Abstract
Candida dubliniensis is closely related to Candida albicans; however, it is responsible for fewer infections in humans and is less virulent in animal models of infection. C. dubliniensis forms fewer hyphae in vivo, and this may contribute to its reduced virulence. In this study we show that, unlike C. albicans, C. dubliniensis fails to form hyphae in yeast extract-peptone-dextrose (YPD) medium supplemented with 10% (vol/vol) fetal calf serum (YPDS medium). However, C. dubliniensis filaments in water plus 10% (vol/vol) fetal calf serum (WS), and this filamentation is inhibited by the addition of peptone and glucose. Repression of filamentation in YPDS medium could be partly overcome by preculture in synthetic Lee's medium. Unlike C. albicans, inoculation of C. dubliniensis in YPDS medium did not result in increased UME6 transcription. However, >100-fold induction of UME6 was observed when C. dubliniensis was inoculated in nutrient-poor WS medium. The addition of increasing concentrations of peptone to WS medium had a dose-dependent effect on reducing UME6 expression. Transcript profiling of C. dubliniensis hyphae in WS medium identified a starvation response involving expression of genes in the glyoxylate cycle and fatty acid oxidation. In addition, a core, shared transcriptional response with C. albicans could be identified, including expression of virulence-associated genes including SAP456, SAP7, HWP1, and SOD5. Preculture in nutrient-limiting medium enhanced adherence of C. dubliniensis, epithelial invasion, and survival following coculture with murine macrophages. In conclusion, C. albicans, unlike C. dubliniensis, appears to form hyphae in liquid medium regardless of nutrient availability, which may account for its increased capacity to cause disease in humans.
Candida dubliniensis is the closest known relative of Candida albicans, the predominant fungal pathogen of humans (27, 28). Epidemiological evidence has shown that C. albicans is more prevalent in the human population as a commensal of the oral cavity and is responsible for more infections (both oral and systemic) than C. dubliniensis (10, 13, 15). C. albicans is responsible for approximately 60% of cases of candidemia, whereas C. dubliniensis accounts for fewer than 2% of cases (13). Evidence from animal infection models also suggests that C. dubliniensis is less virulent than C. albicans (26, 29). Following oral-intragastric inoculation, C. dubliniensis strains are more rapidly cleared from the gastrointestinal tract than C. albicans strains and are less able to establish disseminated infection (26). Following tail vein inoculation in the systemic mouse model of infection, only a small number of C. dubliniensis isolates have been shown to establish disseminated infections, and most studies conclude that C. dubliniensis isolates are generally less virulent than C. albicans isolates (1, 29).
Virulence studies have associated the reduced capacity of C. dubliniensis to establish infection with a reduced ability to undergo the transition from yeasts to hyphae (1, 26). In the oral-intragastric infection model, C. dubliniensis cells in the stomach and kidney were found to be in the yeast form only, while using the same models C. albicans cells were found to be in both the yeast and hyphal forms (26). Asmundsdottir et al. (1) also noted that C. dubliniensis produced significantly fewer hyphae than C. albicans following dissemination to the liver and kidney in mice. In vitro, C. dubliniensis forms true hyphae less efficiently than C. albicans in response to serum, pH shifts in Lee's medium, and CO2 and in certain defined media such as RPMI 1640 medium (16, 26). Poor hypha production has also been observed in C. dubliniensis in vitro during coculture with murine macrophages and during infection of reconstituted human oral epithelial tissues (16, 24). This results in an inability of C. dubliniensis to evade macrophage killing and limited invasion of epithelial surfaces.
Although C. dubliniensis produces true hyphae less efficiently than C. albicans, C. dubliniensis can produce abundant pseudohyphae and chlamydospores on certain solid media (27). Recently, Staib and Morschhauser (25) demonstrated that the propensity for C. dubliniensis to form large numbers of chlamydospores on these media was due to species-specific downregulation of the NRG1 repressor. Further studies have shown that downregulation of the NRG1 transcript is also required for efficient production of true hyphae in C. albicans in response to serum (22). We have shown that under conditions where C. dubliniensis fails to filament, for example, following phagocytosis by murine macrophages, this species does not downregulate NRG1, whereas C. albicans responds to these conditions by shutting down NRG1 transcription (16). Deletion of the NRG1 gene in C. dubliniensis can partly offset the failure of this species to filament in vitro and leads to more efficient production of hyphae in response to serum and CO2 and during coculture with murine macrophages (16).
In this study, we have examined in detail the environmental signals required for filamentation in C. dubliniensis. We have shown that nutrient-rich conditions inhibit efficient hypha formation by suppressing UME6 expression in C. dubliniensis. This study also includes the first description of a C. dubliniensis-specific microarray that we used to generate a transcript profile for C. dubliniensis true hyphae. The effects of inducing hypha formation in C. dubliniensis under these conditions on the ability to infect reconstituted oral epithelial tissues and to evade macrophage killing were also examined.
MATERIALS AND METHODS
Candida strains and culture conditions.
All Candida strains were routinely cultured on yeast extract-peptone-dextrose (YPD) agar at 37°C. For liquid culture, cells were grown with shaking (at 200 rpm) in YPD broth at 30°C or 37°C, as indicated in the figure legends (9). Genotypes of strains used in this study are listed in Table S1 in the supplemental material. Liquid culture was also carried out at 30°C in the liquid medium of Lee et al. (14) supplemented with 400 mM arginine, 0.001% (wt/vol) biotin, and trace metals (0.2 mM ZnSO, 0.25 mM CuSO, 1 mM FeCl, 1 mM MgCl, and 1 mM CaCl). Where indicated in Fig. 5b and 7f, Lee's medium was buffered to pH 5.0 or pH 7.2 with 0.1 M potassium phosphate buffer. Supplementation of Lee's and other media with peptone was carried out with bacteriological peptone (Oxoid). Peptone supplementation up to 2% (wt/vol) did not significantly alter the pH of Lee's medium or serum. Hyphal induction was carried out in liquid YPD plus 10% (vol/vol) fetal calf serum (YPDS) or in sterile Milli-Q H2O supplemented with 10% (vol/vol) fetal calf serum (WS) at 37°C. The proportion of germ tubes or hyphae in each culture was assessed at intervals by microscopic examination of an aliquot of culture with a Nikon Eclipse 600 microscope (Nikon U.K., Surrey, United Kingdom).
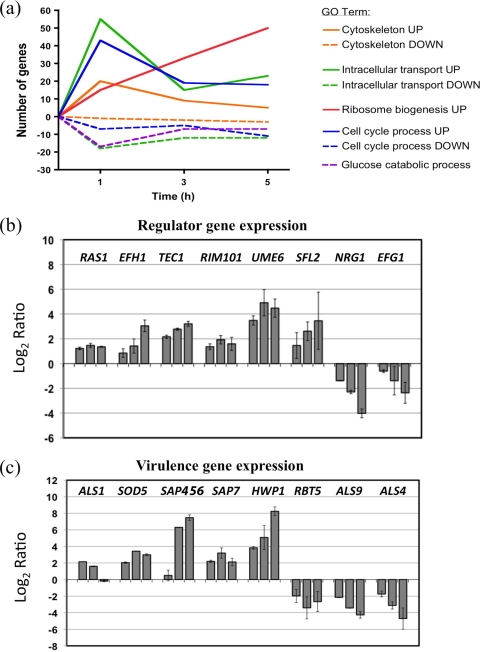
Fig. 5.
(a) Graphical representation of the changes in expression in selected Gene Ontology (GO) groups during filamentation in C. dubliniensis. The total number of genes up- or downregulated 2.5-fold in each group is shown at each time point. (b) Microarray expression of selected regulators of filamentous growth during hypha formation in C. dubliniensis Wü284 in WS medium. Columns for each gene show expression levels relative to preculture cells at (left to right) 1, 3, and 5 h postinoculation. (c) Microarray expression of selected virulence-associated genes during hypha formation in C. dubliniensis Wü284 in WS medium. Columns for each gene show expression levels relative to preculture cells at (left to right) 1, 3, and 5 h postinoculation. Error bars in panels b and c represent standard deviations from the mean generated in Genespring GX11 from two distinct oligonucleotide probes per gene in four biological replicate experiments.
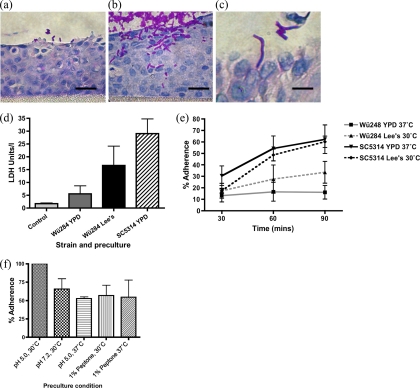
Fig. 7.
Interaction of C. dubliniensis Wü284 with reconstituted human oral epithelium (RHE) following 24 h of incubation. (a) Photomicrograph of C. dubliniensis yeast cells at the surface of RHE following preculture in YPD medium at 37°C. Scale bar, 25 μm. (b) Localized invasion of the surface of the RHE by C. dubliniensis following preculture in Lee's medium at pH 4.5 and 30°C. Scale bar, 25 μm. (c) High-magnification photomicrograph of a hyphal C. dubliniensis cell penetrating the surface of the RHE, following preculture in Lee's medium at pH 4.5 and 30°C. Scale bar, 10 μm. (d) Damage to the RHE tissues estimated by measurement of lactate dehydrogenase (LDH) release in control (uninfected) tissues, tissues infected with cells pregrown in YPD medium, and tissues infected with cells pregrown in Lee's medium at pH 4.5 after 24 h of incubation. (e) Adherence of C. dubliniensis Wü284 and C. albicans SC5314 to TR146 monolayers over time. Adherence was determined in cells precultured in YPD medium at 37°C and in Lee's medium at 30°C and is expressed as the percentage of adherent CFU relative to the inoculum. Error bars represent standard deviation from the mean of three replicate experiments. (f) Examination of adherence of C. dubliniensis Wü284 precultured in various modifications of Lee's medium, including medium buffered to pH 5.0 and pH 7.2, incubated at 37°C, or supplemented with peptone. Error bars represent standard deviations from the mean of three replicate experiments.
Genetic manipulation of C. dubliniensis.
Ectopic expression of C. albicans UME6 (CaUME6) in C. dubliniensis was achieved using plasmid pCaUme6-3, containing UME6 under the control of a doxycycline-inducible promoter (32). The expression cassette was released from pCaUme6-3 by ApaI and PmlI digestion and was used to transform strains Wü284 and CDM10 by electroporation, as described previously (16). Plasmid pNRG1 was generated from plasmids pNIM1 and pTET42 (18). NRG1 was removed from pTET42 as an SalI/BglII fragment and ligated to SalI/BglII-digested pNIM1 to generate pNRG1. The expression cassette was released from pNRG1 by SacII and KpnI digestion and was used to transform Wü284 and CDM10 by electroporation, as described previously (16). Integration of pNIM1 derivatives at the ADH1 locus was confirmed by PCR.
In order to create strains harboring a PECE1-GFP fusion, we used the integrating vector pCDRI (16). A derivative of this plasmid was created by inserting yeast enhanced green fluorescent protein (yEGFP) fused to the actin terminator on a HindIII/MluI fragment to create pGM175. An ECE1 promoter fragment from bases −1 to −921 was amplified from C. albicans SC5314 with primers ECEAF (GTACGGGCCCAAGAGTCTCATTCAGATAACG) and ECEXR (GCATCTCGAGTTTAACGAATGGAAAATAGTTG) and cloned upstream of yEGFP following digestion of both fragments with ApaI and XhoI. The plasmid was linearized within the CDR1 region and used to transform C. albicans SC5314, C. dubliniensis Wü284, and the nrg1Δ derivative CDM10 as described previously (16). Ectopic integration in the CDR1 gene was confirmed by Southern hybridization.
Transcriptional profiling with oligonucleotide microarrays.
A set of 5,999 open reading frames (ORFs) from the CD36 genome was used to design a C. dubliniensis expression microarray. Two unique 60-mer oligonucleotides specific for each ORF were designed using the Agilent eArray probe design tool. Each 60-mer was printed in quadruplicate on glass slides by Agilent technologies. To examine the hyphal transcript profile of C. dubliniensis strain Wü284, the strain was grown for 18 h in Lee's medium (pH 4.5) at 30°C with shaking, washed in sterile H2O, and inoculated in 200 ml of H2O plus 10% (vol/vol) fetal calf serum to a density of 2 × 106 cells/ml. Samples (50 ml) were removed for RNA preparation at 1, 3, and 5 h postinoculation. To examine the effects of cell density changes, nutrient depletion, a shift to 37°C, and a shift to alkaline pH, identical 18-h Lee's medium cultures were washed and inoculated at 2 × 106 cells/ml in (i) fresh Lee's medium (pH 4.5) at 30°C, (ii) 10% (vol/vol) Lee's medium (pH 4.5) at 30°C, (iii) Lee's medium (pH 4.5) at 37°C, and (iv) Lee's medium (pH 7.2) at 30°C. RNA was extracted from these cultures following 3 h of incubation under each condition. To identify NRG1-regulated genes in C. dubliniensis, RNA was extracted from Wü284 and its nrg1Δ derivative, CDM10, following growth to an optical density at 600 nm (OD600) of 1.0 in YPD broth at 30°C. For RNA preparation, cell pellets were snap-frozen in liquid N2 and disrupted using Mikro-Dismembrator S system (Sartorius Stedim Biotech, Göttingen, Germany). RNA was prepared using TRI-Reagent (Sigma Chemical Co.) according to the manufacturer's instructions. Poly(A) mRNA was then isolated using a Sigma Genelute mRNA isolation kit. A 200-ng aliquot of mRNA was labeled with Cy5 or Cy3 using an Agilent Two-Color Low RNA Input Linear Amplification Kit PLUS, according to the manufacturer's instructions. Hybridization and washing of the arrays were carried out using an Agilent Gene Expression Hybridization Kit and Gene Expression Wash Pack according the manufacturer's instructions. For each condition, four biological replicate experiments were performed, including two dye swap experiments. Slides were scanned using a GenePix personal 4100A scanner (Axon), and data were extracted using GenePix Pro, version 6.1 (Axon). Spots were flagged absent if the signal was less than background +1 standard deviation in both fluorescent channels. Raw data were exported to GeneSpring GX11, and signals for each replicate spot were background corrected and normalized using Loess normalization. Log2 fluorescence ratios were generated for each replicate spot and averaged. Oligonucleotides were excluded from analysis if >50% of replicates in each condition were flagged absent. Genes differentially expressed across all conditions were identified by analysis of variance (ANOVA) with a Student Newman Keuls (SNK) posthoc test in Genespring GX11. A total of 7,107 oligonucleotide probes were significantly differentially expressed, with a corrected P value (Benjamini-Hochberg false discovery rate [FDR]) of ≤0.05. Hierarchical clustering was used to compare gene expression under each condition using the default settings in Genespring GX11. Some individual samples (sera from 1 h, 3 h, and 5 h) were also analyzed using a one-sample t test in order to identify genes exhibiting significant differential expression (2-fold or greater) from preculture cells. All P values were adjusted using the Benjamini-Hochberg multiple correction test to limit false differential gene expression, and oligonucleotides with P values of ≤0.015 were selected for analysis.
The C. albicans hypha-induced gene set used in this study included the hypha-regulated genes identified by Nantel et al. (17) and by Kadosh and Johnson (12). Additional C. albicans hypha-regulated genes were identified in the data set of Kadosh and Johnson (12) following analysis of the data set with GeneSpring GX11. These additional genes were included if they exhibited significant (>2-fold) regulation (t test, P ≤ 0.01) in the 2-h and 3-h data sets (12).
Real-time PCR analysis of gene expression.
Cultures for RNA preparation for quantitative reverse transcription-PCR (QRT-PCR) were set up in identical fashion to those used for microarray analysis. RNA for QRT-PCR was isolated using an RNeasy Mini-kit (Qiagen). Cells were disrupted using a FastPrep bead beater (Bio101). RNA samples were rendered DNA free by incubation with Turbo-DNA free reagent (Ambion, Austin, TX). cDNA synthesis was carried out as described by Moran et al. (16). Primers used in this study are listed in Table S2 in the supplemental material and were designed using Primer Express software, version 1.5 (Applied Biosystems, Foster City, CA). These primers yielded single, specific amplimers from genomic DNA and cDNA templates. Primer pairs for UME6 and NRG1 were selected that yielded similar amplification efficiencies as the TEF1 primer pair against a serial dilution of template DNA. Real-time detection of amplimers was carried out using a Power SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) and an ABI 7500 sequence detector, with separate reactions performed for each gene. Gene expression levels were normalized against the expression levels of the constitutively expressed TEF1 gene in the same cDNA sample.
Epithelial adhesion and invasion studies.
Adherence of Candida strains to monolayers of the oral epithelial cell line TR146 was determined using the assay of Rotrosen et al. (20). Monolayers of TR146 cells were cultured in six-well tissue culture dishes in complete medium (CM), which consisted of Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). A suspension of 2 × 102 yeast cells per ml was prepared in CM, and 1 ml was added to triplicate wells and incubated at 37°C in 5% (vol/vol) CO2 for 30, 60, or 90 min. The same suspension was also plated on YPD agar to enumerate CFU in the starting inoculum. Following incubation, nonadherent cells were removed from the monolayer by washing with 10 ml of phosphate-buffered saline (PBS). The monolayer was then overlaid with 2 ml of YPD agar and incubated at 37°C overnight. The number of colonies present on the monolayers relative to the starting inoculum was determined, and results are expressed as percentage adherence. Statistical analysis of the data was performed using ANOVA in Prism, version 4.0 (GraphPad Software).
Invasion of reconstituted human oral epithelial (RHE) tissue of TR146 cells was determined using RHE tissues purchased from Skinethic Laboratories (Nice, France) and used as described previously (23, 26). The release of lactate dehydrogenase (LDH) from epithelial cells into the cell culture medium was measured to quantify the extent of epithelial cell damage using a CytoTox 96 nonradioactive cytotoxicity assay (Promega Corp., Madison, WI) as described by Moran et al. (16).
Macrophage cell culture and infection with Candida.
Infection of the murine macrophage-like cell line RAW264.7 with Candida isolates was carried out as described by Moran et al. (16). Evaluation of yeast cell proliferation in coculture with macrophages was assessed after 18 h of incubation using an XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] dye reduction assay (Sigma-Aldrich), also described by Moran et al. (16).
Microarray data accession number.
Results from all 32 microarrays have been submitted to the Gene Expression Omnibus (GEO) archive under accession number GSE20537.
RESULTS
Effect of nutrient concentration on hypha formation in C. dubliniensis.
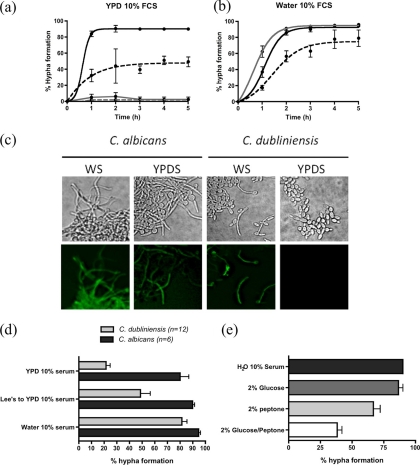
Previous studies have examined the transcript profile of C. albicans hyphae when they are induced in YPD medium supplemented with 10% (vol/vol) fetal calf serum (YPDS) at 37°C (12, 17). In this study, we wished to compare the transcript profiles of C. dubliniensis Wü284 hyphae induced in YPDS medium. However, preliminary experiments demonstrated that C. dubliniensis did not produce sufficient numbers of true hyphae under these conditions over a period of 5 h (Fig. 1a). This differential filamentation phenotype was confirmed with an additional 11 C. dubliniensis isolates and five C. albicans isolates (Fig. 1d). On average, 21% (range, 9 to 43%) of yeast cells in C. dubliniensis YPDS cultures produced germ tubes or filaments following 2 h of incubation (Fig. 1d). In contrast, 80% (range, 61 to 95%) of cells in C. albicans cultures produced germ tubes or filaments under the same conditions (Fig. 1d). Previous studies by Stokes et al. (26) demonstrated that water supplemented with 10% fetal calf serum (WS) was a more potent inducer of C. dubliniensis hyphae. Under these conditions, C. dubliniensis Wü284 was approximately 90% hyphal after 3 h of incubation (Fig. 1b). Eleven additional C. dubliniensis isolates exhibited significantly increased rates of filamentation in WS medium compared to filamentation in YPDS medium, whereas the rate of filamentation in six C. albicans isolates was similar in both media (Fig. 1d). Induction of hypha-specific gene expression was examined by observing induction of yEGFP expression from the CaECE1 promoter in both species. C. albicans produced fluorescent hyphae in WS and YPDS media, whereas cells of C. dubliniensis produced fluorescence only in WS medium (Fig. 1c).
Fig. 1.
(a) Hypha formation in YPD medium plus 10% FCS (YPDS) by C. dubliniensis Wü284 (gray lines) and C. albicans SC5314 (black lines) following preculture in YPD medium at 30°C (solid lines) or 37°C (dashed lines). (b) Enhanced filamentation of C. dubliniensis Wü284 in water plus 10% fetal calf serum (WS) following preculture in YPD medium at 30°C (solid black line), YPD medium at 37°C (dashed black line), or in Lee's medium at pH 4.5 and 30°C (gray line). Error bars correspond to standard deviations in at least three replicate experiments. A sigmoidal curve was fitted to the data for visualization using Prism, version 4.0 (GraphPad Software, Inc.). (c) Examination of induction of GFP expression from the hypha-specific ECE1 promoter in C. albicans and C. dubliniensis in YPDS and WS media. (d) Average percent hypha formation in C. dubliniensis (12 isolates) and C. albicans (6 isolates) at 37°C in YPD medium plus 10% serum (YPDS), in YPDS following preculture in Lee's medium at pH 4.5, and in water plus 10% serum (WS). Error bars correspond to the standard error of the mean. (e) Filamentation of C. dubliniensis Wü284 in WS medium supplemented with 2% peptone, 2% glucose, or both peptone and glucose.
These data suggest that efficient filamentation in C. dubliniensis requires nutrient depletion. We investigated whether the addition of nutrients present in YPD medium such as glucose or peptone to C. dubliniensis incubated in WS medium could inhibit filamentation. The addition of 2% (wt/vol) glucose to WS cultures had no significant effect on the rate of filamentation of C. dubliniensis Wü284 (Fig. 1e). However, a reduction in filamentation was observed upon the addition of 2% (wt/vol) peptone, and a greater effect was observed when WS medium was supplemented with both glucose and peptone (Fig. 1e). As the addition of a nitrogen source (peptone) determined whether filamentation could proceed in C. dubliniensis, we carried out further experiments to characterize whether different nitrogen sources could mediate this effect. Addition of whole protein (bovine serum albumin [BSA]) to C. dubliniensis WS cultures elicited similar effects as peptone but did not affect filamentation by C. albicans. Ammonium sulfate (100 mM) had no effect on filamentation of C. albicans but resulted in an ∼50% reduction in filamentation by C. dubliniensis. Addition of specific amino acids (up to 10 mM methionine, tryptophan, histidine, or proline) to WS medium did not affect filamentation of either species.
Preculture in Lee's medium at pH 4.5 enhances filamentation in C. dubliniensis.
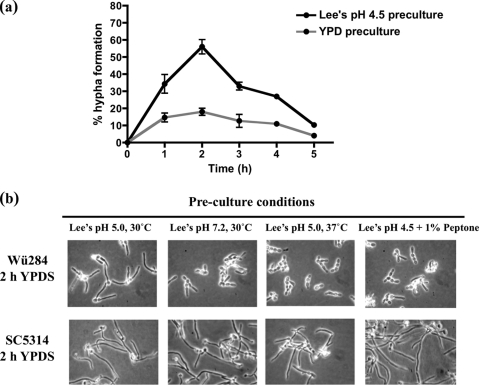
We investigated whether preculture in Lee's medium, a peptone-free synthetic medium, could affect subsequent filamentation of C. dubliniensis in YPDS medium. Cells precultured in Lee's medium (pH 4.5) at 30°C showed a greater capacity to form true hyphae than cells precultured in YPD medium (pH 5.6), also at 30°C (Fig. 2a). Following preculture in Lee's medium, approximately 56% of cells were observed to produce germ tubes (Fig. 2a). However, budding growth resumed after several hours of incubation, indicating that Lee's medium preculture alone could not maintain hyphal elongation under these conditions (Fig. 2a). We examined whether the pH shift, the temperature shift, or the nutrient composition of Lee's medium was responsible for this phenotype. Preculture in Lee's medium at 37°C or in Lee's medium buffered to pH 7.2 could inhibit filamentation in strain Wü284, indicating that a pH and temperature shift was required (Fig. 2b). However, we also showed that addition of 1% peptone to Lee's medium could inhibit subsequent filamentation by Wü284 in YPDS medium, indicating that the medium composition also played a role (Fig. 2b). In C. albicans SC5314, the addition of peptone (1%) to the Lee's preculture medium could not inhibit filamentation in YPDS medium (Fig. 2b), whereas preculture of C. albicans at 37°C in Lee's medium increased the numbers of pseudohyphae relative to true hyphae (Fig. 2b).
Fig. 2.
(a) Filamentation rate of strain Wü284 in YPDS medium at 37°C following preculture in YPD broth at 30°C or following preculture in Lee's medium at pH 4.5 and 30°C. Error bars correspond to standard deviations in three replicate experiments. (b) Photomicrographs showing typical morphologies of C. dubliniensis Wü284 and C. albicans SC5314 following 2 h of incubation in YPD medium plus 10% (vol/vol) FCS following preculture in Lee's medium. Cells were precultured for 24 h in modified Lee's medium, buffered to pH 5.0 or 7.2 with 0.1 M potassium phosphate buffer or supplemented with 1% (wt/vol) peptone.
Lee's medium preculture enhanced filamentation in 10 of 12 additional C. dubliniensis isolates examined, exhibiting an average rate of filamentation of 48% following 2 h of incubation in YPDS medium (Fig. 1d). Analysis of six independent C. albicans isolates showed that Lee's medium preculture also enhanced filamentation in YPDS medium by approximately 10% in these isolates relative to cells precultured in YPD medium (Fig. 1d).
Regulation of UME6 and NRG1 transcription.
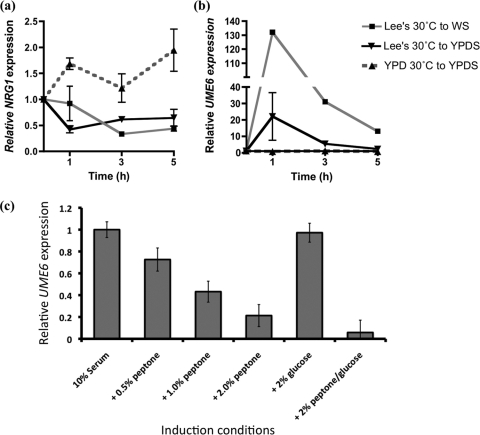
Previous studies have shown that in C. albicans, filamentation in YPDS medium is associated with downregulation of NRG1 transcript levels and increased expression of UME6 (16). Examination of NRG1 transcript levels in C. dubliniensis in YPDS medium demonstrated that NRG1 transcript levels increased following 1 h of incubation in YPDS medium at 37°C (Fig. 3a). However, inoculation of cells precultured in Lee's medium resulted in a transient drop in NRG1 transcript levels by approximately 50% following 1 h (Fig. 3a). Inoculation of C. dubliniensis in WS medium yielded a 70% decrease in NRG1 transcript levels by 3 h (Fig. 3a), similar to the decreases observed during filamentation of C. albicans in YPDS medium (data not shown). Analysis of UME6 transcript levels in C. dubliniensis in YPDS medium revealed no significant change (Fig. 3b). However, when cells were precultured in Lee's medium (pH 4.5), we observed an ∼30-fold increase in UME6 expression in YPDS medium (Fig. 3b). In addition, we observed >100-fold induction of UME6 in C. dubliniensis following inoculation in WS medium (Fig. 3b).
Fig. 3.
Real-time PCR analysis showing relative levels of NRG1 transcript (a) and relative levels of UME6 transcript (b) in C. dubliniensis incubated in serum-containing medium. Expression levels were normalized to TEF1 expression levels in each sample. C. dubliniensis was cultured in WS medium following preculture in Lee's medium at 30°C, in YPDS medium following preculture in Lee's medium at 30°C, and in YPDS medium following preculture in YPD medium at 30°C. Error bars represent standard deviations of results from three replicate RNA preparations. In the case of UME6 expression in WS medium, representative data from one replicate are shown; additional experiments all showed >100-fold induction at 1 h. (c) Relative expression of UME6 in C. dubliniensis in WS medium supplemented with additional nutrients. Cells were precultured in YPD medium at 30°C and inoculated in WS medium alone or supplemented with the indicated concentrations of peptone or glucose. UME6 expression levels were normalized to TEF1 expression levels in the same sample.
Addition of peptone to WS cultures showed that peptone could decrease UME6 expression in C. dubliniensis in a concentration-dependent manner, with 2% (wt/vol) peptone reducing UME6 expression by approximately 80%. Glucose (2% wt/vol) alone did not significantly decrease UME6 expression although the combination of glucose and peptone had an additive effect on UME6 expression.
Overexpression of UME6 enhances filamentation in C. dubliniensis.
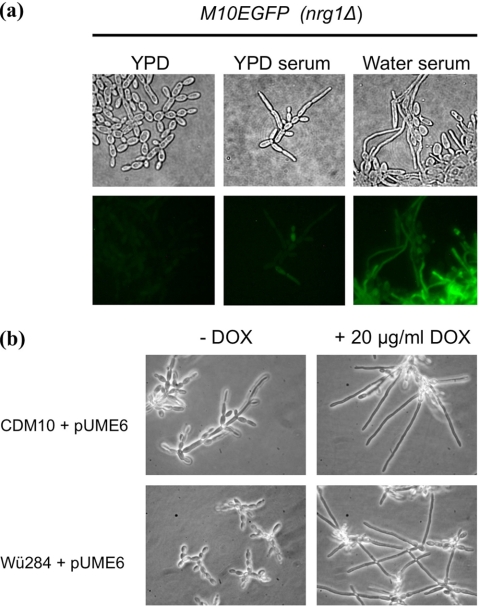
We further investigated the roles of NRG1 and UME6 in hypha formation in the C. dubliniensis nrg1Δ mutant CDM10. Previously, we have shown that the nrg1Δ strain, unlike the wild type, forms hyphae in response to CO2 and filaments more rapidly in response to serum in water (16). In this study, a derivative of CDM10 harboring a PECE1-GFP promoter fusion (M10EGFP) formed elongated filaments in YPDS medium; however, these filaments possessed the characteristic constrictions of pseudohyphae (Fig. 4a). Strain M10EGFP was weakly fluorescent in YPD and YPDS media (Fig. 4a), whereas in WS medium the same strain emitted strong fluorescence and formed masses of true hyphae (Fig. 4a). We tested whether overexpression of UME6 from a doxycycline-inducible promoter could enhance true hypha production by CDM10 in YPDS medium. Addition of 20 μg/ml doxycycline promoted conversion of pseudohyphae to true hyphae in this strain (Fig. 4b). Similarly, introduction of the same construct in the parent isolate Wü284 could promote the formation of true hyphae in YPDS medium (Fig. 4b).
Fig. 4.
(a) Photomicrographs showing morphology of a derivative of the C. dubliniensis nrg1Δ mutant harboring a PECE1-GFP construct (CDM10). Top panel shows morphology in YPD, YPDS, and WS media. Lower panel shows levels of fluorescence expressed from the PECE1-GFP fusion under each condition. (b) Morphology of CDM10 and Wü284 derivative strains harboring plasmid pCaUME6, containing the UME6 gene under the control of a doxycycline-inducible promoter. Morphology is shown following 3 h of incubation in YPDS medium with (+) or without (−) 20 μg/ml doxycycline (DOX).
We also tested whether constitutive NRG1 expression from the doxycycline-inducible promoter could prevent filamentation. Constitutive expression of NRG1 in Wü284 and CDM10 could block pseudohypha formation in YPDS medium. However, expression of NRG1 from this promoter was not sufficient to block true hypha formation in WS medium (data not shown).
Transcript profiling of C. dubliniensis in serum.
This study has shown that under nutrient depleted conditions, C. dubliniensis can form hyphae as effectively as C. albicans. In order to determine whether C. dubliniensis hyphae can express the same range of virulence-associated factors as C. albicans hyphae, we carried out whole-genome transcript profiling of C. dubliniensis during growth in WS medium. Samples were analyzed at 1 h, 3 h, and 5 h postinoculation in WS medium. Within 1 h, we observed a 2.5-fold or greater change in transcription in 1,095 genes relative to preculture cells (t test, P < 0.015) (see Table S3 in the supplemental material). This corresponds to 18% of the genome. Analysis of the upregulated genes (n = 526) for significant shared Gene Ontology (GO) terms identified large groups of genes associated with transport (102 genes), organelle organization (73), the cell cycle (44), and translation (43) (Fig. 5a). Many of these genes were associated with processes known to be involved in hyphal development, such as the assembly of actin cables (TPM2, ARF3, MEA1, ARP9, and YEL1), Spitzenkörper assembly (MLC1), and GTPases with roles in actin organization (RSR1, RAC1, RDI1, and RHO3) (see Fig. S1 in the supplemental material). These data also highlighted some processes not previously associated with hypha formation, such as downregulation of vacuolar metabolism, including vacuolar protein catabolysis (8 of 10 annotated genes) (see Fig. S1), suggesting a shutdown in autophagic processes. However, increased expression of genes with roles in vacuolar biogenesis and inheritance was also observed (VAM3, YPT7, YPT72, and YKT6) (see Fig. S1). Reorganization of membrane lipid structure was indicated by a significant decrease in sphingolipid metabolism (9 of 25 annotated genes) (see Fig. S1). Reorganization of the cell surface was indicated by an increase in expression of genes associated with glycosylphosphatidylinositol (GPI) anchor biosynthesis (DPM1, MCD4, and orf19.538) and glycosylation (PMI1, PMT2, PMT5, ALG5, ALG6, ALG7, GFA1, DPM1, orf19.2298, and orf19.7426).
Within 1 h, significant upregulation of RAS1, an upstream regulatory element of the cyclic AMP-protein kinase A (cAMP-PKA) pathway was detected (Fig. 5b). Regulation of several transcriptional regulators of filamentous growth was also observed, including that of EFH1, TEC1, and UME6 (Fig. 5b). Induction of the pH regulator RIM101 was also observed. Downregulation of EFG1 and the transcriptional repressor NRG1 was also observed by 1 h (Fig. 5b). We also observed increased expression of Cd36_54430, the putative orthologue of CaSFL2, a novel regulator of hypha production that we have previously shown to be uniquely expressed by C. albicans during infection of oral epithelial tissues in vitro (Fig. 5b) (24).
By 3 h, approximately 90% of cells in WS medium produced true hyphae. At this time point, 345 genes exhibited a >2.5-fold induction and 348 exhibited a >2.5-fold decrease in expression, relative to the preculture cells (t test, P ≤ 0.015) (see Table S4 in the supplemental material). A significant proportion of the genes upregulated at 3 h were orthologous to C. albicans genes annotated with the GO term pathogenesis (n = 20; P ≤ 0.044), including the secreted proteinase SAP7 and C. dubliniensis SAP4-SAP5-SAP6 (CdSAP4,5,6), the single C. dubliniensis orthologue of C. albicans SAP4, SAP5, and SAP6 (Fig. 5c). We also observed induction of the predicted GPI-anchored proteins SOD5, HWP1, and ALS1 and downregulation of the orthologues of ALS4, ALS9, and RBT5 (Fig. 5c).
Environmental regulation of gene expression in C. dubliniensis.
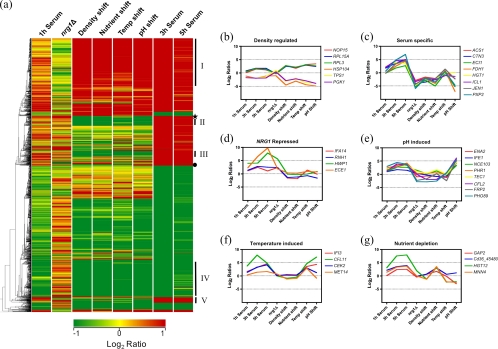
In order to understand how different environmental stimuli shaped the transcriptional response to growth in 10% serum, we also analyzed the transcript profile of C. dubliniensis following a change in cell density, a shift to 37°C, nutrient depletion (10% [vol/vol] Lee's medium) or a shift to alkaline pH (pH 7.2). None of these conditions alone could induce morphogenesis in C. dubliniensis. Although both UME6 and NRG1 exhibited regulation under the conditions examined, the changes did not reach the levels seen in WS cultures, indicating perhaps that multiple environmental signals are required to alter their expression levels sufficiently to allow filamentation to proceed (see Fig. S2 in the supplemental material). We carried out ANOVA to identify differentially regulated transcripts (P ≤ 0.05) and visualized the results using hierarchical clustering. From this analysis we could identify two large clusters of genes regulated by changes in cell density (Fig. 6a). Cluster I genes (n = 163) were induced in all experiments involving a change in cell density and were significantly enriched for genes encoding ribosomal subunits or proteins involved in ribosome biogenesis (Fig. 6b). Cluster IV (n = 167) included genes downregulated by cell density changes and was significantly enriched for genes involved in glycolysis and trehalose biosynthesis (Fig. 6b). Three clusters of serum-specific genes could also be identified (clusters II, III, and V) (Fig. 6a), and these were largely involved in metabolism of alternative carbon sources (ECI1, ICL1, and PXP2) (Fig. 6c) and nutrient transport (e.g., HGT1 and JEN1) (Fig. 6c). These data show that growth in WS medium resulted in a switch from carbohydrate catabolism to fatty acid oxidation and the glyoxylate cycle for energy production (see Fig. S3 in the supplemental material). Smaller clusters of genes were identified that were induced by alkaline pH or relief of NRG1 repression (Fig. 6d and e). The regulation of genes in response to nutrient-depleted Lee's medium and the temperature shift was more complex (Fig. 6f and g). Some transcripts exhibited clear temperature induction (MET14 and CEK2) (Fig. 6f) or nutrient depletion induction (GAP2 and MNN4) (Fig. 6g). Other transcripts responded to several conditions (e.g., HGT12 was NRG1 repressed and nutrient regulated, whereas CFL11 was induced by both temperature and pH).
Fig. 6.
(a) Hierarchical cluster analysis showing expression patterns of differentially regulated genes in C. dubliniensis (ANOVA, P ≤ 0.05) induced 2-fold or greater in WS medium at 3 h. Clustering was carried out in Genespring GX11 using default hierarchical clustering parameters. Colors refer to log2 ratio values as depicted in the bar legend. Conditions include a change in cell density (density shift), a switch to 10% (vol/vol) Lee's medium (nutrient shift), a switch to growth at 37°C (temperature shift), a shift to pH 7.5 (pH shift), or expression in an nrg1Δ background. Solid bars to the right (labeled I, II, III, IV, and V) indicate major clusters of coregulated genes (see text). The star shows the location of the major group of NRG1-regulated genes, and the circle shows the position of the main pH-regulated group. (b to g). Graphs showing expression plots of representative genes indentified from clusters in panel a.
Comparison of the C. albicans and C. dubliniensis hypha-regulated gene sets.
We compared the list of C. dubliniensis hypha-expressed genes (see Table S3 in the supplemental material) with a list of genes regulated during hypha formation in serum by C. albicans (see Material and Methods). We identified a core set of 65 hypha-induced genes in both species (Table 1). Sixty-seven genes were found to be downregulated by both species (Table 2). This analysis could identify common sets of cell surface, stress response, and regulatory genes induced or repressed in hyphae of both species. The specific transcriptional response of C. dubliniensis to WS medium was largely associated with the nutrient-poor conditions used and included genes of the glyoxylate cycle and fatty acid beta-oxidation (Fig. 6c; see also Fig S3). Increased expression of several species-specific hypothetical genes in C. dubliniensis could also be detected (Cd36_41370, Cd36_63200, and Cd36_65070) as well as downregulation of a putative glutamate decarboxylase (Cd36_10760) and a predicted ORF (Cd36_34790).
Table 1.
Selected genes of C. albicans and C. dubliniensis commonly upregulated during hypha formation in response to seruma
| Category | GeneDB no. | CGD no.b | Fold changec |
Name | Descriptiond | |
|---|---|---|---|---|---|---|
| C. albicans | C. dubliniensis | |||||
| Cell surface/secreted | Cd36_43360 | orf19.1321 | 71 | 48 | HWP1 | Hyphal wall protein |
| Cd36_52240 | orf19.4255 | 5.9 | 4.8 | ECM331 | GPI-anchored protein | |
| Cd36_64370 | orf19.5760 | 4.3 | 5.0 | IHD1 | GPI-anchored protein | |
| Cd36_63420 | orf19.5542 | 75 | 28* | SAP456 | Secreted aspartyl proteinase | |
| Cd36_43260 | orf19.3374 | 87 | 52 | ECE1 | Secreted cell elongation protein | |
| Cd36_44230 | orf19.3829 | 7.7 | 8.8 | PHR1 | GPI-anchored protein | |
| Stress response | Cd36_60850 | orf19.85 | 9.8 | 9.1 | GPX1 | Glutathione peroxidase |
| Cd36_15620 | orf19.2060 | 11.2 | 15.9 | SOD5 | Copper-zinc superoxide dismutase | |
| Cd36_33470 | orf19.3710 | 8.6 | 3.4* | YHB5 | Protein related to flavohemoglobins | |
| DNA replication | Cd36_23200 | orf19.201 | 3.5 | 8.0 | CDC47 | DNA helicase |
| Cd36_20640 | orf19.5487 | 5.7 | 10.8 | CDC46 | Part of ARS replication complex | |
| Cd36_21620 | orf19.1901 | 3.3 | 4.4 | MCM3 | Part of ARS replication complex | |
| Cd36_63950 | orf19.5597 | 2.5 | 4.7 | POL5 | DNA polymerase V, 5-prime end | |
| Cd36_41670 | orf19.4616 | 5.2 | 9.5 | POL30 | Accessory for DNA polymerase delta | |
| Cytoskeleton | Cd36_03010 | orf19.3013 | 5 | 4.7 | CDC12 | Septin |
| Cd36_29930 | orf19.548 | 2.5 | 2.9 | CDC10 | Septin | |
| Cd36_11250 | orf19.5265 | 9.5 | 9.7 | KIP4 | Kinesin heavy chain homolog | |
| GTPase | Cd36_81390 | orf19.1702 | 14.8 | 2.8 | ARF3 | GTP-binding ADP-ribosylation factor |
| Cd36_18700 | orf19.815 | 2.7 | 2.8* | DCK1 | DOCK180 protein | |
| Cd36_71380 | orf19.6573 | 5.3 | 3.8 | BEM2 | Bud emergence protein | |
| Cd36_24270 | orf19.1760 | 2.6 | 2.2 | RAS1 | Small monomeric GTPase | |
| Cd36_84970 | orf19.5968 | 3.4 | 2.8* | RDI1 | Rho GDP dissociation inhibitor | |
| Cd36_73140 | orf19.6705 | 7.2 | 11 | YEL1 | Conserved hypothetical protein | |
| Secretion | Cd36_86230 | orf19.7409 | 3.3 | 2.1 | ERV25 | Component of ER- derived vesicles |
| Cd36_40670 | orf19.4181 | 4.2 | 3.3 | SPC2 | Subunit of signal peptidase complex | |
| Cd36_72140 | orf19.6476 | 3.0 | 3.8 | AVL9 | Conserved Golgi protein | |
| Cd36_51450 | orf19.586 | 3.9 | 2.1 | ERV46 | Component of ER-derived vesicles | |
| Glycosylation | Cd36_07530 | orf19.5073 | 4.3 | 2.3 | DPM1 | Dolichol-P-mannose synthesis |
| Cd36_60365 | orf19.1203.1 | 9.0 | 3.3 | DPM2 | Regulator of dolichol-P-mannose | |
| Cd36_32420 | orf19.1843 | 2.3 | 2.3 | ALG6 | Glucosyltransferase | |
| Cd36_02340 | orf19.2937 | 8.4 | 5.5 | PMM1 | Phosphomannomutase | |
| Cd36_23720 | orf19.1390 | 3.9 | 3.7 | PMI1 | Mannose-6-phosphate isomerase | |
| Transcription factors | Cd36_81290 | orf19.1715 | 5.5 | 4.9 | IRO1 | Transcription factor |
| Cd36_01290 | orf19.3328 | 3.9 | 2.0 | HOT1 | Osmostress transcription factor | |
| Cd36_05880 | orf19.1822 | 21 | 4.3 | UME6 | Regulator of filamentation | |
| Kinases/phosphatases | Cd36_08920 | orf19.4809 | 4.3 | 4.2 | ERG12 | Mevalonate kinase |
| Cd36_40980 | orf19.4698 | 2.6 | 4.7 | PTC8 | Serine/threonine phosphatase | |
| Cd36_42970 | orf19.2678 | 3.3 | 2.8 | BUB1 | Protein kinase in mitosis checkpoint | |
Ribosomal proteins were excluded.
CGD, Candida Genome Database.
Expression relative to yeast cells. C. albicans values are taken from the data of Kadosh and Johnson (12) except those marked with an asterisk, which are taken from Nantel et al. (17).
ARS, autonomously replicating sequence; ER, endoplasmic reticulum.
Table 2.
Genes of C. albicans and C. dubliniensis commonly downregulated during hypha formation in response to serum
| Category | GeneDB no. | CGD no.a | Fold changeb |
Common name | Description | |
|---|---|---|---|---|---|---|
| C. albicans | C. dubliniensis | |||||
| Cell surface | Cd36_64800 | orf19.1097 | 14.3 | 7.1 | CdALS21 | Agglutinin-like sequence protein |
| Cd36_65010 | orf19.1097 | 14.3 | 10.0 | CdALS22 | Agglutinin-like sequence protein | |
| Cd36_64610 | orf19.4555 | 7.1 | 6.7 | ALS4 | Agglutinin-like sequence protein | |
| Cd36_26450 | orf19.2531 | 5.0 | 2.5 | CSP37 | Cell surface protein | |
| Cd36_51670 | orf19.575 | 3.8 | 3.7 | HYR5 | Similar to HYR1 | |
| Cd36_29770 | orf19.532 | 3.7 | 3.8 | RBR2 | Hypothetical protein | |
| Cd36_43810 | orf19.5305 | 12.5 | 5.0 | RHD3 | Conserved protein repressed in hyphal development | |
| Cd36_22720 | orf19.3618 | 80.0 | 10.0 | YWP1 | Putative cell wall protein | |
| Cd36_23050 | orf19.220 | 25.0 | 7.1 | PIR1 | Cell wall structural constituent with tandem repeats | |
| Transport | Cd36_20820 | orf19.23 | 4.0 | 10.0 | RTA3 | Putative transporter or flippase upregulated during the acquisition of azole resistance |
| Cd36_28130 | orf19.2425 | 5.3 | 2.4 | HGT18 | Putative glucose transporter | |
| Cd36_29200 | orf19.473 | 1.7* | 2.6 | TPO4 | Spermidine transporter | |
| Cd36_27990 | orf19.2849 | 33.3 | 2.0 | AQY1 | Aquaporin | |
| Cd36_27190 | orf19.3749 | 2.0 | 4.5 | IFC3 | Peptide transporter | |
| Cd36_41090 | orf19.4679 | 5.0 | 4.5 | AGP2 | Amino acid permease | |
| Cd36_71860 | orf19.6514 | 3.0 | 2.5 | CUP9 | Copper homeostasis | |
| Cd36_35530 | orf19.7666 | 2.2 | 16.7 | SEO3 | Permease | |
| Cd36_83640 | orf19.6956 | 5.9 | 5.3 | DAL9 | Allantoate permease | |
| Mitochondrial | Cd36_17750 | orf19.5805 | 5.9 | 12.5 | DLD3 | Mitochondrial d-lactate ferricytochrome c oxidoreductase |
| Cd36_41790 | orf19.4602 | 7.1 | 4.2 | MDH1 | Mitochondrial malate dehydrogenase | |
| Cd36_01500 | orf19.3353 | 9.1 | 6.3 | CIA30 | Possible complex I intermediate associated protein | |
| Cd36_60630 | orf19.3656 | 2.0 | 2.0 | COX15 | Cytochrome oxidase assembly factor | |
| Transcription factor | Cd36_84590 | orf19.5924 | 5.6 | 2.5 | ZCF31 | Conserved hypothetical protein |
| Cd36_12210 | orf19.4941 | 1.7* | 2.4 | TYE7 | Basic helix-loop-helix transcription factor | |
| Cd36_73890 | orf19.7150 | 2.0 | 5.3 | NRG1 | Transcriptional repressor | |
| Cd36_06830 | orf19.4438 | 33.3 | 14.3 | RME1 | Zinc-finger transcription factor | |
| Cd36_52720 | orf19.4318 | 1.7 | 3.0 | MIG1 | Transcriptional regulator | |
| Stress response | Cd36_80290 | orf19.5437 | 1.7* | 2.3 | RHR2 | dl-Glycerol-3-phosphatase |
| Cd36_01850 | orf19.4526 | 100.0 | 33.3 | HSP30 | Plasma membrane heat shock protein | |
| Cd36_01930 | orf19.3664 | 9.1 | 2.6 | HSP31 | Membrane heat shock protein | |
| Cd36_10070 | orf19.2344 | 33.3 | 4.0 | ASR1 | Similar to heat shock proteins | |
| Glutamate metabolism | Cd36_01650 | orf19.4543 | 10.0 | 3.0 | UGA22 | Succinate-semialdehyde dehydrogenase |
| Cd36_10950 | orf19.1153 | 5.3 | 2.9 | GAD1 | Glutamate decarboxylase | |
| Cd36_45660 | orf19.4716 | 2.0* | 7.1 | GDH3 | NADP-glutamate dehydrogenase | |
The specific response of C. albicans included several predicted GPI-anchored proteins, including RBT4, PGA54, and PGA55 (see Table S5 in the supplemental material). Nine of the C. albicans-specific genes had no direct orthologue in C. dubliniensis (i.e., genes without BLAST matches in C. dubliniensis or where the top BLAST hit in C. dubliniensis was not reciprocal). These included EED1, SAP4, SAP5, ALS3, and HYR1 and several members of the C. albicans telomeric TLO gene family (Table 3). The transcriptional regulator BCR1 was also induced in C. albicans, which may contribute to the concomitant upregulation of the BCR1-regulated genes HYR1, ALS3, GCN1, and orf19.6079.
Table 3.
C. albicans-specific genes expressed during hyphal development
| ORF no. | Common name | Description |
|---|---|---|
| orf19.5716 | SAP4 | Secreted aspartyl proteinase |
| orf19.5585 | SAP5 | Secreted aspartyl proteinase |
| orf19.4975 | HYR1 | Predicted GPI anchored cell wall protein |
| orf19.1816 | ALS3 | ALS family; role in epithelial adhesion, endothelial invasiveness |
| orf19.7561 | EED1 | Protein required for filamentous growth and for escape from epithelial cells |
| orf19.7544 | TLO1 | Member of a family of telomere-proximal genes |
| orf19.4054 | TLO12 | Member of a family of telomere-proximal genes |
| orf19.7127 | TLO16 | Member of a family of telomere-proximal genes |
| orf19.3074 | TLO10 | Member of a family of telomere-proximal genes |
Can stimulation of hypha formation in C. dubliniensis result in tissue damage?
We wished to determine, using simple infection models, whether induction of hyphae can increase the invasive potential of C. dubliniensis. Previous studies have demonstrated that C. dubliniensis, in contrast to C. albicans, does not invade reconstituted oral epithelium tissue when it is precultured in YPD medium at 37°C (16, 24, 26). These findings were confirmed here when C. dubliniensis Wü284 was inoculated on the surface of RHE cultures following preculture in YPD medium at 37°C. Cells grown under these conditions remained exclusively in the yeast phase and attached poorly to the surface of the tissue. Penetration of the tissue by filaments did not occur (Fig. 7a). In contrast, when C. dubliniensis cells precultured in Lee's medium, pH 4.5, at 30°C were inoculated on the tissue, we observed a mixture of morphologies (yeasts, pseudohyphae, and some true hyphae), and the cells adhered more closely to the surface of the epithelial tissue (Fig. 7b). In addition, localized invasion was observed by hyphae and pseudohyphae at 24 h postinfection (Fig. 7b and c). When a quantitative assessment of epithelial damage was made by measuring the release of lactate dehydrogenase from epithelial cells, we observed a significant increase in damage caused by cultures incubated at 30°C in Lee's pH 4.5 medium compared to YPD-grown cultures (Fig. 7d). Increased cell damage was also recorded in RHE infections with C. dubliniensis strain CD36 following preculture in Lee's medium (7.0 ± 0.6 LDH U/liter) relative to YPD medium (5.2 ± 0.2 LDH U/liter).
These data suggest that the difference in tissue damage and invasion elicited by C. dubliniensis cells grown in YPD medium and Lee's medium may be due to differences in adherence. We carried out a more detailed investigation of the adhesion of C. dubliniensis to TR146 cell monolayers over 90 min. Within 30 min of inoculation, 10 to 20% of yeast cells had adhered to the monolayer (Fig. 7e). Adherence of C. albicans SC5314 increased by 60 min, and this was independent of preculture conditions and corresponded with germ tube formation by C. albicans (Fig. 7e). In contrast, only C. dubliniensis cells precultured in Lee's medium at 30°C exhibited an increase in adherence over time (Fig. 7e). The difference in adherence at 90 min was highly significant (P < 0.01, by two-way ANOVA). An additional C. dubliniensis strain, CD36, was also shown to exhibit increased adhesion to TR146 monolayers following preculture in Lee's medium (see Fig. S4 in the supplemental material). In additional experiments, we altered the preculture conditions in order to determine the role of the temperature shift, the pH shift, and the nutrient composition of Lee's medium in this phenotype (Fig. 7f). Preculture at 37°C or at pH 7.2 reduced adhesion by 48% and 35%, respectively (Fig. 7f). The addition of 1% (wt/vol) peptone to the preculture medium also significantly reduced adhesion by 43% (P < 0.05, ANOVA). Preculture at 37°C with peptone did not have any significant additive effect on adhesion (P > 0.05).
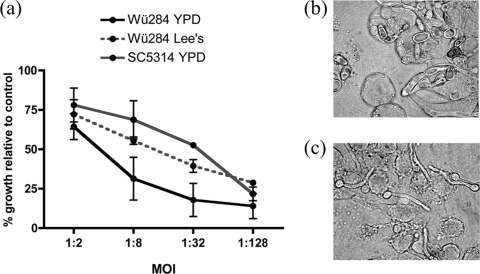
In addition, we have previously observed that C. dubliniensis is engulfed and killed more efficiently than C. albicans by RAW264.7 murine macrophages (16). This phenotype was associated with the inability of C. dubliniensis to filament and destroy the macrophage. However, preculture of C. dubliniensis in Lee's medium at pH 4.5 at 30°C led to an increase in the rate of filamentation following phagocytosis by murine macrophages compared to cells grown in YPD medium at 37°C (Fig. 8). Assessment of candidal growth in coculture with the macrophage cells demonstrated that cells grown Lee's medium at pH 4.5 could proliferate to a significantly greater level than cells grown in YPD medium (Fig. 8a). No difference in proliferation was noted with C. albicans cultures pregrown in YPD medium at 37°C or Lee's medium grown at 30°C (data not shown).
Fig. 8.
Survival of C. dubliniensis Wü284 following coculture with murine RAW264.7 macrophages. (a) Proliferation of viable Candida cells was assayed using an XTT dye reduction assay following 18 h of coculture at several multiplicities of infection (MOI; Candida CFU to macrophages). Wü284 cells precultured in Lee's medium at pH 4.5 exhibited significantly greater proliferation at MOIs of 1:8 and 1:32. Morphology of C. dubliniensis Wü284 cells grown in YPD medium (b) and in Lee's medium at pH 4.5 (c) following 5 h of incubation with murine RAW264.7 macrophages. Error bars represent standard deviations from the means of three replicate experiments.
DISCUSSION
In our attempts to generate a hyphal transcript profile for C. dubliniensis, we initially encountered problems in inducing ∼100% hyphal growth in liquid medium with this species. This led us to carry out a thorough investigation of the environmental conditions that favor the yeast-to-hypha transition in C. dubliniensis in liquid medium. Nutrient depletion was found to be the most important requirement for filamentation of C. dubliniensis in liquid medium. Highly efficient filamentation was observed in C. dubliniensis when a nutrient-poor inducing medium (water plus 10% [vol/vol] FCS) was used, and this could be suppressed by the addition of peptone and glucose. Although nutrient limitation has been shown to induce hypha formation in C. albicans in liquid and solid media, this species still filaments efficiently in nutrient-rich YPD medium in the presence of a shift to alkaline pH at 37°C (5). In C. dubliniensis, a shift from YPD medium to nutrient-rich YPDS medium (pH ∼7.5) could not induce significant morphological changes. However, filamentation of C. dubliniensis was partly induced in YPDS medium when this species was precultured in synthetic Lee's medium. This Lee's medium induction could also be suppressed by the addition of 1% peptone to the preculture medium. These data indicate that nutrient-sensing mechanisms, specifically those that sense complex mixtures of peptides, may somehow suppress pH- and temperature-induced filamentation in C. dubliniensis. We have shown that the mechanism of inhibition involves suppression of UME6 induction. Addition of peptone to WS medium could inhibit filamentation in C. dubliniensis and suppressed UME6 induction in a concentration-dependent manner. We also observed induction of NRG1 transcription in C. dubliniensis following inoculation in YPDS medium, and this may also play a significant role in preventing filamentation in this medium as Saville et al. (22) have shown that induced NRG1 transcription can prevent filamentation in YPDS medium by C. albicans. In previous studies we have hypothesized that the lack of filamentation observed in C. dubliniensis in certain media may be due to lack of NRG1 downregulation (16). However, in the present study, examination of the nrg1Δ mutant in YPDS medium showed that although removal of Nrg1 repression could enhance filamentation in this medium, the mutant still exhibited pseudohyphal characteristics and exhibited only moderate fluorescence from a PECE1-GFP fusion, suggesting an additional mechanism of nutrient repression (Fig. 3a). Induction of UME6 expression from a doxycycline-inducible promoter promoted true hypha formation in the nrg1Δ mutant in YPDS medium (Fig. 3b). In addition, overexpression of UME6 in the wild-type strain could also induce filamentation in YPDS medium, indicating that differential expression of UME6 may be the key reason for reduced filamentation of C. dubliniensis in these media. In C. albicans, it has been shown that UME6 may also play a role in suppressing NRG1 transcription during filamentation, and the differential expression of NRG1 observed in C. dubliniensis may also be UME6 dependent (2, 8). Unexpectedly, constitutive expression of NRG1 from the doxycycline-inducible promoter could not prevent hypha formation in WS medium, suggesting that high-level UME6 expression may also affect NRG1 function posttranscriptionally.
To further examine the response of C. dubliniensis in a nutrient-poor medium, we examined the transcript profile of Wü284 grown in 10% (vol/vol) Lee's medium. Nutrient depletion induced expression of genes involved in amino acid, carbohydrate, and iron uptake (GAP2, HGT12, and FET3). In C. albicans, expression of the hexose transporter HGT12 is induced by glucose limitation, and the glucose sensor Hgt4 mediates this induction (7). In addition, HGT4 is required for filamentation under some conditions (spider medium) in C. albicans (7). However, low glucose stimulation is not essential for filamentation in C. albicans as HGT4 mutants form filaments normally in glucose-rich YPDS medium (7). It is also unlikely that HGT4 signaling is required for filamentation of C. dubliniensis in WS medium as addition of 2% glucose to WS medium did not significantly inhibit filamentation in C. dubliniensis. Repression of filamentation was more apparent when C. dubliniensis was exposed to a complex mixture of peptides or whole protein (BSA), indicating that nitrogen-sensing mechanisms may be important. Growth in WS medium was associated with induction of several general amino acid permeases (GAP2, GAP4, CAN2, and CAN3), suggesting that this medium is amino acid limiting. However, addition of amino acids to WS medium did not affect filamentation in C. dubliniensis, indicating that cell surface amino acid sensors such as SSY1 may not be involved in repression of filamentous growth (6). However, high concentrations of ammonium sulfate (100 mM) could reduce filamentation. Ammonium sulfate has been shown to inhibit filamentation of C. albicans on solid medium via the Mep2 sensor; however, as shown here and previously by Biswas and Morschhauser (4), this effect is not seen in C. albicans in the presence of serum. However, ammonium sulfate-mediated repression of filamentation may be serum independent in C. dubliniensis. Interestingly, the most repressive effects on filamentation were observed when WS medium was supplemented with a mixture of both peptone and glucose, suggesting that either a combination of sensing mechanisms or possibly a more general nutrient-sensing mechanism is involved. It has recently been shown in C. albicans that an orthologue of the general nutrient sensor Tor1 can modulate NRG1 expression in spider medium (3). In addition, it has also been shown that a C. albicans MDS3 mutant can form hyphae only in the presence of the Tor1 inhibitor rapamycin (30). We are currently assessing whether C. dubliniensis Tor1 could play novel role in nutrient sensing and filamentation.
The transcript profiling data presented here also indicate important roles for pH, temperature, and cell density changes in activating the transcription of hypha-specific genes in C. dubliniensis. The transcript profiling data presented here show a key role for the pH response in activating the filamentous growth regulators SFL2, UME6, TEC1, and RIM101 (5, 8). UME6 was also found to be NRG1 repressed, whereas TEC1 also exhibited induction due to cell density changes. Temperature changes also induced EFH1 and CPH1. These data show that induction of filamentation under the conditions examined in C. dubliniensis involves multiple environmental signals.
The microarray data presented here highlighted some novel processes regulated during filamentation in C. dubliniensis and identified a strong core transcriptional response shared with C. albicans. The data show rapid induction of genes involved in regulating polarized growth, including genes involved in actin polymerization, vesicle transport, and septin formation. The data also provide evidence for processes not previously described during the morphological switch. This includes evidence for changes in lipid composition, with a shutdown in transcription of genes involved in sphingolipid synthesis and an increase in fatty acid biosynthesis gene expression. Changes in vacuole function are also indicated with an increase in expression of genes involved in vacuolar biogenesis and inheritance and decreases in expression of vacuolar proteases, suggesting that the vacuole plays a structural rather than metabolic role in hyphae. Comparison of this transcript profile with previously published studies of gene expression in C. albicans allowed us to identify a core transcriptional response to filamentation in both species which consists of 132 genes regulated 2-fold or greater (12, 17). This strongly conserved core response supports the hypothesis that a specific program of transcriptional changes may be essential for filamentation to proceed in both species, in addition to posttranscriptional events. Induction of several secreted and cell wall-associated proteins was specific to C. albicans under the conditions examined, and these included RBT4, PGA54, and PGA55. Several species-specific genes were also induced in C. albicans, including HYR1, ALS3, and EED1. C. albicans expresses three SAP genes, SAP4, SAP5, and SAP6 during filamentation, whereas C. dubliniensis possesses only one orthologue of these genes, termed CdSAP456, which is also induced during hyphal growth (11, 21). However, secreted aspartyl proteinase (SAP) activity in C. dubliniensis may be supplemented by SAP7 expression, which exhibited an 8-fold increase in expression. C. albicans also expresses the putative invasin ALS3 (19). However, we did not identify any compensatory expression of ALS genes in C. dubliniensis although orthologues of C. albicans ALS2, ALS4, and ALS9 all exhibited decreased expression during hyphal growth.
Overall, transcript profiling revealed that C. dubliniensis hyphae express a number of genes associated with virulence, suggesting that induction of filamentation in C. dubliniensis could promote tissue invasion. Recently, Spiering et al. concluded that the reduced virulence of C. dubliniensis in the RHE model was a result of a failure to initiate filamentation and the specific transcriptional program associated with this (24). In the present study we have shown that induction of UME6 expression in C. dubliniensis by preculturing in Lee's medium at 30°C could enhance filamentation in the RHE model. This resulted in greater attachment of C. dubliniensis cells to the tissue surface and localized invasion of the epithelium. We have never previously identified RHE invasion in a wild-type strain of C. dubliniensis (16, 24, 26). Examination of adhesion of C. dubliniensis to TR146 monolayers demonstrated that this adherent phenotype could be partly inhibited by the addition of peptone to the preculture medium, as well as by removing the pH or temperature shift. However, the level of damage to the RHE tissues was still significantly lower than that routinely observed when tissues are infected with C. albicans. There may be several reasons for this. First, the transition following Lee's preculture is largely short-lived, and by 24 h most cells have reverted to budding growth. Second, although C. dubliniensis can be induced to form hyphae, the absence of several C. albicans-specific hypha-associated genes (ALS3, SAP5, HYR1, and EED1) may also attenuate the virulence of this species (19, 21, 31). Studies are under way to determine if these genetic differences are crucial to the greater pathogenicity of C. albicans.
Finally, this study suggests that the ability of C. albicans to form filaments at alkaline pH, irrespective of nutrient availability, may enable it to colonize and infect a wider range of niches relative to C. dubliniensis. C. dubliniensis may have lost or perhaps failed to acquire this morphological flexibility since the divergence of the two species. The genome sequence of C. dubliniensis suggests that, due to gene loss and pseudogenization, C. dubliniensis may be undergoing niche specialization. It may be possible that reduced filamentation is part of this specialization process and that it may even be of benefit to C. dubliniensis in certain niches, particularly where tissue damage, inflammation, and attraction of the of host's defenses is unfavorable.
Supplementary Material
ACKNOWLEDGMENTS
Plasmids pNIM1 and pTET42 were obtained from the Joachim Morschhäuser Institut für Molekulare Infektionsbiologie, Universität Wurzburg, and plasmid pCaUme6-3 was generated by Arnold Bito (Department of Cell Biology, University of Salzburg). We thank Jan Walker at St. James's Hospital Dublin for fixation and staining of the RHE tissue sections. We also thank the anonymous reviewers whose helpful comments greatly enhanced the manuscript.
This work was supported by the Irish Health Research Board (research grant RP/2004/235) and by Science Foundation Ireland (Programme Investigator grant number 04/IN3/B463).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Asmundsdottir L. R., ErlendsdÛttir H., Agnarsson B. A., Gottfredsson M. 2009. The importance of strain variation in virulence of Candida dubliniensis and Candida albicans: results of a blinded histopathological study of invasive candidiasis. Clin. Microbiol. Infect. 15:576–585 [DOI] [PubMed] [Google Scholar]
- 2.Banerjee M., Thompson D. S., Lazzell A., Carlisle P. L., Pierce C., Monteagudo C., Lopez-Ribot J. L., Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastidas R. J., Heitman J., Cardenas M. E. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas K., Morschhauser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649–669 [DOI] [PubMed] [Google Scholar]
- 5.Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brega E., Zufferey R., Mamoun C. B. 2004. Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3:135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown V., Sexton J. A., Johnston M. 2006. A glucose sensor in Candida albicans. Eukaryot. Cell 5:1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlisle P. L., Banerjee M., Lazzell A., Monteagudo C., Lopez-Ribot J. L., Kadosh D. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher P. J., Bennett D. E., Henman M. C., Russell R. J., Flint S. R., Shanley D. B., Coleman D. C. 1992. Reduced azole susceptibility of Candida albicans from HIV-positive patients and a derivative exhibiting colony morphology variation. J. Gen. Microbiol. 138:1901–1911 [DOI] [PubMed] [Google Scholar]
- 10.Jabra-Rizk M. A., Johnson J. K., Forrest G., Mankes K., Meiller T. F., Venezia R. A. 2005. Prevalence of Candida dubliniensis fungemia at a large teaching hospital. Clin. Infect. Dis. 41:1064–1067 [DOI] [PubMed] [Google Scholar]
- 11.Jackson A. P., Gamble J. A., Yeomans T., Moran G. P., Saunders D., Harris D., Aslett M., Barrell J. F., Butler G., Citiulo F., Coleman D. C., de Groot P. W., Goodwin T. J., Quail M. A., McQuillan J., Munro C. A., Pain A., Poulter R. T., Rajandream M. A., Renauld H., Spiering M. J., Tivey A., Gow N. A., Barrell B., Sullivan D. J., Berriman M. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and C. albicans. Genome Res. 10:2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kibbler C. C., Seaton S., Barnes R. A., Gransden W. R., Holliman R. E., Johnson E. M., Perry J. D., Sullivan D. J., Wilson J. A. 2003. Management and outcome of blood stream infections due to Candida species in England and Wales. J. Hosp. Infect. 54:18–24 [DOI] [PubMed] [Google Scholar]
- 14.Lee K. L., Buckley H. R., Campbell C. C. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148–153 [DOI] [PubMed] [Google Scholar]
- 15.Meiller T. F., Jabra-Rizk M. A., Baqui A., Kelley J. I., Meeks V. I., Merz W. G., Falkler W. A. 1999. Oral Candida dubliniensis as a clinically important species in HIV-seropositive patients in the United States. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 88:573–580 [DOI] [PubMed] [Google Scholar]
- 16.Moran G. P., MacCallum D. M., Spiering M. J., Coleman D. C., Sullivan D. J. 2007. Differential regulation of the transcriptional repressor NRG1 accounts for altered host cell interactions in Candida albicans and Candida dubliniensis. Mol. Microbiol. 66:915–929 [DOI] [PubMed] [Google Scholar]
- 17.Nantel A., Dignard D., Bachewich C., Harcus D., Marcil A., Bouin A. P., Sensen C. W., Hogues H., Van het Hoog M., Gordon P., Rigby T., Benoit F., Tessier D. C., Thomas D. Y., Whiteway M. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y. N., Morschhauser J. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4:1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr., Filler S. G. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotrosen D., Edwards J. E., Jr., Gibson T. R., Moore J. C., Cohen A. H., Green I. 1985. Adherence of Candida to cultured vascular endothelial cells: mechanisms of attachment and endothelial cell penetration. J. Infect. Dis. 152:1264–1274 [DOI] [PubMed] [Google Scholar]
- 21.Sanglard D., Hube B., Monod M., Odds F. C., Gow N. A. R. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65:3539–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saville S. P., Lazzell A. L., Monteagudo C., Lopez-Ribot J. L. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaller M., Korting H. C., Schafer W., Bastert J., Chen W., Hube B. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 34:169–180 [DOI] [PubMed] [Google Scholar]
- 24.Spiering M. J., Moran G. P., Chauvel M., Maccallum D. M., Higgins J., Hokamp K., Yeomans T., D'Enfert C., Coleman D. C., Sullivan D. J. 2010. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot. Cell 9:251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staib P., Morschhauser J. 2005. Differential expression of the NRG1 repressor controls species-specific regulation of chlamydospore development in Candida albicans and Candida dubliniensis. Mol. Microbiol. 55:637–652 [DOI] [PubMed] [Google Scholar]
- 26.Stokes C., Moran G. P., Spiering M. J., Cole G. T., Coleman D. C., Sullivan D. J. 2007. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet. Biol. 44:920–931 [DOI] [PubMed] [Google Scholar]
- 27.Sullivan D. J., Moran G. P., Pinjon E., Al-Mosaid A., Stokes C., Vaughan C., Coleman D. C. 2004. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 4:369–376 [DOI] [PubMed] [Google Scholar]
- 28.Sullivan D. J., Westerneng T. J., Haynes K. A., Bennett D. E., Coleman D. C. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507–1521 [DOI] [PubMed] [Google Scholar]
- 29.Vilela M. M., Kamei K., Sano A., Tanaka R., Uno J., Takahashi I., Ito J., Yarita K., M. M 2002. Pathogenicity and virulence of Candida dubliniensis: comparison with C. albicans. Med. Mycol. 40:249–257 [DOI] [PubMed] [Google Scholar]
- 30.Zacchi L. F., Gomez-Raja J., Davis D. A. 10May2010. Mds3 regulates morphogenesis in Candida albicans through the TOR pathway. Mol. Cell. Biol. doi.10.1128/MCB.01549-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakikhany K., Naglik J. R., Schmidt-Westhausen A., Holland H., Schaller M., Hube B. 2007. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell. Microbiol. 9:2938–2954 [DOI] [PubMed] [Google Scholar]
- 32.Zeidler U., Lettner T., Lassnig C., Muller M., Lajko R., Hintner H., Breitenbach M., Bito A. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9:126–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.