Abstract
The presence of a mannitol cycle in fungi has been subject to discussion for many years. Recent studies have found no evidence for the presence of this cycle and its putative role in regenerating NADPH. However, all enzymes of the cycle could be measured in cultures of Aspergillus niger. In this study we have analyzed the localization of two enzymes from the pathway, mannitol dehydrogenase and mannitol-1-phosphate dehydrogenase, and the expression of their encoding genes in nonsporulating and sporulating cultures of A. niger. Northern analysis demonstrated that mpdA was expressed in both sporulating and nonsporulating mycelia, while expression of mtdA was expressed only in sporulating mycelium. More detailed studies using green fluorescent protein and dTomato fused to the promoters of mtdA and mpdA, respectively, demonstrated that expression of mpdA occurs in vegetative hyphae while mtdA expression occurs in conidiospores. Activity assays for MtdA and MpdA confirmed the expression data, indicating that streaming of these proteins is not likely to occur. These results confirm the absence of the putative mannitol cycle in A. niger as two of the enzymes of the cycle are not present in the same part of A. niger colonies. The results also demonstrate the existence of spore-specific genes and enzymes in A. niger.
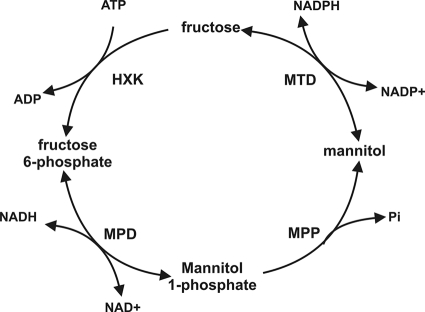
Mannitol has been described as one of the main compatible solutes in fungi (20) and may play a role as a storage carbon source (3) or a protectant against a variety of stresses (10, 16, 20, 22). Mannitol metabolism in fungi has been the subject of study for decades. It was proposed to exist in the form of a cyclic pathway, the mannitol cycle (9). This cycle consists of four steps enabling the conversion of fructose into mannitol and back to fructose (Fig. 1). The main role proposed for this cycle was regenerating NADPH (9, 10). Subsequently, many studies have questioned the existence of a mannitol cycle (reviewed in reference 20), and it has been shown that a mannitol cycle is not involved in NADPH regeneration in Stagonospora nodorum (19), Aspergillus niger (16), and Alternaria alternata (21). However, all enzymes of the cycle were detected in both sporulating and nonsporulating mycelia in A. niger (16), suggesting that a cycle could operate in this fungus. Fungi are able to use mannitol as a sole carbon source but do so in various ways (7).
Fig. 1.
Putative mannitol cycle in fungi as proposed by Hult and Gatenbeck (9). HXK, hexokinase (EC 2.7.1.1); MTD, mannitol dehydrogenase (EC 1.1.1.138); MPD, mannitol-1-phosphate dehydrogenase (EC 1.1.1.17); MPP, mannitol-1-phosphate phosphatase (EC 3.1.3.22).
d-Mannitol plays an important role in germination of Aspergillus conidia. In A. niger (23) and Aspergillus oryzae (8), mannitol accumulates in conidiospores and is utilized during the initial stages of germination. Production of mannitol appears to be largely dependent on mannitol-1-phosphate dehydrogenase (MPD) while mannitol dehydrogenase (MTD) contributes to a lesser extent (16, 19, 20).
In this study we demonstrate that MTD and MPD as well as the expression of the corresponding genes (mtdA and mpdA) are spatially separated in colonies of A. niger. This demonstrates that a mannitol cycle does not exist in this fungus and shows that spores express specific genes that are involved in germination.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The A. niger strains used in this study are listed in Table 1. A. niger strains were grown in minimal medium (MM) or complete medium (CM) (5) with the addition of a carbon source at 30°C. For growth on solid medium, 1.5% agar was added to the medium. When necessary, the medium was supplemented with 0.2 g/liter arginine, 0.2 g/liter leucine, 0.2 g/liter uridine, and/or 1 mg/liter nicotinamide.
Table 1.
Fungal strains used in this study
| Strain(s) | Genotype | Description | Reference or source |
|---|---|---|---|
| N402 | cspA1 | Low-sporulating wild type | 2 |
| NW249 | cspA1 nicA1 leuA1 pyrA6 ΔargB | Transformable strain | 11 |
| UU-A022.2 and UU-A022.4 | cspA1 nicA1 leuA1 pyrA6::pRV459 ΔargB | NW249 containing the mtdAp-H2B-GFP fusion | This study |
| UU-A075.5 and UU-A075.6 | cspA1 nicA1 leuA1 pyrA6 ΔargB::argB pRV908 | NW249 containing the mpdAp-H2B-dTomato fusion | This study |
| UU-A076.5 and UU-A075.6 | cspA1 nicA1 leuA1 pyrA6::pRV459 ΔargB pRV908 | UU-A022.2 containing the mpdAp-H2B-dTomato fusion | This study |
For expression in vegetative and sporulating colonies, all the strains were pregrown in liquid CM containing 1% glucose (CM-G). After 24 h of incubation at 250 rpm and 30°C, 12 to 15 ml of the culture was harvested directly onto a perforated polycarbonate membrane (diameter, 76 mm; pore size, 0.1 μm; Osmonics, GE Water Technologies, Trevose, PA) by suction and placed on top of CM plates containing 1% glucose. To obtain a sporulating mycelium, plates were incubated like this at 30°C for up to 24 h. To obtain a vegetative mycelium, the mycelium was covered with a second perforated polycarbonate membrane to prevent sporulation and also incubated at 30°C for up to 24 h. At the sampling times, the mycelia were harvested, dried between tissue paper, and directly frozen in liquid nitrogen.
For confocal microscopy, strains were grown on sterile microscope slides. The slides were placed in sterile petri dishes, and 2 ml of 1.5% agarose in MM solution containing 1% glucose and selection markers was placed on top of the slides. When the agarose solidified, 20 ml of MM with added glucose and selection markers was poured in each petri dish, and 5 μl of each strain at 500,000 spores per ml was added. The petri dishes were incubated at 30°C overnight to let the spores sink down and attach to the slides. The next day, liquid medium was removed, and the slides were incubated for 24 to 48 h. A coverslip was placed on top of the medium layer, and the slides were used for confocal microscopy.
For the determination of enzyme activities and metabolic levels, strains were grown on CM plates with 1% glucose, and a polycarbonate (PC) membrane (diameter, 76 mm; pore size 0.1 μm; Osmonics, GE Technologies, Trevose, PA) was placed on top. For the vegetative mycelium, 2 μl of a spore suspension was inoculated on the center of the plate and then incubated at 30°C overnight. The next day, a second similar PC membrane was placed on top of the first to prevent sporulation, and the plates were incubated again for 3 to 5 days. At the end of the incubation period, vegetative mycelia grown between two membranes were collected from membranes, dried between tissue paper, and directly frozen in liquid nitrogen. For the sporulating mycelium the same procedure was used except that the second membrane had a pore size of 10 μm (diameter, 76 mm; Osmonics, GE Technologies, Trevose, PA), allowing aerial hyphae and sporulation to develop. To harvest the spores the top membrane was removed from the plate, and spores were collected in an Eppendorf tube and frozen in liquid nitrogen (young spores). Mycelia retained on the first membrane were also harvested, dried, and frozen in liquid nitrogen. Spores were also collected from normal spore plates by adding saline-Tween solution directly to a spore plate with CM medium (mature spores). The suspension was washed twice by centrifugation with cold (2 to 3°C) saline solution. After the last washing step, liquid was removed, and the spore pellet was resuspended in 1 ml of the same solution, transferred to a 1.5-ml Eppendorf tube, and centrifuged for 5 min at 6,000 rpm. The liquid was removed, and the resulting spore pellet was frozen in liquid nitrogen (mature spores).
Molecular biology methods.
General methods (PCR, ligation, digestion, transformation of Escherichia coli DHF5αF, plasmid DNA isolation, and gel electrophoresis) were performed according to standard procedures (17). The mtdA probe fragment and the promoter were amplified with the primer pair mtdA-up (GCAGCAGGCCAGATGTTC) and mtdA-dw (TTGTCCGGGTCATCCTTG) and the pair mtdA-prom-NotI-dw (GCGGCCGCAAATACAGCATATCC) and mtdA-H2B-KpnI-up (GGTACCTGCAGTAGATGATTGTTG), respectively, by PCR using A. niger N402 chromosomal DNA as a template. Similarly, the mpdA probe fragment and the promoter were amplified with the primer pair mpdA-dw (CTCCACAAGGCGGGCTAC) and mpdA-up (CTCGACAGCATCCATCAAGG) and the pair mpdAp-up-HinDIII (AAGCTTGGAAGACTGATCAAAAG) and mpdAp-down-NotI (GCGGCCGCTAACAGTAGAATCTC), respectively.
An mtdA promoter-histone 2B (H2B)-green fluorescent protein (GFP)-trpC terminator fusion construct (pRV459) was obtained by exchanging the agsA promoter fragment from PagsA-H2BGFP (4) for the mtdA promoter using an NotI-KpnI digest. An mpdA promoter-H2B-dTomato-trpC terminator fusion construct (pRV908) was obtained by first ligating the NotI blunt fragment of the mpdA promoter in pRV459 digested with NotI and KpnI (made blunt) and replacing the mtdA promoter with the mpdA promoter, resulting in plasmid pRV907. Second, an NcoI/BamHI fragment containing dTomato was ligated in pRV907 digested with NcoI and BamHI, replacing GFP with dTomato.
Both constructs were transformed into A. niger NW249. For each transformation, 20 transformants were analyzed by fluorescence microscopy, and 2 transformants of each transformation were selected (Table 1). pRV908 was also transformed into strain UU-A022.2, resulting in transformants containing both fusion constructs. Again, 20 transformants were analyzed by fluorescence microscopy, and 2 transformants were selected (Table 1). A. niger transformations were carried out as described previously (12).
RNA isolation and Northern analysis were performed as described previously (6).
Fluorescence microscopy.
For fluorescence microscopy, a Zeiss Axioskop 2 Plus microscope was used with an HBO 100 lamp. The filter sets used were set 09 (fluorescein isothiocyanate [FITC]; excitation, band-pass [BP] 450 to 490; beam splitter, FT 510; emission, long pass [LP] 515) for GFP fluorescence and set 15 (excitation, BP 546/12; beam splitter, FT 580; emission, LP 590) for dTomato fluorescence.
For confocal microscopy, an Axiovert 200 M microscope with an Apochromat 40× (1.3 numerical aperture) water immersion objective and Zeiss LSM 5 Pa confocal laser scanning microscope were used. For GFP, an argon laser was used for excitation, and Haupt-Farb-Teiler (HFT; a dichroic beam splitter) 488-nm and LP 530-nm filters were used. For dTomato location, a helium-neon laser was used for excitation, and HFT 543-nm and LP 560-nm filters were used.
Enzyme assays.
To measure MTD and MPD activity, mycelia or spores were ground using a microdismembrator S (Braun Biotech). Cell extract was prepared by adding 1 ml of extraction buffer (50 mM K2HPO4, 5 mM MgCl2, 5 mM 2-mercaptoethanol, 0.5 mM EDTA) to the powdered biomass. The mixtures were centrifuged for 10 min at 12,000 rpm at 4°C, after which the supernatant was transferred to a new Eppendorf tube and kept on ice during the measurements. Enzyme activities were determined using 100 mM glycine, pH 9.6, with 0.4 mM NAD+ or NADP+ and 1 M mannitol or mannitol-1-phosphate, respectively. Absorbance changes were measured at 340 nm using a spectrometer (Spectronic Unicam UV1). Enzyme activity was calculated using the molar coefficient for NADPH and NADH (for both, ε = 6.22 mM−1 cm−1) and the following formula: activity (U/ml) = [(A/min − Abl/min) ×d × v]/(l × a × ε), where Abl/min is the increase in absorbance per minute before the addition of substrate, A/min is the increase in absorbance per minute after the addition of substrate, a is the sample volume (ml), d is the sample dilution, v is the total volume of the cuvette, and l is the length of the light path (cm). Protein concentrations of intracellular and extracellular samples were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce).
Metabolite determinations.
For metabolite extractions, 5 to 15 mg of fungal tissue (preweighed) was transferred to a 2-ml microcentrifuge tube, 700 μl of −40°C methanol was added, and the tube was shaken by hand. This mixture was frozen in liquid nitrogen, allowed to thaw on ice, and then centrifuged at 20,000 × g for 1 min, after which the supernatant was transferred to a fresh 2-ml microcentrifuge tube. Another 700 μl of −40°C methanol was added to the cell pellet, and the procedure described above was repeated. The supernatants were pooled, and an equivalent volume of 5 mg of biomass (based on the initial weight) from the supernatant was transferred to a fresh microcentrifuge tube. Fifty microliters of 0.2 mg/ml ribitol (internal standard) was added, and the samples were freeze-dried. The dried metabolites were prepared for gas chromatography-mass spectrometry (GC-MS) analysis as previously described (13). Each sample was vortexed briefly and then extracted at 70°C for 15 min with vigorous shaking. Samples were subsequently centrifuged at 20,000 × g for 3 min. The methanol supernatant was reserved, and the pellet was reextracted with 500 μl of H2O and 375 μl of chloroform by shaking vigorously for 5 min at 37°C and then finally centrifuged at 20,000 × g for 3 min. The polar phase was recovered, added to the original methanol supernatant, and lyophilized in a Speedvac concentrator. Methoximation of carbonyl groups was performed by addition of 50 μl of methoxylamine-HCl (20 mg/ml in pyridine) to the dried metabolites, followed by incubation at 30°C for 90 min with shaking. Trimethylsilyl (TMS) esters were then created by the addition of 80 μl of N-trimethylsilyl-N-methyl trifluoroacetamide (MSTFA) and incubation at 37°C for 30 min with shaking. For the GC-MS analysis, samples were injected as 1-μl derivatized metabolites in a 20:1 split ratio. The GC-MS equipment consisted of an Agilent 7680 autosampler, an Agilent 6890 gas chromatograph, and an Agilent 5973N quadrupole mass spectrometer (Agilent, Palo Alto, CA). The GC-MS system was auto-tuned using perfluorotributylamine (PFTBA). A 30-m Varian VF-5ms column with a 10-m integrated Varian EZ-Guard column was used for the gas chromatography (Varian, Palo Alto, CA). The injection temperature was 230°C, the interface temperature was 300°C, and the ion source temperature was 230°C. The carrier gas (helium) flow rate was retention time locked to elute mannitol-TMS at 30.6 min. The temperature gradient consisted of an initial temperature of 70°C, increasing 1°C per minute for 5 min before increasing to a final temperature of 300°C at a temperature ramp rate of 5.6°C per minute. Mass spectra and chromatograms were normalized to the ribitol internal standard and the weight of the sample and analyzed using AnalyzerPro (SpectralWorks Ltd., Runcorn, United Kingdom) employing the MatrixAnalyser function.
RESULTS
Expression of mtdA is dependent on sporulation.
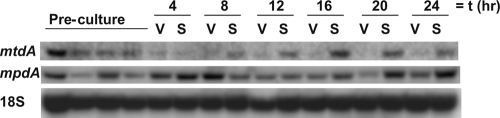
During the analysis of the Trichoderma reesei lxr1 orthologue (mtdA, encoding mannitol dehydrogenase) from A. niger (15), it was observed that this gene was expressed only in liquid culture when wall growth had occurred above the liquid medium. This led to the hypothesis that expression of mtdA is correlated with sporulation. To study this, A. niger was grown overnight in complete medium with 1% d-glucose (CM-G). The resulting mycelium was spread on perforated polycarbonate membranes that had been placed on CM-G agar plates. On 50% of the plates, the mycelium was covered with a second polycarbonate membrane to prevent sporulation. Northern analysis showed that mpdA (encoding mannitol-1-phosphate dehydrogenase) was expressed throughout culturing, irrespective of the presence of the polycarbonate membrane topping the mycelium (Fig. 2). In contrast, expression of mtdA was observed only in samples of sporulating mycelia, and levels correlated with the level of conidiation that had occurred.
Fig. 2.
Expression of mtdA and mpdA in vegetative (V) and sporulating (S) mycelia of A. niger N402. Temporal expression of mtdA and mpdA in cultures that have been grown on solid CM-G medium. Topping these cultures with a perforated polycarbonate membrane prevented formation of conidiophores (V samples), whereas in the absence of such a membrane (S samples), spores were formed starting 16 h after inoculation. The 18S gene was used as a loading control (14). Four independent precultures were mixed to obtain sufficient biomass for the experiment. Expression in each of these is shown in the figure.
Expression of mtdA and mpdA occurs in different parts of a fungal colony.
To localize expression of mtdA and mpdA in colonies of A. niger, two constructs were introduced in A. niger NW249. Construct pRV459 contains the A. nidulans histone 2B (H2B) fused to GFP under regulation of the A. niger mtdA promoter, whereas construct pRV908 contains the A. nidulans H2B fused to dTomato under regulation of the A. niger mpdA promoter. The H2B part of the fusion targets the reporter to the nucleus, ensuring that fluorescence is restricted to the part of the colony where it is expressed.
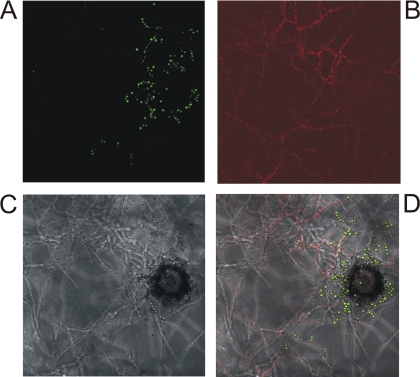
Using fluorescence (see Fig. S1 in the supplemental material) and confocal microscopy (Fig. 3), fluorescence of GFP (expression of mtdA) was observed in the spores, while red fluorescence (expression of mpdA) was detected in the substrate hyphae, demonstrating spatial differentiation of the expression of these two genes. The loose spores observed in the picture were released during the placing of the coverslip.
Fig. 3.
Localization of expression of mtdA and mpdA in A. niger UU-A076.5 using confocal microscopy. (A) GFP under the control of the mtdA promoter. (B) dTomato under the control of the mpdA promoter. (C) Bright-field image. (D) Overlay of the three images.
MtdA and MpdA activities are spatially differentiated in A. niger colonies.
Differential spatial expression profiles do not exclude the possibility that the corresponding enzymes may be active in the same part of the colony as enzymes may be transported passively or actively through the mycelium. To determine whether the enzyme activities were localized in the same part of the colony as the expression of the corresponding genes, MtdA and MpdA activities were determined in different parts of the colony. This demonstrated that MpdA activity is significantly higher in the vegetative mycelia of both sporulating and nonsporulating colonies than in young and mature conidiospores (Table 2). Sporulation induces mycelial MpdA activity as the activity is significantly higher in sporulating mycelia than in nonsporulating mycelia.
Table 2.
Mannitol cycle activities and relative intermediate concentrations in different tissues of A. niger N402
| Tissue | MTD activity (mU/mg) | MPD activity (mU/mg) | Mannitol relative abundancea | Mannitol-1- phosphate relative abundancea |
|---|---|---|---|---|
| Nonsporulating mycelium | 0.0 ± 0.0 | 68.3 ± 0.0 | 4.8 ± 1.9 | 4.4 ± 2.3 |
| Sporulating mycelium | 12.5 ± 5.5 | 453.9 ± 9.5 | 6.3 ± 2.0 | 11.7 ± 5.0 |
| Conidiospores (young) | 19.8 ± 0.0 | 2.8 ± 2.8 | ND | ND |
| Conidiospores (mature) | 236.0 ± 12.2 | 18.6 ± 4.6 | 16.8 ± 5.7 | 42.4 ± 9.9 |
Relative abundances were calculated by dividing the area of the peak representing either mannitol or mannitol-1-phosphate by that of the internal technical standard (ribitol). ND, not determined.
No MtdA activity was detected in nonsporulating mycelia, while similarly low levels of MtdA activity were detected in sporulating mycelia and young conidiospores. A significant increase in MtdA activity was observed in mature conidiospores. The low levels of MTD in mycelia and of MPD in spores are likely due to difficulties in obtaining absolutely pure spore or mycelium samples from sporulating colonies.
Relative mannitol and mannitol-1-phosphate levels were also determined in the same samples and demonstrated a significant increase in both mannitol and mannitol-1-phosphate levels in mature spores compared to both nonsporulating and sporulating mycelia (Table 2).
DISCUSSION
The presence of a mannitol cycle in fungi has been a topic of debate for several decades. While all enzyme activities involved in the putative cycle could be measured simultaneously in mycelia of several fungi, data obtained in recent years support the absence of this cycle (20). In this study we analyzed the localization of two key enzymes of the putative mannitol cycle, MTD and MPD, and demonstrated that both the expression of the corresponding genes as well as the activity of these enzymes are localized in different parts of A. niger colonies, which further supports the absence of a cycle. Expression of mtdA and MTD activity were detected only in spores. In contrast, expression of mpdA and MPD activity were detected only in vegetative mycelia, but MPD activity increased in vegetative mycelia during sporulation. This suggests that expression of mtdA is dependent on developmental processes and that expression of mpdA is positively affected by these processes. These results correlate with the presence of AbaA sites (1) in the promoters of both genes (data not shown). While mature spores are commonly considered to be dormant, detection of gene expression and enzyme activity may suggest that gene expression and metabolic activity occur in these spores. However, we cannot exclude the possibility that gene expression occurred mainly in immature spores and that the fluorescence observed in the spores originates from GFP produced during maturation of the spores. Similarly, production of MtdA could have occurred during maturation of the spores, after which the enzyme was stored in the mature spores and would become active only during germination or when the spores were disrupted. The latter hypothesis fits with the observation that, upon germination, mannitol levels in spores very rapidly decrease (23) and with earlier data in A. oryzae, where no synthesis of MTD was observed during germination (8). The absence of mtdA expression in the vegetative mycelium demonstrates that specific genes are expressed during sporulation and in specific parts of the mycelium. Absence of mpdA expression in the spores indicates that there is, in fact, a spatial separation between vegetative genes and sporulation genes, suggesting a tight regulation of the two groups. Future studies involving whole-genome expression analysis of these two parts of the colony will indicate how many genes are affected by this regulatory system and whether genes are also expressed in both parts of the colony.
The spatial differentiation of these two enzymes of the putative mannitol cycle would suggest that the cycle does not exist in A. niger. Surprisingly, though, mannitol and mannitol-1-phosphate levels were both higher in spores than in vegetative mycelia. This could suggest that the intermediates of the cycle are transported from one part of the mycelium to the other to enable completion of the cycle. However, since levels of both intermediates were higher in sporulating mycelia than in vegetative mycelia, we feel that it is more likely that both intermediates streamed from the vegetative mycelium into the conidiophores and the spores.
Previously it was reported that deletion of mpdA in A. niger results in the absence of mannitol in a vegetative mycelium (16), but spores retained 30% of the wild-type level of mannitol. Several studies of other fungi have suggested that mannitol can also be formed through the action of MTD (21, 22) and that, in fact, mannitol utilization can also occur through MPD (19). Our study demonstrated that MTD is not present in the vegetative mycelium, which can explain the absence of mannitol in this part of the colony and the presence of mannitol in spores.
Our study suggests a clear spatial and enzymatic division between mannitol formation and utilization in A. niger. Mannitol formation occurs in the vegetative hyphae through the pathway that includes mannitol-1-phosphate dehydrogenase, while mannitol utilization occurs in germinating spores through the pathway that includes mannitol dehydrogenase. This would support a role for mannitol as a storage carbon source, as suggested previously (20), that provides the required energy during germination before other metabolic systems take over. It also correlates with mannitol as a protectant molecule in spores as it is degraded upon germination. Additional studies of the pathway are needed to further clarify the biological functions of mannitol and the mannitol-related pathways.
In conclusion, the data described in this paper, in combination with previous studies in which a role in NADPH regeneration could not be detected in Aspergillus (16, 18), provide strong support for the absence of a mannitol cycle in A. niger. Our study also demonstrates that gene expression and intracellular enzyme activities are highly differentiated in different parts of fungal colonies, suggesting a regulatory mechanism to ensure the presence of the required metabolic functions in different parts of the colony. The expression and activity of mtdA demonstrate that spores are not dormant with respect to metabolic activity and that some genes are exclusively expressed in spores.
Supplementary Material
ACKNOWLEDGMENTS
R.P.D.V. and G.A-O. were supported by grants (07063 and 07938, respectively) of the Dutch Foundation for Applied Science (STW).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Andrianopoulos A., Timberlake W. E. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos C. J., Debets A. J. M., Swart K., Huybers A., Kobus G., Slakhorst S. M. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437–443 [DOI] [PubMed] [Google Scholar]
- 3.Corina D. L., Munday K. A. 1971. Studies on polyol function in Aspergillus clavatus: a role for mannitol and ribitol. J. Gen. Microbiol. 69:221–227 [DOI] [PubMed] [Google Scholar]
- 4.Damveld R. A., Franken A., Arentshorst M., Punt P. J., Klis F. M., van den Hondel C. A. M. J. J., Ram A. F. J. 2008. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178:873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries R. P., Burgers K., van de Vondervoort P. J. I., Frisvad J. C., Samson R. A., Visser J. 2004. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl. Environ. Microbiol. 70:3954–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries R. P., van de Vondervoort P. J. I., Hendriks L., van de Belt M., Visser J. 2002. Regulation of the α-glucuronidase encoding gene (aguA) from Aspergillus niger. Mol. Gen. Genet. 268:96–102 [DOI] [PubMed] [Google Scholar]
- 7.Dijksterhuis J., de Vries R. P. 2006. Compatible solutes and fungal development. Biochem. J. 399:e3–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horikoshi K., Iida S., Ikeda Y. 1965. Mannitol and mannitol dehydrogenases in conidia of Aspergillus oryzae. J. Bacteriol. 89:326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hult K., Gatenbeck S. 1978. Production of NADPH in the mannitol cycle and its relation to polyketide formation in Alternaria alternata. Eur. J. Biochem. 88:607–612 [DOI] [PubMed] [Google Scholar]
- 10.Hult K., Veide A., Gatenbeck S. 1980. The distribution of the NADPH regenerating mannitol cycle among fungal species. Arch. Microbiol. 128:253–255 [DOI] [PubMed] [Google Scholar]
- 11.Jalving R., van de Vondervoort P. J. I., Visser J., Schaap P. J. 2000. Characterization of the kexin-like maturase of Aspergillus niger. Appl. Environ. Microbiol. 66:363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusters-van Someren M. A., Harmsen J. A. M., Kester H. C. M., Visser J. 1991. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20:293–299 [DOI] [PubMed] [Google Scholar]
- 13.Lowe R. G. T., Lord M., Rybak K., Trengove R. D., Oliver R. P., Solomon P. S. 2008. A metabolomic approach to dissecting osmotic stress in the wheat pathogen Stagonospora nodorum. Fungal Genet. Biol. 45:1479–1486 [DOI] [PubMed] [Google Scholar]
- 14.Melchers W. J. G., Verweij P. E., van den Hurk P., van Belkum A., de Pauw B. E., Hoogkamp-Korstanje A. A., Meis J. F. G. M. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metz B., de Vries R. P., Polak S., Seidl V., Seiboth B. 2009. The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a D-mannitol dehydrogenase and is not involved in L-arabinose catabolism. FEBS Lett. 583:1309–1313 [DOI] [PubMed] [Google Scholar]
- 16.Ruijter G. J. G., Bax M., Patel H., Flitter S. J., van de Vondervoort P. J. I., vanKuyk P. A., de Vries R. P., Visser J. 2003. Mannitol is required for stress tolerance in Aspergillus niger conidiospores. Eukaryot. Cell 2:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 18.Singh M., Scrutton N. S., Scrutton M. C. 1988. NADPH generation in Aspergillus nidulans: is the mannitol cycle involved? J. Gen. Microbiol. 134:643–654 [DOI] [PubMed] [Google Scholar]
- 19.Solomon P. S., Waters O. D., Jorgens C. I., Lowe R. G., Rechberger J., Trengove R. D., Oliver R. P. 2006. Mannitol is required for asexual sporulation in the wheat pathogen Stagonospora nodorum (glume blotch). Biochem. J. 399:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomon P. S., Waters O. D., Oliver R. P. 2007. Decoding the mannitol enigma in filamentous fungi. Trends Microbiol. 15:257–262 [DOI] [PubMed] [Google Scholar]
- 21.Velez H., Glassbrook N. J., Daub M. E. 2007. Mannitol metabolism in the phytopathogenic fungus Alternaria alternata. Fungal Genet. Biol. 44:258–268 [DOI] [PubMed] [Google Scholar]
- 22.Voegele R. T., Hahn M., Lohaus G., Link T., Heiser I., Mendgen K. 2005. Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae. Plant Physiol. 137:190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witteveen C. F. B., Visser J. 1995. Polyol pools in Aspergillus niger. FEMS Microbiol. Lett. 134:57–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.