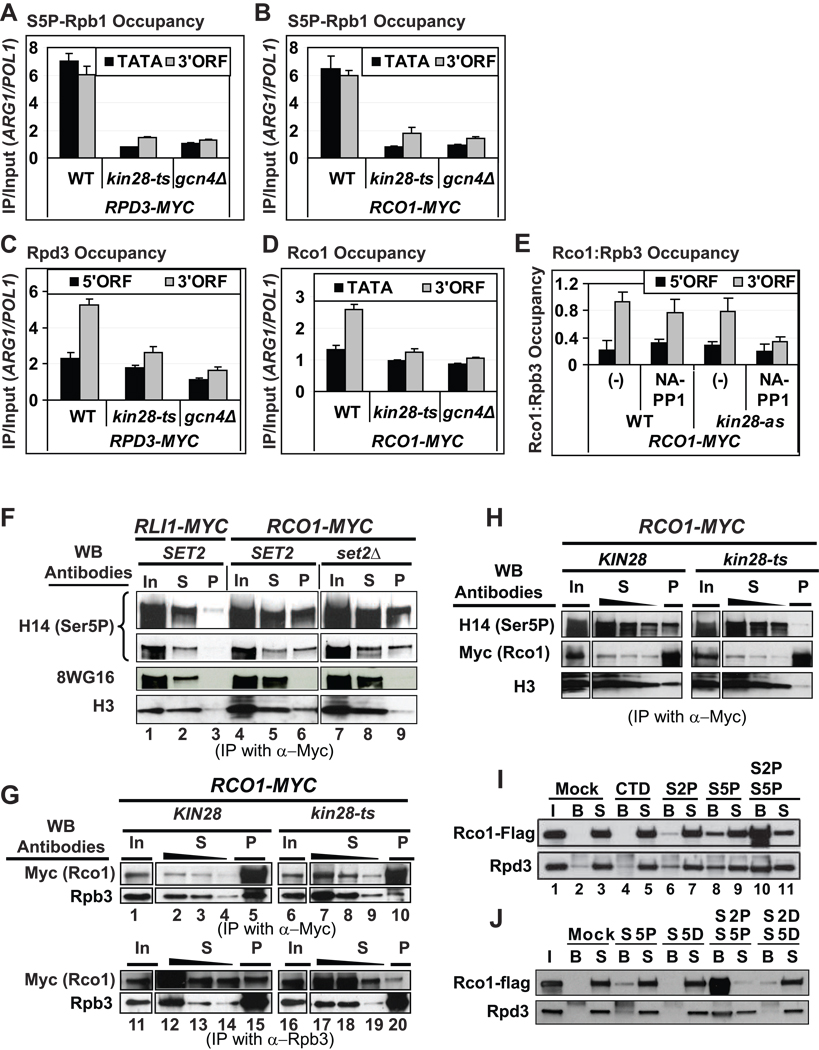
Having found that co-transcriptional recruitment of Rpd3C(S) is independent of Set2 and Eaf3, we asked whether phosphorylation of the Pol II CTD is involved. Kinase CDK7/Kin28 of TFIIH phosphorylates Ser5 of the heptad repeats in the CTD to stimulate recruitment of various factors involved in elongation (Phatnani and Greenleaf, 2006), including mRNA capping enzyme (Cho et al., 1997), Set1 (Ng et al., 2003), SAGA (Govind et al., 2007), Paf1C (Qiu et al., 2006), and Bur1 (Qiu et al., 2009). In accordance with previous results (Govind et al., 2007; Pascual-Garcia et al., 2008; Qiu et al., 2006), ChIP analysis with antibodies against Ser5-phosphorylated Rpb1 (Ser5P) revealed that Ser5P occupancies in promoter and ORF locations are strongly reduced in the kin28-ts16 mutant at the restrictive temperature of 37°C (Fig. 2A). This reduction occurred without any decrease in occupancy of total Pol II, as indicated by ChIP analysis of Rpb3 (Fig. S1B). Importantly, Rpd3 occupancy in the 3’ ORF was substantially reduced in the kin28-ts16 cells at 37°C, to nearly the same extent as in gcn4Δ cells (Fig. 2C). Similar results were obtained for the Rpd3C(S) subunit Rco1 (Fig. 2B & D, S1C).
Fig. 2. Co-transcriptional recruitment of Rpd3(S) is stimulated by Kin28-dependent CTD phosphorylation.
(A–D) ChIP analyses of Ser5-phosphoryated Rpb1 (S5P-Rpb1) (A–B), Rpd3-myc (C), and Rco1-myc (D) occupancies in WT, kin28-ts16 or gcn4Δ strains harboring RPD3-myc (A & C) or RCO1-myc (B & D). Cells were cultured in SC at 25°C, transferred to 37 °C for 30 min prior to treatment with SM for another 30 min, and then subjected to ChIP. (E) ChIP analyses of Rco1-myc and Rpb3 in KIN28 RCO1-myc and kin28-as RCO1-myc strains cultured in SC medium lacking Ile and Val and treated with NA-PP1 at 12 µM for 42 min and with SM for the last 30 min of the NA-PP1 treatment. (F) WCEs of WT or set2Δ strains harboring RLI1-myc or RCO1-myc cultured in YEPD were immunoprecipitated with anti-myc antibodies and immune complexes were probed by Western blot (WB) with antibodies against Ser5P (H14), hypophosphorylated Rpb1 (8WG16), or histone H3. Input (In), supernatant (S) and pellet (P) fractions are marked. Two exposures are shown for the H14 blots. (G) Top two panels: WCEs of isogenic KIN28 or kin28-ts16 strains harboring RCO1-myc, cultured in YEPD at 25°C and transferred to 37 °C for 1 h, were immunoprecipitated with anti-myc antibodies and immune complexes probed with the indicated antibodies. Bottom panels: same as the top except that anti-Rpb3 antibodies were used for immunoprecipitation. (H) WCEs from KIN28 RCO1-myc or kin28-ts16 RCO1-myc cells cultured as in (G) were immunoprecipitated with anti-myc antibodies and probed with the indicated antibodies. (I) Biotinylated CTD peptides (1.5µg) unphosphorylated (CTD) or phosphorylated on Ser5 (S5P), Ser2 (S2P), or both residues (S2P S5P) were adsorbed to streptavidin-coated magnetic beads and incubated at 4°C with reconstituted Rpd3C(S) expressed in insect cells. Mock: incubations with beads alone. Bound (B) and unbound, supernatant proteins (S) were subjected to SDS-PAGE and Western analysis with anti-Flag or Rpd3 antibodies. (J) Same as (H) except including CTD peptides with Asp replacing Ser2 or Ser5. All error bars represent the SEM. For Rpb3 occupancies in kin28-ts see Fig. S1B–C.
We conducted similar experiments using the kin28-as mutant, whose kinase activity can be chemically inactivated by the inhibitor NA-PP1 (Liu et al., 2004). We showed previously that treatment of kin28-as cells with NA-PP1 reduces Gcn4p–dependent occupancy of Ser5P in the ARG1 ORF with little effect on Rpb3 occupancy (Qiu et al., 2006). Findings from other labs confirm that inhibiting Kin28 activity in kin28-as cells has relatively little effect on Pol II occupancy or transcription (Hong et al., 2009; Kanin et al., 2007). Importantly, inhibition of kin28-as with NA-PP1 reduced Rco1-myc occupancy without decreasing Rpb3 levels and, hence, reduced the Rco1:Rpb3 ratio in the ARG1 3’ORF (Fig. 2E). Together, these findings suggest that co-transcriptional recruitment of Rpd3C(S) is stimulated by Pol II CTD phosphorylation by Kin28.
To provide independent evidence for this last conclusion, we asked whether Rpd3C(S) is physically associated with Ser5P Pol II in vivo. To this end, we immunoprecipitated myc-tagged Rco1 and probed immune complexes with antibodies against Ser5P (H14), hypophosphorylated Rpb1 (8WG16), or histone H3. Ser5P, but not hypophosphorylated Rpb1, coimmunoprecipitated with myc-tagged Rco1, and not with an unrelated myc-tagged protein involved in protein synthesis (Rli1) (cf. pellet fractions in lanes 3, 6, and 9 of top three panels in Fig. 2F). The interaction between Rco1 and Ser5P was unaffected by set2Δ (Fig. 2F, Ser5P, lanes 6 vs. 9). By contrast, coimmunoprecipitation of Rpb3 with myc-Rco1 was greatly reduced by kin28-ts16 (Fig. 2G, top, Rpb3 panel, lane 10 vs. 5). The amount of myc-Rco1 that coimmunoprecipitated with Rpb3 was likewise reduced by kin28-ts16 (Fig. 2G, bottom, Rco1 panel, lane 15 vs. 20). These results support the idea that Rpd3C(S) is recruited by Kin28-phosphorylated Pol II in a manner independent of Set2.
Interestingly, histone H3 also specifically coimmunoprecipitates with myc-Rco1, but in contrast to the Rco1-Ser5P interaction, Rco1-H3 association was reduced by set2Δ (Fig. 2F, bottom panel, lane 6 vs. 9). These findings fit well with in vitro demonstrations that interaction of Rpd3C(S) with nucleosomes is enhanced by methylated H3-K36 (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Li et al., 2007a). Importantly, Rco1 interaction with H3 is also enhanced by Kin28, being reduced in the kin28-ts16 mutant (Fig. 2H, bottom panel). This finding implies that association of Rpd3 with Ser5P is necessary for its interaction with methylated nucleosomes. Together, these results suggest the intriguing model that Rpd3C(S) is recruited to transcribed coding sequences by Kin28-phosphorylated Pol II, but the Eaf3 and Rco1 subunits of Rpd3C(S) must interact with H3-K36Me2/3 for stable association with nucleosomes and histone deacetylation by Rpd3.
To test whether interaction between Rpd3C(S) and the phosphorylated Pol II is direct, we reconstituted a recombinant Rpd3C(S) using a baculovirus expression system, and the reconstituted complex was purified via FLAG-tagged Rco1. We then examined binding of this complex to biotinylated peptides containing 3 CTD repeats phosphorylated on Ser2, Ser5, or both residues, plus a non-phosphorylated CTD peptide. As shown in Fig. 2I, both Flag-tagged Rco1 and Rpd3 showed no binding above background to unphosphorylated peptides, much greater binding to peptides singly-phosphorylated on Ser5 versus Ser2, and the strongest binding to Ser2-, Ser5- diphosphorylated peptides. To establish the specificity of this interaction, we tested binding of purified Rpd3C(S) to peptides containing aspartate in place of Ser5 or both Ser2,Ser5. The Asp-substituted peptides showed little binding of Rpd3C(S) at the same concentrations where Ser5P, or Ser2P,Ser5P peptides showed moderate or strong binding, respectively (Fig. 2J). These results eliminate the possibility that binding of Rpd3C(S) to phosphorylated peptides merely reflects non-specific interaction with negatively charged residues. Together, the results in Fig. 2I–J support the idea that Rpd3C(S) is recruited directly by the Rpb1 CTD phosphorylated on Ser5 and Ser2.
Co-transcriptional recruitment of Hos2 and Hda1 to ARG1 is independent of histone methylation
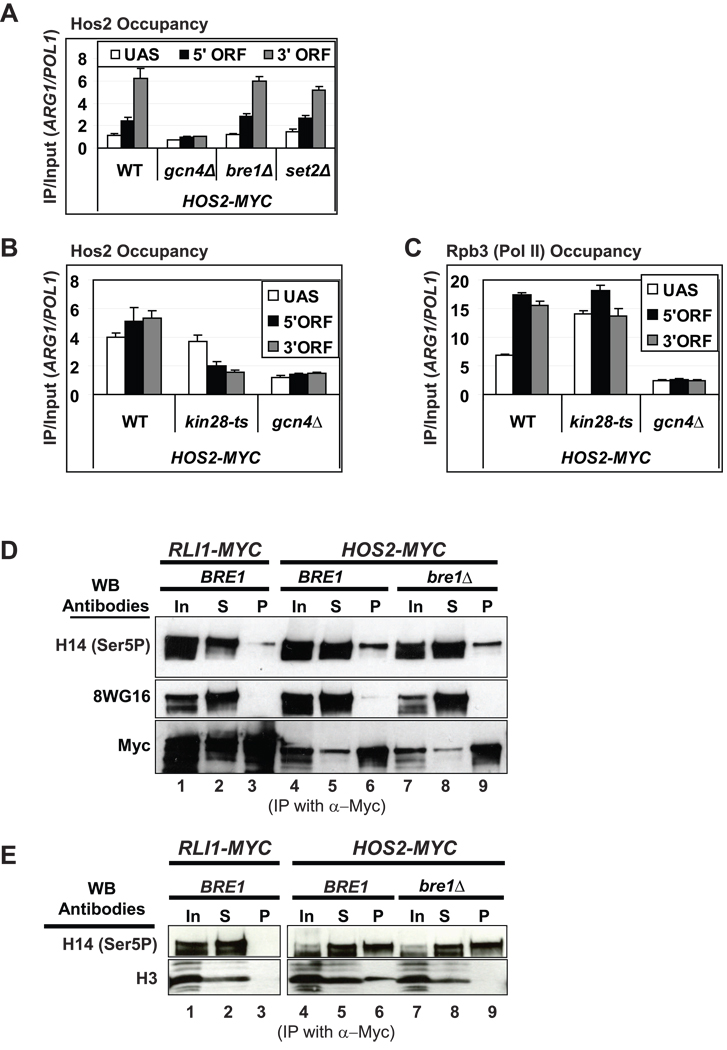
The HDA Hos2 is also recruited to ARG1 coding sequences dependent on Gcn4 (Fig. 3A) (Govind et al., 2007), and we found that deletion of the TATA element reduces Hos2 occupancy of the ARG1 ORF (data not shown). It was shown recently that recruitment of Hos2/Set3C to YEF3 coding sequences is mediated by interaction of the plant homeodomain (PHD) in Set3 with H3-K4me2 produced by Set1 (Kim and Buratowski, 2009). Unexpectedly, however, we found no effect of the bre1Δ mutation on Hos2 occupancy at ARG1 (Fig. 3A), suggesting that recruitment of Set3C to this gene is independent of H3-K4 methylation. As expected, elimination of SET2 also has little effect on Hos2 occupancy at ARG1 (Fig. 3A).
Fig. 3. Co-transcriptional recruitment of Hos2 is stimulated by Kin28.
(A–C) Occupancies of Hos2-myc and Rpb3 at ARG1 measured by ChIP under Gcn4-inducing conditions (D–E) WCEs were immunoprecipitated with anti-myc antibodies and immune complexes probed with the antibodies listed on the left. All error bars represent the SEM.
We examined whether Set3C recruitment to ARG1 might follow a mechanism similar to that elucidated above for Rpd3C(S) involving Kin28. Supporting this possibility, myc-Hos2 occupancy at ARG1 was reduced by the kin28-ts16 mutation (Fig. 3B) without a significant reduction in occupancy of Rpb3 (Pol II) (Fig. 3C). Moreover, myc-Hos2 specifically coimmunoprecipitates with Ser5P, but not hypophosphorylated Rpb1, and Hos2-Ser5P association is retained in bre1Δ cells lacking H3-K4me2/3 (Fig. 3D, cf. lanes 3, 6, and 9). Importantly, H3 also coimmunoprecipitates with Hos2 and this interaction is abolished by bre1Δ (Fig. 3E). These results are consistent with the idea that Set3C is recruited by phosphorylated Pol II but is dependent on Bre1/Set1-dependent dimethylation of H3-K4 for interaction with nucleosomes. Thus far, we have been unable to demonstrate association of Set3C, affinity-purified from a HOS2-TAP strain, with phosphorylated CTD peptides (data not shown). Hence, it is possible that Set3C associates with elongating Pol II indirectly, through some other component of the elongation complex that binds directly to the Ser5-phosphorylated CTD.
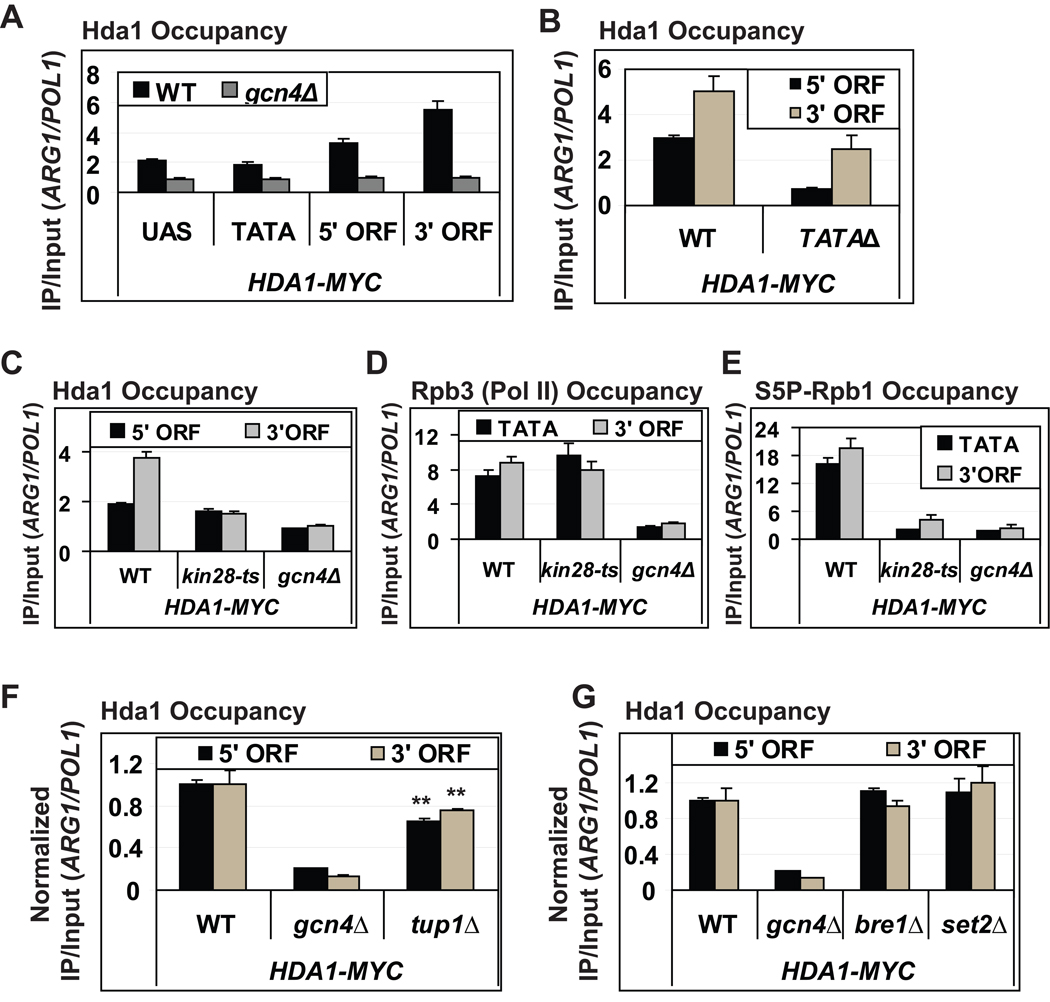
It was shown previously that HDAs Hda1 and Rpd3 cooperate in regulating acetylation levels and transcriptional control of PHO5 (Vogelauer et al., 2000). Hence, we tested whether Hda1 is also recruited co-transcriptionally to ARG1 dependent on Kin28. Indeed, we observed Gcn4-dependent association of Hda1 with ARG1 that was greatest at the 3’ end (Fig. 4A) and diminished by deletion of the TATA element (Fig. 4B). As described above for Rpd3C(S) and Set3C, Hda1 occupancy was reduced in kin28-ts16 cells in parallel with the decreased Ser5P content of Pol II (Fig. 4C–E). Consistent with previous evidence that recruitment of Hda1 at certain genes requires Tup1 (Robyr et al., 2002; Wu et al., 2001), we observed a small, but significant reduction in Hda1 occupancies at both ends of the ARG1 ORF in tup1 Δ cells (Fig. 4F). Elimination of BRE1 or SET2 had no impact on Hda1 occupancy at ARG1 (Fig. 4G), however, indicating that H3-K4me2/3 and H3-K36me2/3 are dispensable for Hda1 recruitment. We conclude that co-transcriptional recruitment of Hda1 to ARG1 is weakly dependent on Tup1, but resembles the recruitment of Rpd3C(S) and Set3C at this gene in showing strong dependence on Kin28. All together, our data indicate the importance of phosphorylated Pol II, and the non-essential role of H3 methylation, in co-transcriptional targeting of HDAs to the ARG1 coding sequence.
Fig. 4. Co-transcriptional recruitment of Hda1 toARG1 is stimulated by Kin28.
(A–B & F–G) Occupancies of Hda1-myc at ARG1 measured by ChIP. **; p value calculated using student’s t test is < 0.005. (C–E) Occupancies of Hda1 (C) Rpb3 (Pol II; D) and S5P-Rpb1 (E) measured by ChIP. All error bars represent the SEM.
Cooperation between multiple HDAs in histone deacetylation and co-transcriptional histone eviction at ARG1
We provided evidence previously that recruitment of SAGA to coding sequences, and attendant H3 acetylation by Gcn5, facilitates co-transcriptional eviction of nucleosomes from coding sequences. Thus, gcn5Δ dampens the eviction of H3 from the GAL1 ORF during induction by galactose. By contrast, gcn5Δ did not affect H3 levels at induced ARG1 (Govind et al., 2007), and we speculated that the multiple HDAs recruited to this gene might impede histone eviction by counteracting the KAT activity of Gcn5. If so, we would expect to find that H3 eviction from ARG1 would be revealed in mutant cells lacking these HDAs.
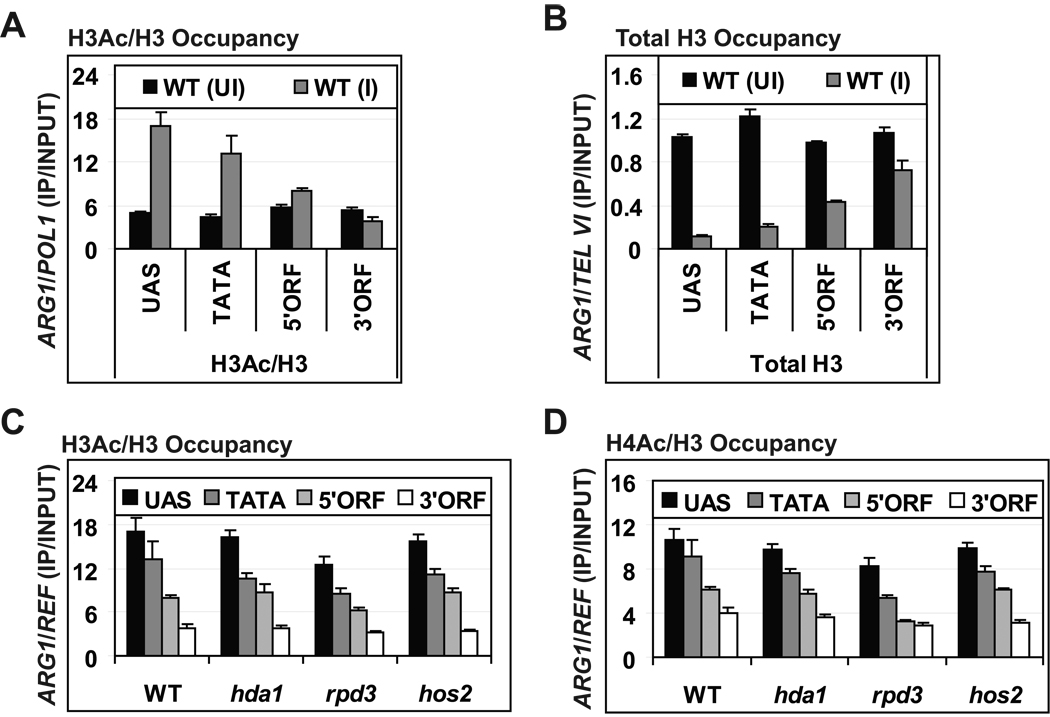
To test this, we first investigated whether the HDAs Rpd3, Hos2, and Hda1 make independent contributions to H3 and H4 acetylation at ARG1. In agreement with previous results (Govind et al., 2007), the H3Ac/H3 ratio increases dramatically in the UAS and promoter but is relatively unchanged or even decreases in the ARG1 ORF during induction by Gcn4 (Fig. 5A; uninduced (UI) vs. induced (I)). Total H3 shows the inverse pattern, being reduced in the UAS/promoter but not in the ORF on induction by Gcn4 (Fig. 5B). Similar results were obtained for acetylated H4 (Fig. 5D, WT lanes and data not shown). The inverse relationship between acetylation and histone occupancy supports the idea that histone eviction is enhanced by histone acetylation across ARG1.
Fig. 5. Multiple HDAs regulate co-transcriptional H3 and H4 acetylation in the ARG1 ORF.
(A–B) Occupancies of H3Ac, H4Ac or total H3 at ARG1 determined by ChIP in WT cells uninduced (UI) or induced (I) for Gcn4 and the ratio of H3Ac to H3 (A) or H3 (B) occupancies were calculated. H3Ac/H3 ratios (C) or H4Ac/H3 ratios (D) were determined by ChIP for WT and the indicated single HDA mutants. POL1 and TEL VI regions were used to normalize total H3, and acetylated H3 and H4 occupancies at ARG1, respectively. All error bars represent the SEM. Occupancies of H3Ac and H4Ac in double deletion mutants are shown in Fig. S2.
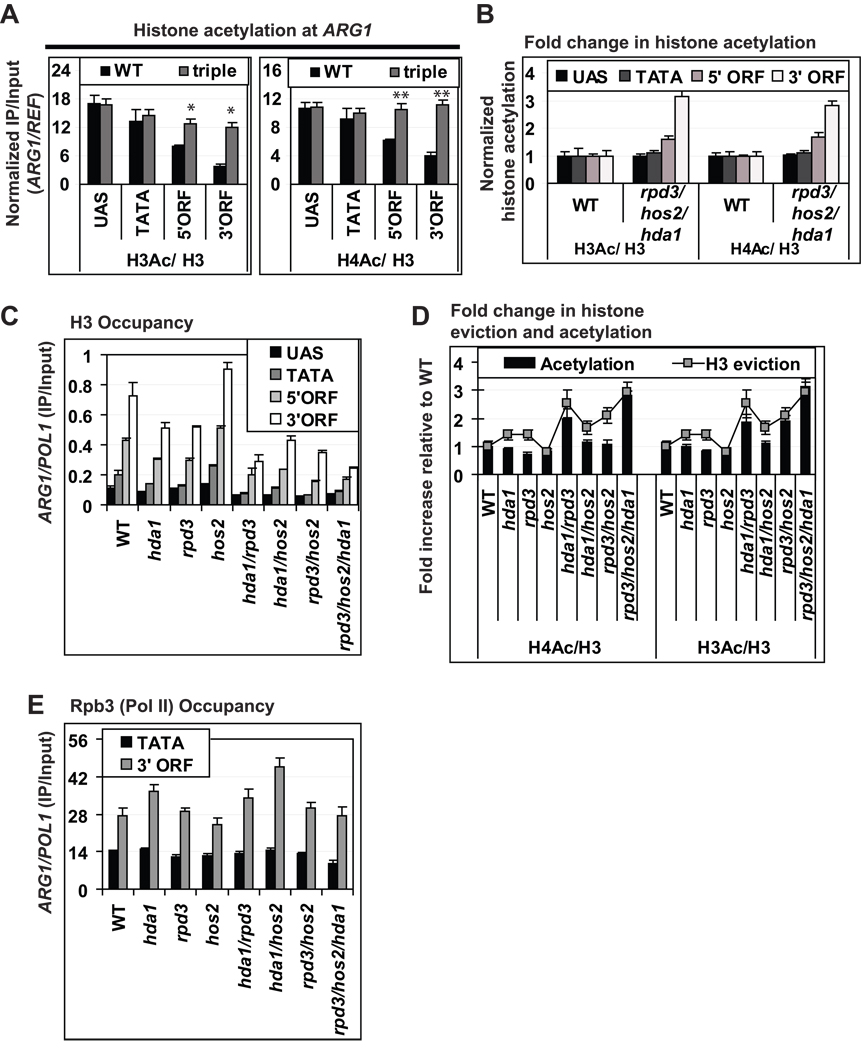
We then examined the effects on histone acetylation of eliminating the HDAs Hda1, Hos2 and Rpd3 in single, double or triple mutants. None of the mutants lacking single HDAs displayed any significant increase in H3Ac/H3 (Fig. 5C) or H4Ac/H3 (Fig. 5D) at any location at ARG1. In the three different double mutants, there were no significant changes in the H3Ac/H3 or H4Ac/H3 levels in the UAS or TATA region (Fig. S2A & C). However, statistically significant increases in the H3Ac/H3 or H4Ac/H3 ratios in the 5’ ORF were found in the double mutants lacking Hos2 (Fig. S2B & D), and the double mutants lacking Rpd3 displayed moderately higher H3Ac/H3 or H4Ac/H3 ratios in the 3’ ORF (Fig. S2B & D).
Importantly, the mutant lacking all three HDAs exhibits substantial (∼3-fold) increases in H3Ac/H3 and H4Ac/H4 ratios in the ARG1 3’ORF (Fig. 6A, both panels, 3’ORF), greater than observed in any double mutant. The same was generally true for the 5’ORF (Fig. 6A, both panels, 5’ORF). Thus, the three HDAs have overlapping functions in deacetylating H3 and H4 in ARG1 coding sequences during activation by Gcn4. As illustrated in Fig. 6B, the HDAs exert their greatest influence in the 3’ORF and have little effect on histone acetylation in the UAS or promoter, consistent with the fact that the UAS/TATA are already the most highly acetylated regions of induced ARG1 in WT cells (Fig. 5A & D).
Fig. 6. Direct correlation between co-transcriptional acetylation and eviction of histones in ARG1 coding sequences.
(A) H3Ac/H3 ratios (left) or H4Ac/H3 ratios (right) at ARG1 determined by ChIP for WT and the rpd3Δ hos2Δ hda1Δ triple mutant. *; p value < 0.01 and **; p value < 0.005. (B) H3Ac/H3 and H4Ac/H3 ratios in (A) were normalized to the values in WT cells at each location at ARG1. (C) H3 occupancies at ARG1 measured by ChIP. (D) H3Ac/H3 and H4Ac/H3 ratios determined in (C) and normalized to the WT ratios plotted as a histogram (Acetylation), along with the fold increase in H3 eviction as a line graph (H3 eviction) calculated as the reciprocal of the mutant total H3 occupancies normalized to the WT H3 occupancy. (E) Rpb3 (Pol II) occupancies at ARG1 determined by ChIP. All error bars represent the SEM.
Quantification of total H3 in the HDA mutants reveals that these three HDAs also act redundantly to suppress histone eviction from the coding sequences during ARG1 activation. The hda1 Δ and rpd3Δ single mutants show moderate reductions in H3 occupancy, especially in the ORF, whereas the hos2Δ strain is almost indistinguishable from WT (Fig. 6C, 1st four strains). All three double mutants display marked reductions in H3 occupancy, and the triple mutant shows the largest effect of all, particularly in the 3’ORF of ARG1 (Fig. 6C, last four strains). The graph in Fig. 6D illustrates the robust correlation between levels of H3Ac/H3 or H4/Ac and histone eviction in the 3’ORF for the set of HDA mutants we examined. The increased H3 eviction in the HDA mutants cannot be attributed to increased transcription, because their Pol II (Rpb3) occupancies in the 3’ ORF (and TATA region) of ARG1 vary little from the WT level (Fig. 6E). These Pol II levels correlated well with the ARG1 mRNA levels in these mutants determined by Northern analysis (data not shown). Together, these results indicate that the HDAs act to minimize co-transcriptional histone eviction by preventing the increased acetylation of H3 and H4 that would otherwise occur during transcriptional activation.
Distinct patterns of HDA function at different genes
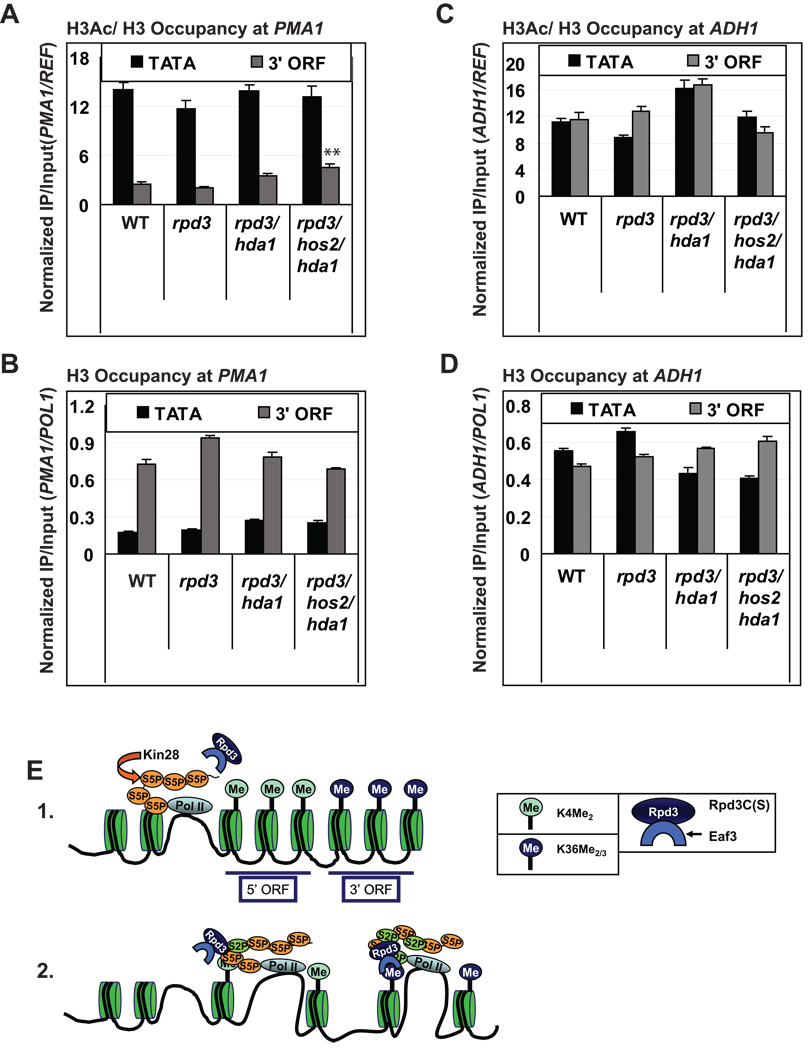
To determine whether the strong reduction in histone occupancy in the induced ARG1 ORF in the triple HDA mutant is a general phenomenon, we examined the effects of HDA mutations on histone acetylation and occupancy at constitutively transcribed PMA1 and ADH1. Similar to induced ARG1, in WT cells PMA1 exhibits a higher level of H3 acetylation and lower total H3 occupancy at the promoter versus the 3’ORF (Fig. 7A and 7B, WT cells, TATA vs. 3’ORF). Unlike ARG1, however, inactivating Rpd3, Hos2 and Hda1 in the triple mutant produced only a small (∼50%) increase in the H3Ac/H3 ratio and no significant increase in H3 eviction (Fig. 7A and 7B, WT vs. triple mutant). Presumably, either KAT complexes are relatively sparse or inactive in the PMA1 ORF, or another HDA besides these three is sufficient for deacetylation at this gene.
Fig. 7. Histone acetylation and H3 occupancy at constitutively expressed genes, and a model for Rpd3C(S) recruitment by the phospho-CTD and methylated H3 tails.
(A–D) Occupancies of H3Ac/H3 (A and C, **; p value < 0.005) and H3 (B and D) at PMA1 (A and B) and ADH1 (C and D) determined by ChIP. (E) Model for two-stage co-transcriptional recruitment of Rpd3C(S). (1) Pol II is phosphorylated on Ser5 of the CTD by Kin28 during promoter clearance. Previous rounds of transcription leave ORF nucleosomes methylated on H3-K4 by Set1, more heavily towards the 5’ end, and on H3-K36 by Set2, more heavily towards the 3’ end. (2) Elongating Ser5-phosphorylated Pol II is phosphorylated on Ser2 by Bur1 and Ctk1, enabling efficient recruitment of Rpd3C(S) by direct interaction with Ser2-, Ser5-diphosphorylated CTD repeats. Subsequently, Rpd3C(S) is transferred from the phospho-CTD to H3 tails by interaction of H3-K36me2 with the chromodomain in Eaf3 and PHD finger in Rco1 (not depicted) for subsequent histone deacetylation. All error bars represent the SEM.
The ADH1 gene differed from ARG1 and PMA1 in showing high H3Ac/H3 and total H3 levels in both promoter and 3’ORF sequences in WT cells (Fig. 7C–D, WT). Moreover, inactivation of the HDAs produced little change in either parameter (Fig. 7C–D, triple mutant vs. WT). The high-level acetylation in the ADH1 ORF in WT cells (Fig. 7C) could reflect abundant KAT complexes or a paucity of HDAs in the ADH1 coding sequences. The high-level histone occupancy in the ADH1 ORF implies either the absence of factors required for evicting acetylated nucleosomes or an elevated level of histone chaperones that efficiently replace nucleosomes in the wake of elongating Pol II, compared to the situation at ARG1. In any event, the pronounced, overlapping functions of Rpd3, Hos2, and Hda1 in deacetylating histones and impeding co-transcriptional histone eviction uncovered at ARG1 is not characteristic of all highly transcribed genes.
DISCUSSION
Previous evidence suggested that co-transcriptional recruitment of HDACs Rpd3C(S) and Set3C was driven primarily by their interactions with methylated histone H3 tails. Our results confirm the previous findings (Carrozza et al., 2005) that deleting SET2 increases histone acetylation at STE11 and FLO8 coding sequences (Fig. 1J). In addition, by coimmunoprecipitation studies we observed Set2-enhanced association of Rpd3C(S) subunit Rco1 with bulk H3 in native chromatin, consistent with requirements for the Eaf3 chromodomain and Rco1 PHD finger in association of Rpd3C(S) with nucleosomes or H3 tails methylated on H3-K36 by Set2 (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Li et al., 2007a). Thus, our data support the model that binding of Rpd3C(S) to H3-K36me2/3 is important for histone deacetylation of coding sequences in a subset of yeast genes.
Surprisingly, however, we found that co-transcriptional recruitment of Rco1 to the ARG1 ORF was unaffected by elimination of Set2 or Eaf3. There was previous evidence that recruitment of Eaf3 to a transcribed coding region was moderately enhanced by Set2 and H3-K36me2/3, and was strongly dependent on the Eaf3 CD (Carrozza et al., 2005). However, Eaf3 occupancy could reflect the presence of Rpd3C(S) or NuA4, as Eaf3 is also a component NuA4, which occupies transcribed ARG1 sequences (Ginsburg et al., 2009). Considering recent evidence that Rpd3C(S) interacts with methylated H3-K4 versus H3-K36me2/3 in certain settings (Pinskaya et al., 2009), it is noteworthy that we found no reduction in Rpd3C(S) occupancy at ARG1 in a set1Δ set2Δ double mutant (Fig. 1F). Thus, it appears that efficient recruitment of Rpd3C(S) to induced ARG1 does not require either of these H3 methylation marks.
Our finding that set2Δ does not impair association of bulk Ser5-phosphorylated Rpb1 with Rco1 (Fig 2F) suggests that the ChIP results we obtained for induced ARG1 could be typical of most highly transcribed genes, and that recruitment of Rpd3C(S) is not generally dependent on H3-K36me2/3. Strong support for this possibility is provided by recent findings from the Robert laboratory that eliminating Set2 has little effect on Rpd3C(S) occupancies at the majority of actively transcribed genes that normally contain this HDAC in the coding regions (Simon Drouin and Francois Robert, unpublished observations).
Interestingly, we found evidence that CTD phosphorylation is critical in recruiting Rpd3C(S) to coding regions. Recruitment of Rco1 to the ARG1 ORF was reduced by impairing CDK7/Kin28 in both kin28-ts16 and kin28-as cells, in a manner that cannot be explained by decreased Pol II occupancy. Furthermore, Rpd3C(S) specifically associates with bulk Ser5-phosphorylated Rpb1, but not with hypophosphorylated Rpb1, and Rco1’s association with bulk histones, as well as with Pol II, was impaired in a kin28-ts16 mutant. Finally, reconstituted Rpd3C(S) preferentially binds to CTD peptides phosphorylated on Ser5, and even more strongly to Ser5,Ser2-diphosphoryated peptides, but not to CTD peptides containing negatively charged Asp residues.
These findings and our ability to detect Rpd3C(S) at induced ARG1 in set2Δ cells suggest that Rpd3C(S) interacts with elongating Pol II and that its recruitment to transcribed coding sequences is more dependent on the phosphorylated CTD than on methylated H3-K36, and that association of Rpd3C(S) with the phospho-CTD stimulates its interaction with methylated histones. Hence, we propose that Rpd3C(S) is recruited to sites of transcription by Ser5-phosphorylated, elongating Pol II, and only its subsequent association with nucleosomes and attendant deacetylation function requires interaction of Eaf3 and Rco1 with K36-methylated H3 tails (see model in Fig. 7E).
Our finding that Rpd3S(C) binds more tightly to Ser5,Ser2-diphosphorylated CTD peptides compared to those phosphorylated only on Ser5 is consistent with the fact that Set2 contains a CTD-interaction domain that also binds preferentially to diphosphorylated CTD peptides (Kizer et al., 2005). Thus, Ser5,Ser2-diphosphorylated CTD repeats, which likely persist in elongating Pol II throughout the coding sequences of many genes (Phatnani and Greenleaf, 2006), would be the optimum substrate for recruiting both Rpd3C(S) and its activating H3 methyltransferase, Set2.
For Set3C, it was shown that eliminating Set1 or inactivating the Set3 PHD finger increased histone acetylation at the 5’ ends of multiple genes, and that stable association of Set3C with native nucleosomes or H3 peptides depends on Set1 and H3-K4me2. Furthermore, recruitment of Hos2/Set3C to YEF3 coding sequences required Set1 and the PHD in Set3 (Kim and Buratowski, 2009). Our finding that association of Hos2 with bulk H3 in native chromatin is eliminated by bre1Δ, which blocks Set1 function, is consistent with these previous findings. However, we found that bre1Δ did not reduce Hos2 occupancy at induced ARG1, nor abolish association of bulk Ser5P with Hos2. In addition, we found that Hos2 coimmunoprecipitated with Ser5P, but not with hypophosphorylated Rpb1. Our findings suggest that recruitment of Set3C to sites of transcription is not critically dependent on its physical association with methylated H3 tails, even though its ability to deacetylate nucleosomes is enhanced by H3-K4me2. Consistent with our results, Robert et al. have observed that the absence of H3-K4 methylation in set1 Δ cells has relatively little impact on genome-wide occupancies of Set3 analyzed by ChIP, including the coding sequences of YEF3 (Simon Drouin and Francois Robert, unpublished observations). Accordingly, the two-stage recruitment proposed above for Rpd3C(S) likely also applies to Set3C, except that the role of Kin28 and the phospho-CTD is probably indirect.
We speculate that the discrepancy between our results and those of Kim and Buratowski (2009) regarding the requirement for Set1 in Set3C recruitment might reflect differences in the epitope tags on the Set3C subunits, or the culture conditions or ChIP protocols, that in our experiments would enhance cross-linking of Set3C to components of elongating Pol II complexes, which in turn cross-link to nucleosomes or DNA. This would reduce the need for Set3C cross-linking to methylated histones for immunoprecipitation of transcribed coding sequences with tagged Set3C subunits. Under the conditions of Kim and Buratowski’s ChIP assays, by contrast, the proven interaction of Set3C with H3-K4me2 would be critical for efficient detection of Set3C in transcribed coding sequences.
The association of HDACs with elongating Pol II uncovered here is expected to enhance deacetylation of nucleosomes in transcribed regions simply by increasing the local concentration of HDACs in the vicinity of methylated nucleosomes. We speculate that coupling the initial recruitment of HDACs to elongating Pol II rather than methylated H3 could also serve to prevent excessive deacetylation of coding sequences after they become transcriptionally repressed but still harbor the methylated H3 deposited prior to repression. In addition, our finding that methylated histone tails are not critical for recruiting HDACs to sites of transcription opens up the possibility that interaction with methylated histones allosterically activates HDAC function.
A number of studies have shown that inactivation of a single HDAC or HDA, including Rpd3C(S), Set3C, or Hda1, was sufficient to increase acetylation in the coding sequences of various genes (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Li et al., 2007a; Li et al., 2007b; Vogelauer et al., 2000). It is striking, therefore, that none of the mutants lacking only one HDA increased H3 or H4 acetylation at induced ARG1, and eliminating all three simultaneously was necessary to observe a pronounced increase in acetylation at both the 5’ or 3’ ends of the ARG1 ORF. Thus, Rpd3, Hos2 and Hda1 have highly overlapping functions in deacetylating histones at ARG1. Because we found that Rpd3C(S), Rpd3C(L), Hos2, and Hda1 are co-transcriptionally recruited to induced ARG1, they probably all contribute directly to H3/H4 deacetylation at this gene.
Importantly, we observed a strong correlation between the increases in H3/H4 acetylation per nucleosome and the degree of histone eviction in the 3’ ORF of ARG1 that occurs during its induction by Gcn4 among single, double and triple HDA mutants (Fig. 6D). These findings strongly support the notion that co-transcriptional nucleosome eviction is enhanced by histone acetylation. Consistent with our results, inactivation of Set3C and attendant increased acetylation of 5’ ORF regions was often associated with lower H3 density (Kim and Buratowski, 2009). Furthermore, we found previously that H3 acetylation in GAL1 coding sequences by Gcn5 contributes to the pronounced co-transcriptional eviction of nucleosomes that occurs on galactose induction of this gene (Govind et al., 2007).
The increased nucleosome eviction associated with histone hyperacetylation in the triple HDA mutant could involve a higher rate of nucleosome disassembly, or a lower rate of nucleosome reassembly in the wake of elongating Pol II. Acetylation could facilitate nucleosome disassembly by reducing electrostatic interactions of the histone tails with DNA (Protacio et al., 2000), or by enhancing recruitment of chromatin remodeling complexes bearing bromodomains, without increasing the number of Pol II molecules in the coding sequences. Indeed, nucleosome remodeling complex SWI/SNF stimulates nucleosome eviction in vitro (Gutierrez et al., 2007) and its binding to reconstituted nucleosomes is stimulated by histone acetylation, via the Snf2 bromodomain (Hassan et al., 2002). Furthermore, SWI/SNF occupies transcribed coding sequences and facilitates co-transcriptional nucleosome eviction in the ORF, as well as in the UAS, at SUC2 in vivo (Schwabish and Struhl, 2007). Histone acetylation also enhances recruitment of RSC and its ability to stimulate Pol II elongation through nucleosomes (Carey et al., 2006; Mas et al., 2009), and we found recently that RSC recruitment to transcribed coding sequences is enhanced by NuA4 and SAGA (Ginsburg et al., 2009). The histone chaperone Asf1 stimulates nucleosome disassembly during transcription elongation (Schwabish and Struhl, 2006), which might be attributable to its role in H3-K56 acetylation by Rtt109 (Tsubota et al., 2007), a modification that weakens H3-DNA interaction and promotes nucleosome disassembly at the PHO5 promoter (Williams et al., 2008). Although it is also possible that nucleosome reassembly is reduced by histone hyperacetylation in the triple HDA mutant, we are not aware of any corroborating evidence for this alternative mechanism.
Our finding that recruitment of multiple HDACs is required to prevent excessive nucleosome eviction by elongating Pol II is consistent with previous findings that inactivation of Rpd3C(S) results in activation of cryptic promoters (Carrozza et al., 2005; Li et al., 2007b), a phenomenon first described in spt6 and spt16 mutants (Kaplan et al., 2003; Mason and Struhl, 2003) that are defective in nucleosome reassembly (Kaplan et al., 2003; Schwabish and Struhl, 2004; Sims et al., 2004). We have not observed activation of any cryptic promoters at ARG1 in the HDAC mutants (data not shown), but this might be attributable to the high density of elongating Pol II molecules traversing the ORF during its induction by Gcn4, or the lack of any cryptic promoters within the ARG1 coding region. It seems likely that the unchecked nucleosome eviction in coding regions occurring in the absence of all three HDAs will most likely derepress cryptic promoters at certain Gcn4 target genes yet to be identified, perturbing the regulation of their canonical promoters, and might also render the exposed DNA more susceptible to damage and recombination (Merker et al., 2008).
MATERIALS AND METHODS
Yeast strain constructions
All yeast strains are listed in Table S1 and their derivations are described in the Supplementary Materials.
Chromatin immunoprecipitation experiments
Chromatin was prepared from HCHO cross-linked cells, cultured as indicated in the figure legends, and immunoprecipitated using the following antibodies: anti-myc (Roche) for myc-tagged proteins, H14 (Covance) for Ser5-phosphorylated Rpb1, and anti-Rpb3 (Neoclone); following which the extracted DNA was subjected to PCR using the primers in Table S2, as described in Supplementary Information. For each primer set, we optimized the PCR to ensure that the amounts of amplified 33P-labeled products are proportional to amounts of input DNA over the range of concentrations of the relevant sequences in total or immunoprecipitated chromatin.
Coimmunoprecipitation experiments
Whole cell extracts were prepared from cells grown in YPD medium (Sherman et al., 1974) as described previously (Ginsburg et al., 2009) and 1–1.5 mg of WCE were immunoprecipitated using anti-myc, or anti-Rpb3 antibodies, as described in the Supplementary Materials, and immune complexes were subjected to Western analysis with anti-myc, H14, 8WG16 (Covance), or anti-H3 (Abcam ab1791) antibodies.
Peptide binding assay
Five baculoviruses were constructed, each encoding a different subunit of Rpd3S, including Sin3, Ume1, Rpd3 and Eaf3, and FLAG-tagged Rco1, and used to co-infect Sf21 insect cells. The recombinant Rpd3S(C) was purified with anti-FLAG affinity resin and shown to contain stoichiometric amounts of each subunit and to elute from a gel filtration column at a the same volume observed for the native complex (Ruan and Li, unpublished observations). Peptide binding assays were conducted essentially as described previously (Li et al., 2003) with some modifications (Qiu et al., 2009), using biotinylated CTD peptides purchased from AnaSpec.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joe Reese and Steven Hahn for HDA and kin28-as mutant strains and David Stillman for Rpd3 antibodies. This work was supported in part by the Intramural Research Program of the NIH. B.L. is supported by the Welch Foundation (I-1713) and the American Heart Association and is a W.A. "Tex" Moncrief, Jr. Scholar in Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. Embo J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg DS, Govind CK, Hinnebusch AG. The NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009 doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. Embo J. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc Natl Acad Sci U S A. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci U S A. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007a;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007b;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, Workman JL., 3rd The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G, de Nadal E, Dechant R, de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. Embo J. 2009;28:326–336. doi: 10.1038/emboj.2008.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD. The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7:1298–1308. doi: 10.1016/j.dnarep.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M, Vanti M, Chavez S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. Febs J. 2006;273:756–769. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. Embo J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protacio RU, Li G, Lowary PT, Widom J. Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol Cell Biol. 2000;20:8866–8878. doi: 10.1128/mcb.20.23.8866-8878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol. 2005;25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Tomar RS, Dempsey AE, Reese JC. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol. 2007;27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence CW. Methods of yeast genetics. Cold Spring Harbor Laboratory. 1974:61–64. [Google Scholar]
- Sims RJ, Belotserkovskaya R, 3rd, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr Opin Genet Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M. TUP1 utilizes histone H3/H2B–specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.