Abstract
The opportunistic human fungal pathogen Candida albicans is a major cause of nosocomial infections. One of the fundamental features of C. albicans pathogenesis is the yeast-to-hypha transition. Hypha formation is controlled positively by transcription factors such as Efg1p and Cph1p, which are required for hyphal growth, and negatively by Tup1p, Rfg1p, and Nrg1p. Previous work by our group has shown that modulating NRG1 gene expression, hence altering morphology, is intimately linked to the capacity of C. albicans to cause disease. To further dissect these virulence mechanisms, we employed the same strategy to analyze the role of Rfg1p in filamentation and virulence. Studies using a tet-RFG1 strain revealed that RFG1 overexpression does not inhibit hypha formation in vitro or in the mouse model of hematogenously disseminated candidiasis. Interestingly, RFG1 overexpression drives formation of pseudohyphae under yeast growth conditions—a phenotype similar to that of C. albicans strains with mutations in one of several mitotic regulatory genes. Complementation assays and real-time PCR analysis indicate that, although the morphology of the tet-RFG1 strain resembles that of the mitotic regulator mutants, Rfg1p overexpression does not impact expression of these genes.
The opportunistic fungal pathogen Candida albicans is an important cause of human infection, especially in immunocompromised patients such as transplant recipients, chemotherapy patients, and those with HIV/AIDS. Mortality rates from C. albicans systemic infections range from 30 to 50% (44, 45). The ability of this fungus to reversibly convert between yeast, pseudohyphal, and true hyphal morphologies has been tied to the ability of this species to cause disease both in humans and in a murine model of disseminated candidiasis. The capacity to filament is particularly important for pathology in an infection; mutant strains that are locked either in the filamentous form (8, 9, 30) or in the yeast form (27, 38, 40) of growth show reduced virulence in the murine model of systemic candidiasis. Further, filamentous cells predominate in tissues recovered from patients succumbing to candidiasis and exposure to serum at normal body temperatures (37°C) induces the fungus to switch from the yeast to the hyphal form.
Genetic analysis has revealed that hypha formation in C. albicans is controlled by a number of transcription factors, including Efg1p and Cph1p, which can stimulate filamentation and the transcription of hypha-specific genes (6). These transcription factors induce hypha formation in response to environmental signals transduced by different signaling pathways, including the Cph1p-mediated mitogen-activated protein kinase (MAPK) and Efg1p-mediated cyclic AMP/protein kinase A pathways (7). The activity of these transcription factors is essential for both hypha formation and virulence in C. albicans, although under some growth conditions Efg1p is also required to repress hypha formation (43).
Several negative regulators of hypha formation have also been identified, including the DNA binding proteins Nrg1p and Rfg1p and the global transcriptional repressor Tup1p, but the signals and pathways that regulate the activity of these proteins remain poorly understood. The DNA binding protein Nrg1p is a key repressor of hypha formation and acts with Tup1p to suppress hyphal growth and the expression of many hypha-specific genes (10, 17, 23, 28, 30). NRG1 transcription is elevated in yeast cells, and NRG1 mRNA levels must fall for cells to progress from yeast to hyphal forms (10, 28, 30). The proteins Nrg1p, Rfg1p, and Tup1p were originally characterized as repressors of filamentation because strains lacking any one of these proteins grow as either filamentous pseudohyphae or hyphae under yeast growth conditions (9, 10, 22, 24, 30). The nrg1Δ strain forms wrinkled colonies on yeast extract-peptone-dextrose (YPD) plates, and these colonies consist of a mix of yeast and pseudohyphal cells (10, 30). The tup1Δ null strain, however, grows exclusively as pseudohyphae (9). In a previous study, we constructed a C. albicans strain in which expression of NRG1 could be manipulated through the addition or omission of the tetracycline analogue doxycycline (DOX) from the growth medium or drinking water of an infected animal (38). Analysis of this tet-NRG1 strain revealed that overexpression of NRG1 not only inhibited filament formation under every hypha-inducing condition tested (14) but also rendered the fungus avirulent in the murine model of disseminated candidiasis (38). These studies provide compelling evidence linking morphogenetic changes to the ability of C. albicans to cause disease.
The negative regulator Rfg1p also plays an important role in regulating filamentation. This DNA binding protein was originally characterized as a repressor of filamentation because strains lacking Rfg1p form wrinkled colonies on YPD plates under yeast growth conditions with these colonies containing a mixture of yeast and filamentous cells (22, 24). Rfg1p, like Nrg1p, binds specific DNA sequences upstream of several genes and interacts with Tup1p to repress transcription at those sites. Although Rfg1p bears similarity to the Saccharomyces cerevisiae protein Rox1p, which is involved in repressing hypoxic genes, Rfg1p is not involved in regulating this process in C. albicans (22, 24), an important part of C. albicans transcriptional rewiring. Interestingly, exogenous C. albicans RFG1 expressed in S. cerevisiae is able to repress filamentous growth in response to nitrogen starvation conditions (22), reinforcing the observation that, although there have been changes in the regulation of filamentation between these two fungal species, some conservation in the machinery used to achieve filamentous growth remains. Microarray and Northern analyses have helped to define the regulatory targets of the Tup1p, Nrg1p, and Rfg1p repressors, and it appears that about half of Tup1p repression occurs through Nrg1p and Rfg1p together or independently (23). Although Nrg1p and Rfg1p both regulate the expression of several hypha-specific genes, such as ALS3, ECE1, and HWP1, their regulons do not completely overlap (22, 23), indicating a distinct function for each protein in the control of filamentation. This is reinforced by the observation that colonies formed by the double rfg1Δ nrg1Δ null strain are more wrinkled and have a higher proportion of filamentous cells than does either single mutant strain. Furthermore, the fact that the double mutant does not reach the 100% pseudohyphal phenotype of the tup1Δ null strain indicates the involvement of further Tup1p binding partners in the regulation of filamentation (22). The mechanism of Rfg1p regulation and activity remains poorly characterized. A putative Rfg1p binding site has been identified upstream of the hypha-specific genes HWP1 and RBP1, but the hypha-specific cell wall protein-encoding gene ALS3, which is also regulated by Rfg1p, lacks this site (1). It is important to note that, although the primary role of Rfg1p appears to be the repression of filamentous growth under yeast conditions, there is a requirement for Rfg1p to properly form hyphae when C. albicans is grown under nutrient-limiting conditions (22).
To further explore the role of filamentation in C. albicans pathogenesis, we constructed a tet-RFG1 strain in which the gene encoding this reported repressor protein was placed under the control of a tetracycline-regulatable promoter and analyzed for its ability to filament and cause disease during RFG1 overexpression. Since exogenous expression of C. albicans RFG1 was able to repress filamentous growth in S. cerevisiae, we believed that overexpressing RFG1 directly in C. albicans, its native species, might have a similar effect. Here we describe a novel role for Rfg1p in the regulation of pseudohyphal growth and examine the characteristics of this new regulatable tet-RFG1 strain both in vitro and in two different infection models.
MATERIALS AND METHODS
Strains and media.
The yeast strains and the plasmids used in this study are listed in Tables 1 and 2, respectively. Strains were routinely maintained as −80°C frozen stocks and grown on yeast extract-peptone-dextrose (YPD) medium whereas selections for uracil prototrophy were performed on minimal SD plates lacking uridine (39). Expression from the tet promoter was abolished by the addition of 20 μg ml−1 doxycycline to the growth medium. For filamentation assays in liquid media, strains were grown overnight in YPD plus doxycycline at 30°C, washed in sterile phosphate-buffered saline (PBS), and diluted 1:20 into fresh media such as RPMI 1640 supplemented with l-glutamine and buffered with morpholinepropanesulfonic acid (Angus Buffers and Chemicals) and incubated with shaking at 37°C. For filamentation assays on solid media, strains were grown as for liquid filamentation assays, washed in sterile PBS, and counted using a hemocytometer. For growth on solid Spider medium (26), cells were resuspended to a final concentration of 5 × 106 cells ml−1. Aliquots of 2 μl (containing approximately 10,000 cells) were spotted onto Spider plates, which were subsequently incubated at 37°C for 7 days. For growth on synthetic low ammonia dextrose (SLAD) medium (19), approximately 200 cells were spread onto SLAD plates and were then incubated at 30°C for 5 days. For embedded growth, approximately 200 cells were mixed with molten YPD agar and poured onto YPD plates, which were subsequently incubated at 30°C for 3 days. Colonies were examined and photographed using a GL9-280 Stereo Zoom microscope (Jenco) equipped with a digital camera. All morphological tests were performed in at least biological triplicate. For growth curves, overnight cultures were diluted 1:30 in fresh YPD with and without doxycycline and grown with shaking at 30°C. Aliquots were removed at the times indicated, and the optical density at 600 nm (OD600) was measured. All plasmid manipulations were performed with Escherichia coli strain DH5α with selection on Luria-Bertani plates containing 100 μg/ml ampicillin when necessary.
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SC5314 | Wild type | 18 |
| CAF2-1 | URA3/ura3::λimm434 | 16 |
| THE1 | ade2::hisG/ade2::hisG ura3::λimm434/ura3::λimm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 | 32 |
| PMC-X7 | THE1 with RFG1/rfg1::URA3-tetO-RFG1 | This study |
| PMC-X8 | THE1 with RFG1/rfg1::URA3-tetO-RFG1 | This study |
| PMC-X7+empty vector | PMC-X7 with RP10::CIpSATSA | This study |
| PMC-X7+CLB4 | PMC-X7 with RP10::CIpSATSACLB4 | This study |
| YJB6854 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG ARG4::URA3::arg4::hisG/arg4::hisG clb4::ARG4/clb4::HIS1 | 4 |
Table 2.
Plasmids used in this study
Fluorescence microscopy.
C. albicans cells were stained using calcofluor white (Sigma), and nuclei were revealed through the use of Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Labs). A DMR epifluorescence microscope (Leica) was used to visualize fluorescence and capture cell images.
Strain construction and transcriptional analysis.
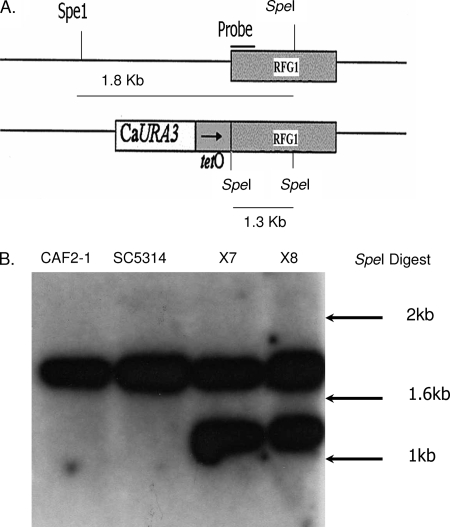
The two independently isolated tet-RFG1 strains PMC-X7 and PMC-X8 were constructed as follows: first, two regions spanning positions −1400 to −499 (RFGA) and positions −21 to +779 (RFGB) relative to the ATG start codon of the RFG1 open reading frame (GenBank accession no. 3642570) were PCR amplified by using the primer pairs RFGA.FOR with RFGA.REV and RFGB.FOR with RFGB.REV (Table 3). These amplification products were digested with KpnI and XhoI or SpeI and SacII at the sites engineered into the primers and ligated sequentially between the KpnI and XhoI or SpeI and SacII sites in the proximal and distal cloning regions of the p97CAU1 plasmid (32) to form plasmid p97RFG1AB. The entire 3.8-kb promoter-replacing construct was then liberated from this plasmid as a KpnI-SacII fragment and transformed into the TR transactivator gene-containing C. albicans strain THE1 (32) using a modified polyethylene glycol-lithium acetate transformation method. Genomic DNA was prepared from several of the Ura+ transformants obtained using the MasterPure yeast DNA extraction kit (Epicentre Biotechnologies), digested with SpeI, transferred to a positively charged Nytran membrane (Whatman), and subjected to Southern blot analysis as previously described (13). The RFGB PCR product (described above) was labeled with [32P]dCTP using Ready-To-Go DNA labeling beads (GE Healthcare) and used as a probe.
Table 3.
Oligonucleotides used for cloning in this study
| Name | Sequencea | Reference |
|---|---|---|
| RFGA.FOR | 5′-CACACATAGGTACCCCAATACAC-3′ | This study |
| RFGA.REV | 5′-CACTTTAAACAGATAAACTCGAGGATATG-3′ | This study |
| RFGB.FOR | 5′-CCACTAGTATTTATTTGCAAACCATCACACC-3′ | This study |
| RFGB.REV | 5′-GTCTACCCAATCCGCGGGAAACTTC-3′ | This study |
| SAT.FOR | 5′-GGGCCCGAGCTCGCATGCCAGCGTC-3′ | This study |
| SAT.REV | 5′-GGACTAGTGATTTCTAGAAGGACCAC-3′ | This study |
| ACTprom.FOR | 5′-GGGCGAATTGGTACCGACGTCGC-3′ | This study |
| ACTprom.REV | 5′-CATACCCTCGAGTTTGAATGATTATATTT-3′ | This study |
| CLB4.FOR | 5′-CCAGAGGTTCACAAGCTTATGCGCGATC-3′ | This study |
| CLB4.REV | 5′-GGGAATTCAATGATGATGGTGGTGGTGACTCTGGGACATTATGTGACG-3′ | This study |
Underlined sequences indicate introduced restriction enzyme sites.
To verify DOX regulation of the modified tet-RFG1 allele, the wild-type (CAF2-1) and modified strains were grown for 3 h in RPMI 1640 at 37°C and total RNA was isolated using a previously published bead beater protocol (34) and then separated through formaldehyde-containing agarose gels. Transfer and hybridization followed the method described above for the Southern blot assay, again using the RFGB PCR product as a probe.
The strain PMC-X7+CLB4 was constructed as follows: plasmid CIpSAT was constructed by replacing the URA3 gene in CIp10 with the SAT1 gene, encoding a protein conferring resistance to nourseothricin. SAT1, along with its ACT1 promoter, was amplified from plasmid pA83 (36) using primers SAT.FOR and SAT.REV. The amplification product was digested with SacI and XbaI at the sites engineered into the primers and ligated between the SacI and XbaI sites in CIp10 (29), thereby replacing the URA3 gene. The sequence upstream of the C. albicans ACT1 gene was amplified from plasmid pA83 using primers ACTprom.FOR and ACTprom.REV. The amplification product was digested with KpnI and XhoI at the sites engineered into the primers and ligated between the KpnI and XhoI sites in CIpSAT, producing CIpSATSA. The CLB4 coding sequence was amplified from genomic DNA using primers CLB4.FOR and CLB4.REV. These amplification products were digested with HindIII and EcoRI at the sites engineered into the primers and ligated between the HindIII and EcoRI sites in CIpSATSA to produce plasmid CIpSATSA-CLB4. Finally, this plasmid was linearized by digestion with StuI and transformed into the tet-RFG1 strain PMC-X7 using a modified electroporation transformation method (25). Nourseothricin-resistant transformants were selected on YPD agar plates containing 200 μg ml−1 nourseothricin (Werner Bioagents, Jena, Germany) as described previously (36). Correct insertion into the RP10 locus was confirmed by PCR.
Murine virulence assay.
For injection, cultures of tet-RFG1 strain PMC-X7 were grown overnight at 25°C in YPD with doxycycline. Cells were harvested by centrifugation and washed three times in sterile pyrogen-free saline. The cells were counted using a hemocytometer, and appropriate dilutions were made so that the required dosage of cells could be injected in a final volume of 200 μl into the lateral tail veins of 6- to 8-week-old female BALB/c mice that had been placed on either 5% sucrose or 5% sucrose containing 2 mg of doxycycline ml−1 3 days prior to infection. Confirmation of the number and viability of cells present in the infecting inocula was performed by plate count. Groups of eight mice were used for each condition. All experiments were performed in accordance with institutional regulations in place at the University of Texas at San Antonio. Mice were allowed a 1-week acclimatization period before experiments were started. Days on which the animals died were recorded; severely moribund animals were humanely sacrificed to minimize suffering and recorded as having died the following day.
Histopathology.
Kidneys excised from deceased or sacrificed mice were fixed in 10% buffered formalin and stored at 4°C until required. After kidneys were embedded in paraffin, tissue slices were removed and stained with Grocott-Gomori methenamine-silver prior to microscopic evaluation (21). The human oral tissue culture models were fixed in 10% buffered formalin and stored at 4°C until required. Tissue slices from paraffin wax-embedded tissue models were stained with Mayer's hematoxylin prior to microscopic evaluation.
Infection of a tissue-engineered 3-dimensional OEMS with C. albicans.
A commercially available human oral epithelium model system (OEMS) (Skinethic Laboratory, Nice, France) was used to investigate the interaction of the mucosal epithelium with C. albicans. Models were maintained according to the manufacturer's instructions. The tet-RFG1 strain PMC-X7 and wild-type CAF2-1 strain were grown overnight as described above, and cells were washed, counted, and resuspended to a final concentration of 5 × 107 CFU/ml. An aliquot of 100 μl of cell suspension was added to each of the models while negative controls were inoculated with PBS. The models were then incubated for 24 or 48 h at 37°C in a 5% CO2 humidified incubator and were fixed in 10% buffered formalin.
Quantitative PCR.
RNA was isolated from C. albicans cells using the MasterPure yeast RNA extraction kit (Epicentre Biotechnologies). RNA was treated with amplification-grade DNase I (Invitrogen) and used for cDNA synthesis with the cMaster RT kit (Eppendorf). The primer sets (Table 4) were used in conjunction with SYBR green PCR master mix (Applied Biosystems) and Twin.tec real-time 96-well PCR plates (Eppendorf) in an ABI 7300 real-time PCR system (Applied Biosystems). Dissociation curves were analyzed for all reactions to verify single peaks/products. Expression levels were analyzed using ABI 7300 system SDS software (Applied Biosystems).
Table 4.
Oligonucleotides used for quantitative real-time PCR
| Name | Sequence | Reference |
|---|---|---|
| ACT1-S | 5′-ATGTGTAAAGCCGGTTTTGCCG-3′ | 41 |
| ACT1-A | 5′-CCATATCGTCCCAGTTGGAAAC-3′ | 41 |
| CLB4_FOR | 5′-CACAAACAAACACCAATCATCACA-3′ | This study |
| CLB4_REV | 5′-ATCGGTTTTCTTTTGCTCTATTTG-3′ | This study |
| RFG1 forward | 5′-AACCCTGAAGTTTCCCGAGAA-3′ | 33 |
| RFG1 reverse | 5′-CAGCAAGATTATTCCAATGTTCCTT-3′ | 33 |
| FKH2_FOR | 5′-GCAAACTCGCTCCAATCAAA-3′ | This study |
| FKH2_REV | 5′-TGCTTGCGTAATCATTGTCG-3′ | This study |
| HSL1_FOR | 5′-GATTGCCGATTTTGGTATGG-3′ | This study |
| HSL1_REV | 5′-TGGTGAAGCATAATGAGGAGAA-3′ | This study |
| GIN4_FOR | 5′-TTATGCTGCTCCAGAAATCGTT-3′ | This study |
| GIN4_REV | 5′-ACCCCACAAGACCAAACATCA-3′ | This study |
| ALS3_FOR | 5′-CAACTTGGGTTATTGAAACAAAAACA-3′ | 31 |
| ALS3_REV | 5′-AGAAACAGAAACCCAAGAACAACCT-5′ | 31 |
| ECE1_FOR | 5′-CCAGAAATTGTTGCTCGTGTTG-3′ | 3 |
| ECE1_REV | 5′-CAGGACGCCATCAAAAACG-3′ | 3 |
| HWP1_FOR | 5′-TCAGCCTGATGACAATCCTC-3′ | This study |
| HWP1_REV | 5′-GCTGGAGTTGTTGGCTTTTC-3′ | This study |
RESULTS
Regulatable strain construction.
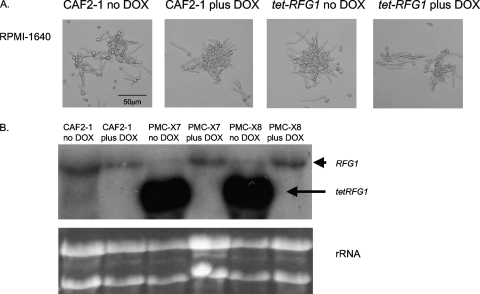
We previously described the construction of an engineered strain in which one copy of NRG1 was placed under the control of a tetracycline-regulatable promoter (38) and showed that regulation of one allele was sufficient to control the phenotype of the strain. A similar strategy was used here to construct a strain regulating RFG1. A small region of the sequence upstream of one allele of RFG1 (position −499 to −21 relative to the ATG start codon) was replaced with the bacterially derived tetO promoter sequence by integration into the genome of the transactivator-containing C. albicans strain THE1. Correct integration of this promoter-modifying fragment at the RFG1 locus was confirmed for two independent transformants, PMC-X7 and PMC-X8, by Southern blot analysis (Fig. 1A). To verify that RFG1 expression could be modulated by doxycycline in the transformed strains, RNA was extracted from the wild-type CAF2-1 and the modified strains grown in RPMI 1640 in the presence or absence of doxycycline. Northern blot analysis revealed that the modified tet-RFG1 allele produced a smaller transcript than did the wild-type copy (Fig. 2B), presumably as a consequence of integration of the tetO promoter very close to the ATG start codon producing a shorter 5′ untranslated leader sequence. This allowed the comparison of transcription from the modified tet-RFG1 allele as well as from the wild-type allele. As predicted, transcription from the modified allele was completely dependent on the presence or absence of doxycycline, and transcription from the remaining unaltered copy of RFG1 did not appear to be affected by the presence of doxycycline (Fig. 2B). Preliminary analysis of the two independently isolated tet-RFG1 strains determined that they were phenotypically indistinguishable, and therefore, all of the subsequent experiments were performed only with the PMC-X7 isolate.
Fig. 1.
Construction of the tet-RFG1 C. albicans strain. (A) The tet-RFG1 strain was produced through integration of an in vitro-produced DNA construct designed to replace the promoter region (−499 to −21) of RFG1 with the bacterially derived tetO sequence. (B) Appropriate integration of this construct at the RFG1 locus was confirmed via Southern blot analysis (PMC-X7 and PMC-X8 represent two independent transformants). Genomic DNA was isolated and digested with SpeI and transferred to a Nytran membrane. In the wild-type control strains CAF2-1 and SC5314, the probe hybridized to a single band of approximately 1.8 kbp representing the endogenous allele. In the transformed strains the probe hybridized to an additional band of approximately 1.3 kbp representing the modified allele. Reference sizes are indicated by arrows.
Fig. 2.
RFG1 overexpression does not inhibit hypha formation under inducing conditions. (A) The effect of RFG1 overexpression was assessed by growing the tet-RFG1 strain in RPMI 1640 at 37°C, in the absence or presence of doxycycline. Both the wild-type CAF2-1 and the modified tet-RFG1 strain formed hyphae. (B) RFG1 transcription in these cultures was examined by Northern blot analysis. RNA was isolated and transferred to a Nytran membrane. The probe was the same as that used for the Southern blot assay. The larger endogenous RFG1 transcript and the smaller tet-RFG1 transcript are indicated by arrows. Ethidium bromide-stained rRNA bands are shown to confirm equal loading of all samples. The tet-RFG1 transcript is absent from the control and from the transformants grown in the presence of doxycycline, indicating doxycycline-dependent regulation of the modified allele.
RFG1 overexpression does not inhibit hypha formation under liquid-inducing conditions.
To determine whether overexpression of the repressor RFG1 would have an inhibitory affect on hypha formation similar to that of NRG1, the tet-RFG1 strain PMC-X7 was grown in medium known to induce hypha formation in C. albicans, in both the presence and the absence of doxycycline. Growth in cell culture medium RPMI 1640 at 37°C strongly stimulates hyphal formation in wild-type strains, and this medium is used for in vitro biofilm formation on polystyrene surfaces (35). As expected, the wild-type CAF2-1 strain formed hyphae when grown in RPMI 1640 at 37°C irrespective of the presence of doxycycline (Fig. 2A). We had predicted that overexpressing RFG1 would inhibit formation of hyphae under inducing conditions, but unexpectedly, the tet-RFG1 strain was still able to respond normally in the absence of doxycycline (Fig. 2A). To assess whether RFG1 overexpression might block hypha formation only in response to a specific environmental stimulus, the strains were grown in additional hypha-inducing liquid media. The tet-RFG1 strain formed hyphae normally, irrespective of the presence of doxycycline, when grown in YPD plus 10% serum at 37°C or in M199 at pH 8 (data not shown), despite expression of RFG1 from the modified allele in the absence of doxycycline (data not shown). This indicates that increased RFG1 transcription is not sufficient to inhibit hypha formation induced by serum or by the Rim101p-mediated pH induction pathway.
RFG1 overexpression does not inhibit filament formation on solid hypha-inducing media.
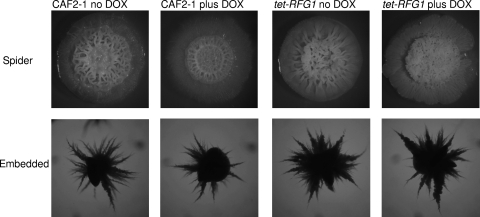
Growth on solid Spider medium is a strong stimulator of hyphal growth in C. albicans (26), and although Rfg1p is required to block hypha formation when strains are grown on YPD plates, a strain lacking Rfg1p was shown to be deficient in hypha formation on Spider medium, although an apparent growth defect made this phenotype difficult to assess (22). When wild-type CAF2-1 is grown on Spider plates at 37°C, highly wrinkled colonies are formed, indicating the presence of hyphal cells (Fig. 3). Overexpressing RFG1 did not appear to affect that ability of the tet-RFG1 strain to form hyphae, since wrinkled colonies were formed in both the presence and the absence of doxycycline. Microscopic examination of cells removed from these colonies revealed that they consisted primarily of hyphal cells (data not shown). Thus, alterations in RFG1 expression did not enhance or impair the ability to form hyphae on solid Spider medium.
Fig. 3.
RFG1 overexpression does not inhibit hypha formation on solid inducing media. For Spider medium, cells were spotted on Spider plates with and without doxycycline and incubated at 37°C for 7 days (upper panels). For embedded conditions, approximately 200 cells were mixed with molten YPD agar, poured onto YPD plates, and incubated at 30°C for 3 days. The wild-type CAF2-1 and the tet-RFG1 strain formed wrinkled colonies on Spider plates and filamentous colonies embedded in YPD irrespective of the presence or absence of doxycycline.
C. albicans cells also form hyphae when grown embedded in solid medium at 30°C in response to low oxygen levels and increased physical pressure (11, 43). These signals are transduced through the transcriptional regulators Czf1p, which is required for hyphal growth under embedded conditions, and Efg1p, which normally promotes hypha formation but appears to play a role in the repression of hypha-specific genes under low-oxygen conditions (20). The CAF2-1 cells embedded in the solid medium began to form hyphae after 48 h growth, and by 72 h, all of the cells had formed hyphal colonies (Fig. 3). The tet-RFG1 strain was able to form hyphae normally in the presence or absence of doxycycline, indicating that under embedded conditions RFG1 overexpression does not block the signals transduced through Efg1p and Czf1p.
Overexpression of RFG1 promotes pseudohyphal growth.
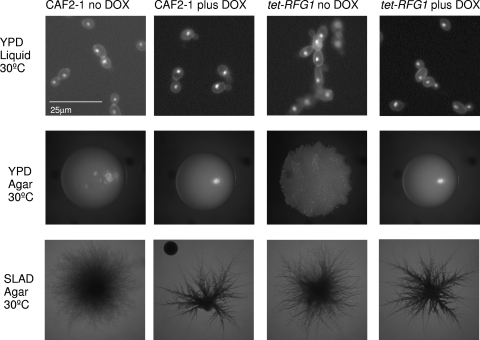
Although overexpressing RFG1 does not inhibit hypha formation, it does influence morphology under yeast growth conditions. When grown in YPD at 30°C in the presence of doxycycline, the tet-RFG1 strain resembles the wild-type strain CAF2-1 and the culture consists entirely of free-floating yeast cells. In the absence of doxycycline (elevated RFG1 expression), however, the tet-RFG1 cells grew exclusively as pseudohyphae (Fig. 4). Visualization of these cells after staining with calcofluor white and DAPI confirmed that these cells were not hyphal but pseudohyphal (Fig. 4). This same phenotype was observed on solid YPD plates, where CAF2-1 formed smooth round colonies, as did the tet-RFG1 strain grown in the presence of doxycycline. When RFG1 levels were elevated, however, the strain formed a wrinkled colony, indicating the presence of filamentous cells (Fig. 4). Microscopic examination revealed that these cells are pseudohyphae rather than hyphae (Fig. 4). Nitrogen-limiting SLAD medium is frequently used to induce pseudohyphal formation in C. albicans, and when strains are incubated on SLAD plates at 30°C, a fraction (less than 5%) of colonies formed by the wild-type CAF2-1 or tet-RFG1 strain in the presence of doxycycline exhibit a smooth center surrounded by a halo of pseudohyphal growth. In the absence of doxycycline, however, greater than 50% of the colonies formed by the tet-RFG1 strain displayed this pseudohyphal morphology (Fig. 4). Although elevated RFG1 levels were unable to block the formation of hyphae, we were able to promote pseudohyphal growth under yeast growth conditions by manipulating RFG1 expression. Previous studies described Rfg1p as a repressor of hyphal growth that is required to block hypha formation under yeast growth conditions (22, 24), while also being necessary for forming hyphae under nutrient-limiting conditions (22). However, Rfg1p may play another, perhaps more significant, role in regulating C. albicans morphology since we have now demonstrated that overexpression of RFG1 can actually drive pseudohyphal growth under yeast conditions.
Fig. 4.
RFG1 overexpression promotes pseudohyphal growth. Strains were grown under standard yeast growth conditions, in liquid YPD (top panels) and on solid YPD agar (middle panels) in the presence of absence of doxycycline. The wild-type CAF2-1 strain grows as round yeast cells in liquid medium and forms smooth colonies on solid medium whereas the tet-RFG1 strain forms chains of elongated pseudohyphal cells in liquid medium and forms wrinkled colonies when RFG1 is overexpressed. Both strains form pseudohyphae when grown on SLAD medium (bottom panels), although the observed frequency of pseudohyphal growth was higher when RFG1 was overexpressed.
Virulence is not affected by modulating RFG1 expression.
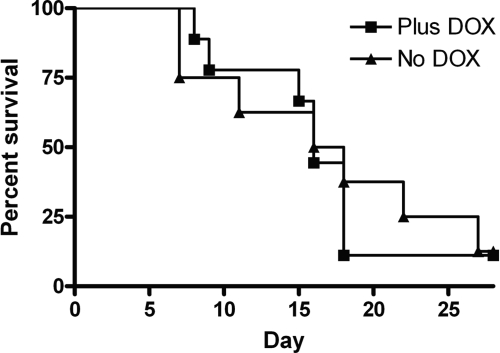
Having demonstrated that RFG1 overexpression alters C. albicans morphology in vitro, we examined its effect on virulence in the murine model of disseminated candidiasis. In this model, strains that are locked in the yeast form or in the hyphal form of growth are avirulent. A strain lacking Rfg1p was previously shown to be avirulent in this disease model (22). Using the tetracycline-regulated promoter system, we are able to manipulate expression of the tet-regulated gene within an animal host by the addition or omission of doxycycline from the drinking water (32). Approximately 3.5 × 105 C. albicans yeast cells of the tet-RFG1 strain were injected into mice that had been placed on either 5% sucrose or 5% sucrose containing 2 mg ml−1 of doxycycline 3 days prior to infection. All the mice, irrespective of the treatment, succumbed to the infection, and there was no significant difference between the virulence of the tet-RFG1 strain in the presence of doxycycline and the virulence in its absence (Fig. 5). We had hypothesized that, similar to our previous results with NRG1, if RFG1 overexpression was able to block filamentation in the host, this might render the strain avirulent. However, elevated RFG1 expression neither impaired nor enhanced virulence, since there was no difference between the survival curves produced in the presence and those produced in the absence of doxycycline.
Fig. 5.
The virulence of the tet-RFG1 strain is unaffected by the presence or absence of doxycycline. Approximately 3.5 × 105 C. albicans yeast cells of the tet-RFG1 strain PMC-X7 were injected into mice that had been placed on either 5% sucrose (No DOX) or 5% sucrose containing 2 mg/ml of doxycycline (Plus DOX). There was no difference in the survival curves produced under the two conditions.
Histopathology of tet-RFG1 strain-infected tissues.
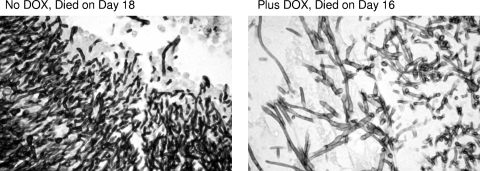
To address the possibility that the modified strain had not affected virulence but had nonetheless altered the morphology of the fungal cells in the animal host, we performed a histological analysis on kidneys recovered from mice that had succumbed to the infection both in the presence and in the absence of doxycycline. Tissues from mice that had died early (7 or 8 days) or died later (16 or 18 days) after infection were examined. This examination revealed the presence of extensive lesions containing mainly hyphal fungal cells irrespective of the presence or absence of doxycycline (Fig. 6). The mice that survived longer showed more extensive mycelial lesions but were otherwise identical to the mice that died earlier (data not shown).
Fig. 6.
Histopathology of tet-RFG1 strain-infected tissues. Examination of kidneys retrieved from mice infected with the tet-RFG1 strain revealed the presence of extensive lesions containing mainly hyphal cells irrespective of the presence or absence of doxycycline.
Overexpression of RFG1 in a reconstituted human oral tissue culture model.
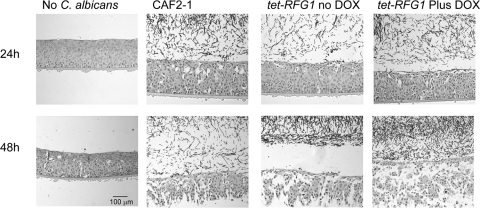
To further examine the influence of RFG1 overexpression on pathogenesis, we used the tet-RFG1 strain to infect a human oral tissue culture model. Overexpressing RFG1 did not appear to have an effect on the ability of the strain to cause damage or to invade the host tissue, as assessed by lactate dehydrogenase (LDH) release (data not shown) and by histopathological examination of sections from the infected models (Fig. 7).
Fig. 7.
Histopathology of the human oral mucosal model. C. albicans strains were incubated with the oral mucosal model for 24 h or 48 h, and the tissue was fixed, stained, and examined microscopically. Both the wild-type CAF2-1 and tet-RFG1 strains produce filaments and cause extensive damage to the tissues. The tet-RFG1 strain is able to damage the tissues to the same extent, even in the presence of elevated RFG1 transcription (no DOX).
Rfg1p does not promote pseudohyphal growth via repression of pseudohypha-associated cell cycle regulators.
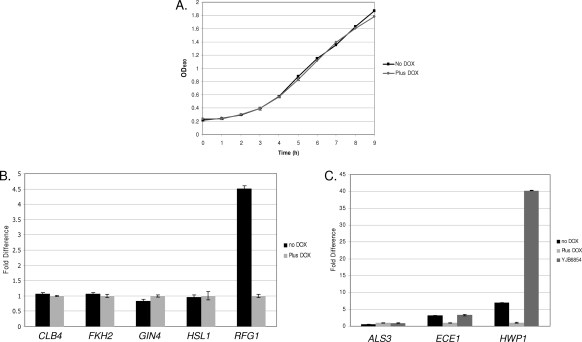
Several mutant C. albicans strains display a morphology similar to that of the tet-RFG1 strain in the absence of doxycycline, forming pseudohyphae under yeast growth conditions. These include strains in which one of several cell cycle regulators of the G2/M transition has been deleted. This suggested that RFG1 overexpression could be perturbing the cell cycle by suppressing one of these regulators and thus leading to the observed pseudohyphal phenotype. As a simple test to determine whether overexpressing RFG1 was affecting cell cycle progression, we therefore compared the growth rates of the tet-RFG1 strain in the presence and absence of doxycycline. There was no discernible difference between the two cultures (Fig. 8A), suggesting that the pseudohyphal growth phenotype of the tet-RFG1 strain grown in the absence of doxycycline is not simply a consequence of RFG1-mediated cell cycle suppression. However, it is known in S. cerevisiae that, when one part of the cell cycle is lengthened, other phases can be shortened to compensate, and thus, the overall length of the cell cycle remains the same (2, 37). For instance, cells of the CLN3-2 strain, which carries a stabilized allele of the G1 cyclin CLN3, have an accelerated entry into Start but do not show a reduced doubling time because the S and G2 phases are lengthened (2). Through such a compensation mechanism, Rfg1p could still be acting by repressing expression of cell cycle regulatory genes without affecting the overall growth rate. One of the proteins governing cell cycle progression, the Fkh2p transcription factor, is required for expression of the G2/M transition regulator CLB4, and the fkh2Δ null strain is constitutively pseudohyphal but remains able to respond to hypha-inducing signals (5). Gin4p is a kinase is required for septin ring formation, and the gin4Δ null strain is also constitutively pseudohyphal but, in this case, unable to form hyphae (46). Deletion of the HSL1 gene, which encodes a septin-associated mitotic kinase, produces a strain which when grown under yeast conditions forms pseudohyphae at low cell density and grows as yeast cells at high density. These yeast cells are able to respond to hypha-inducing stimuli and can form hyphae (42, 46). Quantitative real-time PCR was used to examine whether the striking constitutive pseudohyphal morphology observed as a result of RFG1 overexpression was possibly due to transcriptional repression of one or more of these regulators. The tet-RFG1 strain was grown overnight in the presence of doxycycline, and cells were washed three times in sterile PBS before being diluted 1:20 into fresh YPD in the presence or absence of doxycycline and grown for 6 h at 30°C. While RFG1 expression was elevated in the absence of doxycycline, expression of the other regulatory genes showed no marked change between the two conditions (Fig. 8B). Although the expression of the mitotic regulatory genes varies during the cell cycle, making it more difficult to track such temporal transcriptional changes with certainty in an asynchronous culture, our results suggest a lack of involvement of these genes in the RFG1-mediated pseudohyphal phenotype.
Fig. 8.
(A) Growth curves of the tet-RFG1 strain. The tet-RFG1 strain was grown overnight in the presence of doxycycline, and cells were washed before being diluted 1:30 in fresh YPD or YPD plus DOX. Duplicate cultures were grown with shaking at 30°C, samples were removed at the times indicated, and the OD600 was measured. The strain grew at identical rates regardless of the presence or absence of doxycycline. (B) Quantification of mitotic regulatory gene transcription in the presence of elevated RFG1. The tet-RFG1 strain was grown overnight in the presence of doxycycline, and cells were washed before being diluted 1:20 into fresh YPD and grown for 6 h at 30°C in the presence or absence of doxycycline. At this point the culture grown in the presence of doxycycline consisted entirely of yeast cells, while the culture grown in the absence of doxycycline was approximately 80% pseudohyphal. RNA was isolated from cells harvested from these cultures, and transcript levels were quantified through real-time PCR analysis. Levels were normalized by ACT1 and are expressed relative to the control tet-RFG1 plus doxycycline culture (therefore, its results are 1.0). While RFG1 levels are elevated approximately 4.5-fold in the tet-RFG1 strain grown in the absence of doxycycline, there are only minor differences in transcript levels of the other genes examined. Error bars represent the standard deviations of the threshold cycle (ΔCT) values. (C) Quantification of filament-specific gene transcription in the context of elevated RFG1 expression. The tet-RFG1 and the clb4Δ null (YJB6854) strains were grown, and transcript levels were analyzed as described for panel B. Transcription of these genes followed the same pattern in both the tet-RFG1 strain grown in the absence of doxycycline and the clb4Δ null strain.
Another protein involved in cell cycle progression, and thought to operate at the G2/M transition, is the B-type cyclin Clb4p (4). The clb4Δ null strain, like the tet-RFG1 strain, forms pseudohyphae under yeast growth conditions but is still able to form hyphae when grown in the presence of serum at 37°C (4). To confirm that the filaments produced by the tet-RFG1 strain are pseudohyphae and to determine any further similarities to the clb4Δ strain, we compared the expression of several filament-specific genes in two strains grown under yeast conditions. While no pseudohypha-specific transcripts have yet been described, expression of the genes encoding two components of the hyphal wall, ECE1 and HWP1, has previously been shown to be increased in pseudohyphal cells compared to yeast cells (12). Expression of these genes was elevated in both the clb4Δ strain and the tet-RFG1 strain grown in the absence of doxycycline (Fig. 8C) compared to that in the tet-RFG1 plus doxycycline yeast culture control. Furthermore, transcription of ALS3, which is hypha specific (15), was not elevated in the tet-RFG1 strain filaments, indicating that they are not true hyphae (Fig. 8C) and confirming our microscopic examination (Fig. 4). Although HWP1 was more highly expressed in the clb4Δ strain than in the tet-RFG1 strain, the similarities in the overall patterns of gene expression and morphology between the two strains suggested that Rfg1p might repress CLB4 expression. To test this, we integrated an additional extragenic copy of the CLB4 gene under the control of the ACT1 promoter into the tet-RFG1 strain. Overexpression of CLB4 has previously been demonstrated to slow hypha formation under inducing conditions (4). We therefore tested the functionality of our constitutive CLB4 gene by monitoring the ability of this strain (PMC-X7+CLB4) to filament following inoculation into RPMI 1640 and growth at 37°C. Samples were examined at various time points, and the proportions of yeast and hyphal cells were determined microscopically. The CLB4-containing strain showed delayed hypha formation compared to the PMC-X7+empty vector control strain (Fig. 9A), indicating that our constitutive CLB4 construct was producing functional Clb4p. To test the ability of CLB4 to rescue the pseudohyphal phenotype, the strains were grown overnight in YPD plus doxycycline (to maintain yeast growth), washed, and resuspended in fresh YPD in the presence or absence of doxycycline. These cultures were incubated at 30°C, and cells were examined microscopically at various time points. We observed no marked difference in the rate at which the PMC-X7+CLB4 or control strain formed pseudohyphae (Fig. 9B), indicating that Rfg1p is not promoting pseudohypha formation by repressing CLB4 transcription.
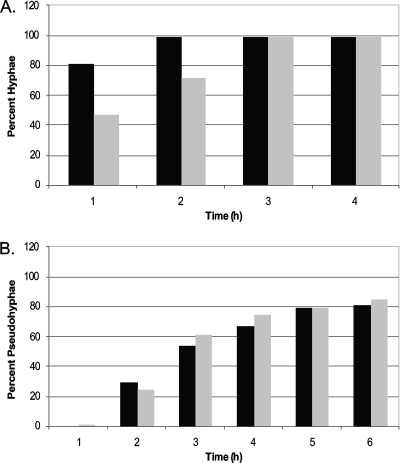
Fig. 9.
CLB4 overexpression does not rescue the pseudohyphal phenotype. (A) The tet-RFG1+empty vector control (black bars) and tet-RFG1+CLB4 (gray bars) strains were grown in RPMI 1640 at 37°C and examined at the indicated time points, and the proportion of hyphal cells was determined microscopically. Note the lag in hypha formation observed in the tet-RFG1+CLB4 strain. (B) The same strains were grown in YPD at 30°C and examined at the indicated time points, and the proportion of pseudohyphal cells was determined microscopically. This figure presents data from a single experiment that was repeated three times with the same results.
DISCUSSION
Here we describe the construction of a novel regulatable tet-RFG1 strain and its characterization both in vitro and in vivo. In the absence of doxycycline, this strain produces elevated levels of RFG1 but, contrary to expectation, is able to filament normally when grown in liquid media or on plates incubated under hypha-inducing conditions. When tested in the murine model of disseminated candidiasis, RFG1 overexpression did not affect the ability to cause disease or to form filaments within infected organs. The tet-RFG1 strain was likewise able to successfully infect and damage a tissue culture model of the human oral mucosa as efficiently as a wild-type strain.
More surprisingly, overexpression of RFG1 was able to promote the formation of pseudohyphae when the strain was grown under yeast conditions, indicating an unexpected and novel role for this protein. Microscopic examination and transcriptional analysis confirmed that these cells were pseudohyphal, not hyphal.
Mutations affecting the cell cycle, particularly the G2/M transition, can result in a pseudohyphal morphology in C. albicans. The resemblance of the cells from a culture where RFG1 was overexpressed to those deleted for several genes involved in cell cycle progression suggested that elevated RFG1 levels may be affecting the G2/M transition. Interestingly, although the cell morphology was altered, the overall growth rate of the cells remained unchanged with or without RFG1 overexpression. In an attempt to determine the mechanism(s) by which RFG1 overexpression causes this pseudohyphal phenotype, we examined the expression of several cell cycle regulatory genes encoding proteins whose absence is known to produce a similar pseudohyphal phenotype. The expression of these genes did not appear to vary significantly in the presence or absence of elevated RFG1 transcription. Furthermore, we attempted to restore a wild-type phenotype through the constitutive expression of one of these genes, CLB4, but this was insufficient to inhibit the pseudohyphal morphology.
While a great deal is known about the signaling pathways and genes involved in C. albicans hyphal development, our understanding of the process of pseudohypha formation is more rudimentary. The observation that defects in several apparently unrelated genes can lead to pseudohypha formation under yeast growth conditions and additionally that some of these mutants can form hyphae (tet-RFG1, clb4Δ, and nrg1Δ mutants) while others cannot (tup1Δ and gin4Δ mutants) suggests that multiple pathways or mechanisms are involved in this process. We have attempted to link our novel RFG1-induced pseudohyphal phenotype to those caused by mutations in several known genes in an attempt to clarify this picture. These experimental results did not reveal a direct connection between RFG1 and any of the genes that we tested but reinforced the multifactorial nature of pseudohypha formation in C. albicans.
ACKNOWLEDGMENTS
The work presented here was funded in part by NIH grant RO3 AI068487-01 to S.P.S.
We thank Alistair Brown for providing plasmid CIp10, Joachim Morschhaüser for providing plasmid pA83, Hironobu Nakayama for providing strain THE1 and plasmid p97CAU1, and Judith Berman for providing strain YJB6854.
Footnotes
Published ahead of print on 23 July 2010.
REFERENCES
- 1.Argimon S., Wishart J. A., Leng R., Macaskill S., Mavor A., Alexandris T., Nicholls S., Knight A. W., Enjalbert B., Walmsley R., Odds F. C., Gow N. A., Brown A. J. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6:682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basco R. D., Segal M. D., Reed S. I. 1995. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5030–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassilana M., Hopkins J., Arkowitz R. A. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4:588–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensen E. S., Clemente-Blanco A., Finley K. R., Correa-Bordes J., Berman J. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16:3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensen E. S., Filler S. G., Berman J. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman J., Sudbery P. E. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 7.Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun B. R., Head W. S., Wang M. X., Johnson A. D. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun B. R., Johnson A. D. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105–109 [DOI] [PubMed] [Google Scholar]
- 10.Braun B. R., Kadosh D., Johnson A. D. 2001. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J. 20:4753–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown D. H., Jr., Giusani A. D., Chen X., Kumamoto C. A. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651–662 [DOI] [PubMed] [Google Scholar]
- 12.Carlisle P. L., Banerjee M., Lazzell A., Monteagudo C., Lopez-Ribot J. L., Kadosh D. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Church G. M., Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleary I. A., Saville S. P. 2010. An analysis of the impact of NRG1 overexpression on the Candida albicans response to specific environmental stimuli. Mycopathologia 170:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman D. A., Oh S. H., Zhao X., Zhao H., Hutchins J. T., Vernachio J. H., Patti J. M., Hoyer L. L. 2009. Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J. Microbiol. Methods 78:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Sanchez S., Mavor A. L., Russell C. L., Argimon S., Dennison P., Enjalbert B., Brown A. J. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 16:2913–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 19.Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 20.Giusani A. D., Vinces M., Kumamoto C. A. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grocott R. G. 1955. A stain for fungi in tissue sections and smears using Gomori's methenamine-silver nitrate technic. Am. J. Clin. Pathol. 25:975–979 [DOI] [PubMed] [Google Scholar]
- 22.Kadosh D., Johnson A. D. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalaf R. A., Zitomer R. S. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler G. A., White T. C., Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 27.Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 28.Murad A. M., d'Enfert C., Gaillardin C., Tournu H., Tekaia F., Talibi D., Marechal D., Marchais V., Cottin J., Brown A. J. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981–993 [DOI] [PubMed] [Google Scholar]
- 29.Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327 [DOI] [PubMed] [Google Scholar]
- 30.Murad A. M., Leng P., Straffon M., Wishart J., Macaskill S., MacCallum D., Schnell N., Talibi D., Marechal D., Tekaia F., d'Enfert C., Gaillardin C., Odds F. C., Brown A. J. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nailis H., Coenye T., Van Nieuwerburgh F., Deforce D., Nelis H. J. 2006. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama H., Mio T., Nagahashi S., Kokado M., Arisawa M., Aoki Y. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68:6712–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver B. G., Silver P. M., Marie C., Hoot S. J., Leyde S. E., White T. C. 2008. Tetracycline alters drug susceptibility in Candida albicans and other pathogenic fungi. Microbiology 154:960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramage G., Saville S. P., Wickes B. L., Lopez-Ribot J. L. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramage G., Vandewalle K., Wickes B. L., Lopez-Ribot J. L. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18:163–170 [PubMed] [Google Scholar]
- 36.Reuss O., Vik A., Kolter R., Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 37.Rupes I. 2002. Checking cell size in yeast. Trends Genet. 18:479–485 [DOI] [PubMed] [Google Scholar]
- 38.Saville S. P., Lazzell A. L., Monteagudo C., Lopez-Ribot J. L. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman F., Fink G. R., Hicks J. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40.Stoldt V. R., Sonneborn A., Leuker C. E., Ernst J. F. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyoda M., Cho T., Kaminishi H., Sudoh M., Chibana H. 2004. Transcriptional profiling of the early stages of germination in Candida albicans by real-time RT-PCR. FEMS Yeast Res. 5:287–296 [DOI] [PubMed] [Google Scholar]
- 42.Umeyama T., Kaneko A., Nagai Y., Hanaoka N., Tanabe K., Takano Y., Niimi M., Uehara Y. 2005. Candida albicans protein kinase CaHsl1p regulates cell elongation and virulence. Mol. Microbiol. 55:381–395 [DOI] [PubMed] [Google Scholar]
- 43.Vinces M. D., Haas C., Kumamoto C. A. 2006. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot. Cell 5:825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viudes A., Peman J., Canton E., Ubeda P., Lopez-Ribot J. L., Gobernado M. 2002. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J. Clin. Microbiol. Infect. Dis. 21:767–774 [DOI] [PubMed] [Google Scholar]
- 45.Wey S. B., Mori M., Pfaller M. A., Woolson R. F., Wenzel R. P. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642–2645 [DOI] [PubMed] [Google Scholar]
- 46.Wightman R., Bates S., Amornrrattanapan P., Sudbery P. 2004. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 164:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]