Abstract
Carnitine palmitoyltransferase II (CPT-II) has a crucial role in the β-oxidation of long-chain fatty acids in mitochondria. We report here the crystal structure of rat CPT-II at 1.9 Å resolution. The overall structure shares strong similarity to those of short- and medium-chain carnitine acyltransferases, although detailed structural differences in the active site region have a significant impact on the substrate selectivity of CPT-II. Three aliphatic chains, possibly from a detergent that is used for the crystallization, were found in the structure. Two of them are located in the carnitine and CoA binding sites, respectively. The third aliphatic chain may mimic the long-chain acyl group in the substrate of CPT-II. The binding site for this aliphatic chain does not exist in the short- and medium-chain carnitine acyltransferases, due to conformational differences among the enzymes. A unique insert in CPT-II is positioned on the surface of the enzyme, with a highly hydrophobic surface. It is likely that this surface patch mediates the association of CPT-II with the inner membrane of the mitochondria.
Keywords: obesity, diabetes, fatty acid metabolism, protein structure
Introduction
Carnitine palmitoyltransferases (CPTs) have crucial roles in the β-oxidation of long-chain fatty acids [1–7]. CPT-I catalyzes the conversion of long-chain acyl-CoA esters to acylcarnitine esters, enabling their transport from the cytosol into the mitochondrial matrix. Once inside the mitochondria, the carnitine esters are converted back to the CoA esters by CPT-II, which can then enter the β-oxidation pathway. Disruptions in the activity of the CPTs are linked to many serious human diseases [3, 4, 6]. CPTs are attractive targets for the development of new therapeutic agents for the treatment of diabetes, obesity, and other human diseases [8–10].
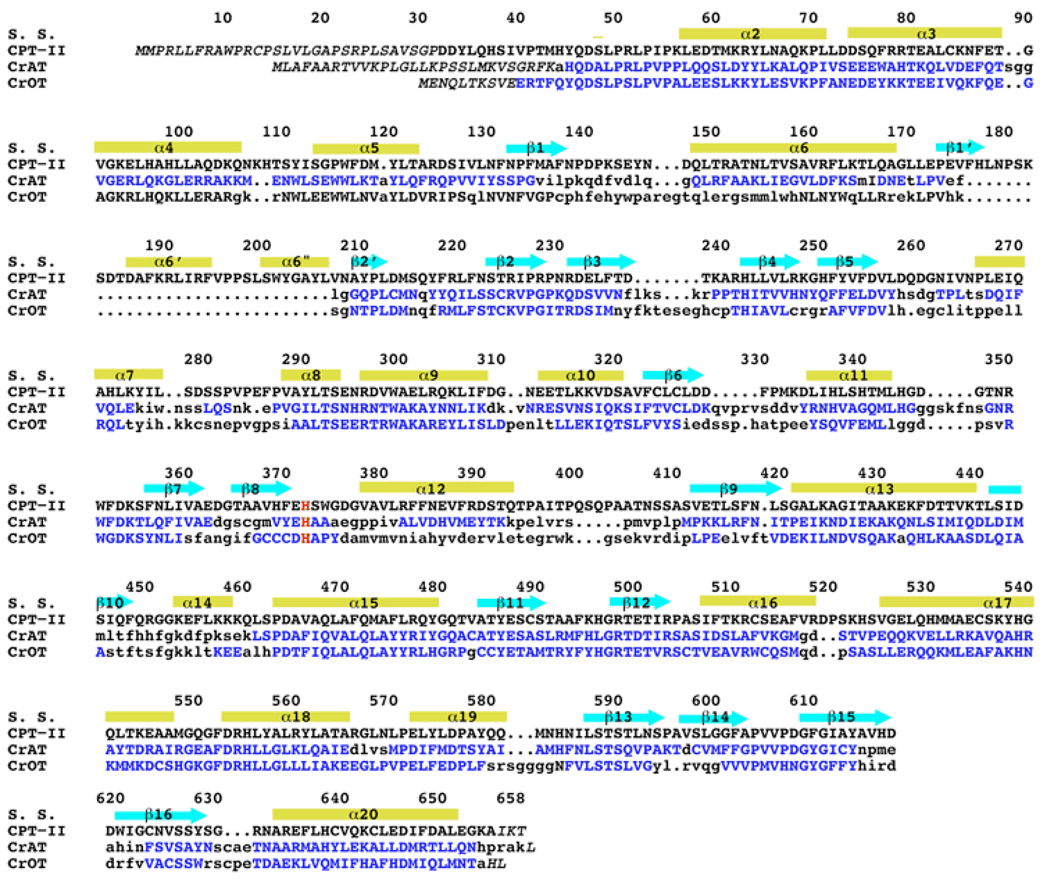
The CPTs belong to the family of carnitine acyltransferases, which also includes carnitine acetyltransferase (CrAT, with a preference for short-chain acyl groups), and carnitine octanoyltransferase (CrOT, with a preference for medium-chain acyl groups). The crystal structures of mammalian CrAT and CrOT have been reported by us and others over the past few years [11–17]. The structures contain two domains, N and C domains, with the same backbone fold, and the active site is located in a tunnel at the interface of the two domains. The carnitine and CoA substrates are bound at opposite sides of the catalytic His residue. CPT-II shares about 35% amino acid sequence identity with these two enzymes (Fig. 1).
Fig. 1.
Structure-based sequence alignment of the carnitine acyltransferases. Residues in CrAT and CrOT that are structurally equivalent to those in CPT-II are shown in blue. The catalytic His residue is shown in red. The secondary structure elements are indicated, and the residue numbers are for rat CPT-II.
We report here the crystal structure of rat CPT-II at 1.9 Å resolution. While the overall structure of CPT-II is similar to that of CrAT and CrOT, detailed structural differences in the active site region have a significant impact on the substrate selectivity of this enzyme. We observed the binding of three aliphatic chains in the structure, two of which are located in the binding sites for carnitine and CoA, respectively. The third aliphatic chain may mimic the long-chain acyl group in the substrate of CPT-II. A unique insert in CPT-II presents an exposed hydrophobic surface and may mediate the association of this enzyme with the inner mitochondrial membrane.
Materials and methods
Residues 32–658 of wild-type rat CPT-II was sub-cloned into the pET24d vector (Novagen) and over-expressed in E. coli at 20°C. The expression construct contains a C-terminal hexa-histidine tag. The soluble protein was purified with nickel-agarose affinity chromatography, anion exchange and gel filtration chromatography. The protein was concentrated to 26 mg/ml in a solution containing 20 mM Tris (pH8.5), 200 mM NaCl, flash-frozen in liquid nitrogen in the presence of 5% (v/v) glycerol, and stored at −80°C. The C-terminal His-tag was not removed for crystallization.
Crystals of rat CPT-II free enzyme were obtained at 21°C by the sitting-drop vapor diffusion method. The reservoir solution contained 100 mM Hepes (pH 7.0), 15% Tacsimate, 16% (w/v) PEG 5000MME, and the protein was at 26 mg/ml concentration. Larger crystals were grown from the same condition by microseeding and using 1% (w/v) benzyldimethyldodecyl ammonium bromide (a detergent) as an additive. The crystals were cryoprotected with the introduction of 15% (v/v) ethylene glycol, and flash-frozen in liquid nitrogen.
X-ray diffraction data were collected on an Mar imaging plate at the X4C beamline of the National Synchrotron Light Source (NSLS). The native data set to 1.9 Å resolution was collected at 100K on the free enzyme crystal. The diffraction images were processed and scaled with the HKL package [18]. The crystals belong to space group P1, with cell parameters of a = 73.77 Å, b = 73.80 Å, c = 90.56 Å, α = 70.65°, β = 73.62°, and γ = 88.06°. There are two molecules in the crystallographic asymmetric unit.
The initial structure of rat CPT-II was determined by the molecular replacement method, with the program COMO [19, 20], using the mouse CrAT structure (PDB entry 1NDF) as the search model [11]. The structure refinement was carried out with the program CNS [21]. Manual adjustment of the atomic model against the electron density was performed with the program O [22]. The crystallographic statistics are summarized in Table 1.
Table 1.
Summary of crystallographic information
| Resolution range (Å) | 30-1.9 |
| Number of observations | 249,051 |
| Rmerge (%)1 | 6.6 (40.2) |
| I/σI | 12.2 (1.9) |
| Redundancy | 2.0 (1.7) |
| Number of reflections | 116,945 |
| Completeness (%) | 86 (71) |
| R factor (%)2 | 18.9 (31.0) |
| Free R factor (%) | 22.1 (31.6) |
| rms deviation in bond lengths (Å) | 0.006 |
| rms deviation in bond angles (°) | 1.2 |
| PDB accession code | 2H4T |
. The numbers in parenthesis are for the highest resolution shell.
Results and discussion
Overall structure
The crystal structure of rat carnitine palmitoyltransferase II (CPT-II) has been determined at 1.9 Å resolution by the molecular replacement method (Table 1). The current atomic model contains residues 31–655 and 31–656 for the two CPT-II molecules in the crystallographic asymmetric unit, respectively, with an R factor of 18.9%. The majority of the residues (93%) are in the most favored region of the Ramachandran plot (data not shown).
The two molecules of CPT-II have the same conformation, with a rms distance of 0.19 Å for all their Cα atoms. They do not have strong contacts with each other, suggesting that the molecules are essentially monomeric in the crystal. Gel filtration and light scattering studies show that purified CPT-II is an oligomer (hexamer or octamer) in solution (unpublished data). It is likely that the use of a detergent (benzyldimethyldodecyl ammonium bromide, BAM) as an additive in the crystallization solution may have disrupted the oligomer, since the CPT-II monomers contain an exposed hydrophobic patch (see below). This detergent is crucial for obtaining crystals of sufficient size and quality for structural studies.
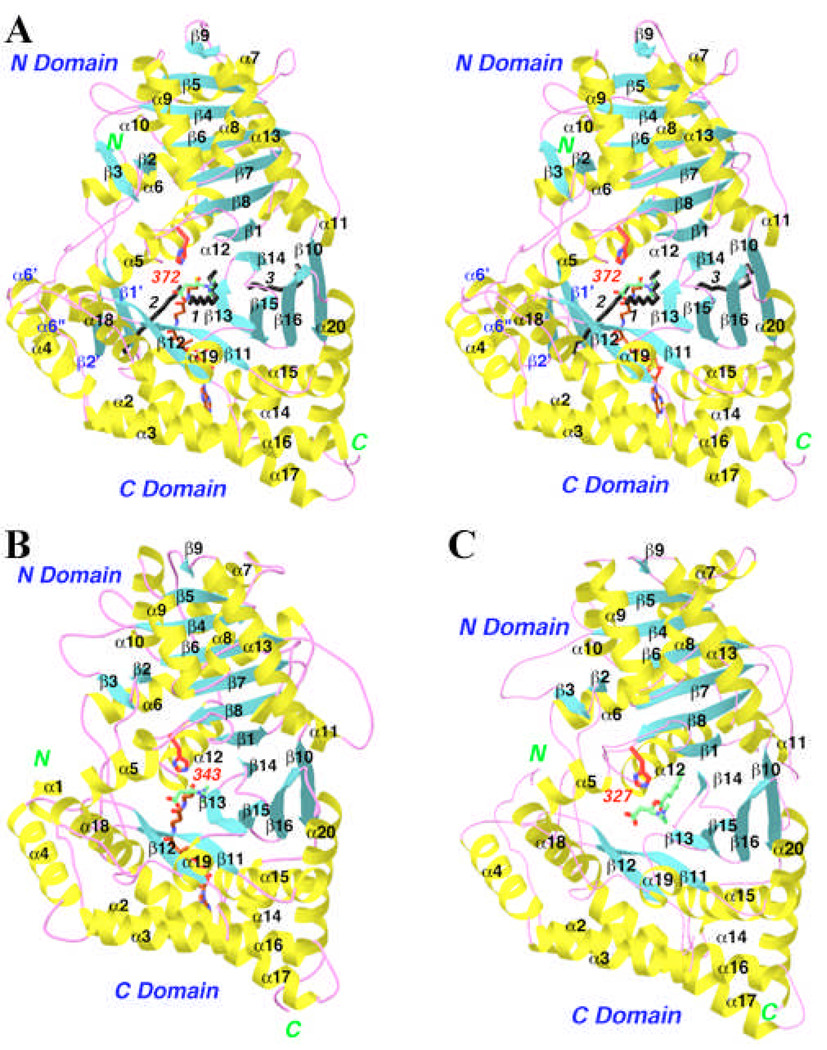
The overall structure of CPT-II is similar to that of CrAT and CrOT that we reported earlier (Figs. 2A–C) [11–13]. The rms distance between equivalent Cα atoms of CPT-II and these other two structures is about 1.2 Å. The secondary structure elements in CPT-II are named using the same system as that in the other two structures (Fig. 1). The structure-based sequence alignment shows that most of the secondary structure elements have highly homologous structures among the three enzymes (but see important exceptions below), while the conformations of the connecting loops are more divergent (Fig. 1).
Fig. 2.
Structure of rat CPT-II. (A). Ribbon representation of the rat CPT-II structure. The α-helices are shown in yellow, β strands in cyan, and the connecting loops in magenta. The catalytic His372 residue is shown in red. The expected positions of carnitine and CoA are shown, in green and brown, respectively. The three aliphatic chains observed in the crystal structure are shown in black and labeled. (B). The structure of mouse CrAT, viewed in the same orientation [11]. Carnitine is shown in green, and CoA in brown for carbon atoms. (C). The structure of mouse CrOT [12]. Octanoylcarnitine is shown in green. Produced with Ribbons [24].
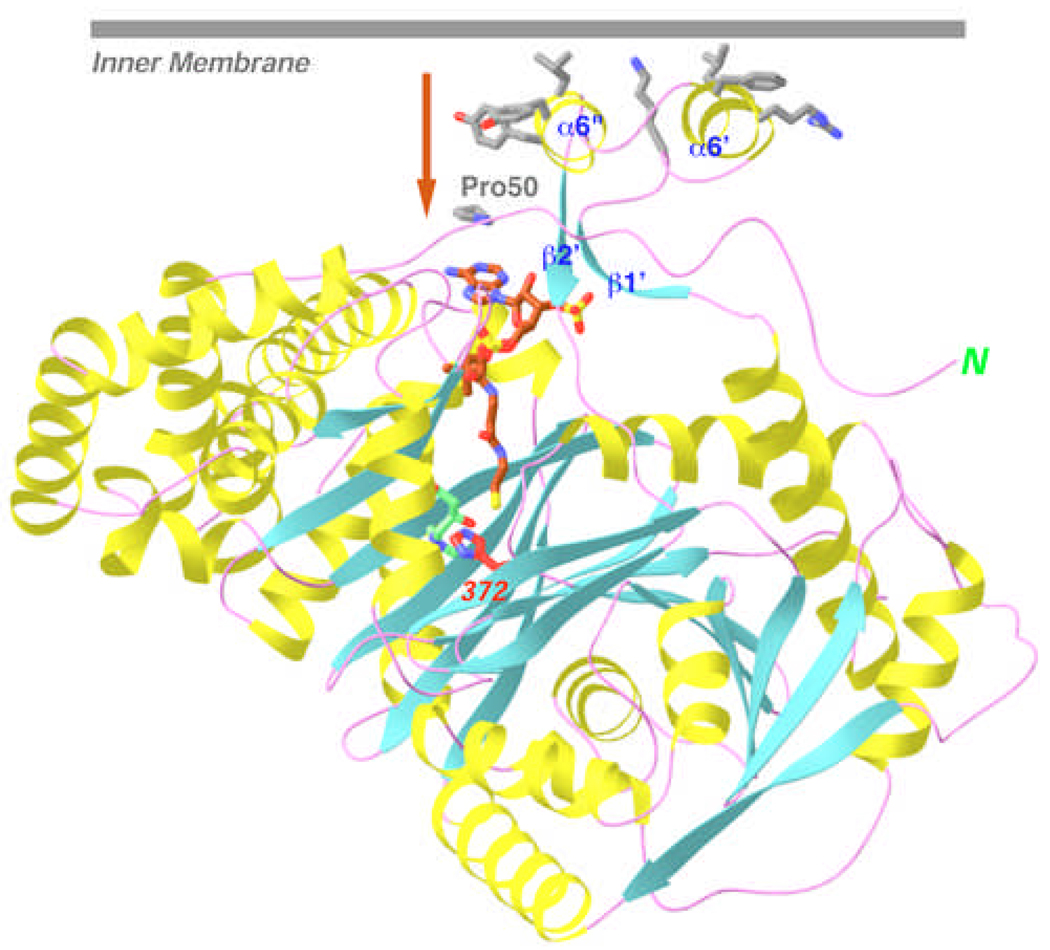
A structural motif for association with the mitochondria
CPT-II contains a unique insertion of 30 residues in the N domain (residues 176–206, Fig. 1), and our structure shows that this segment contains two helices (α6’ and α6”) linked to a small, anti-parallel β-structure (β1’ and β2’) (Fig. 2A). Interestingly, the two helices form a protrusion on the surface of CPT-II, and their exposed face is rather hydrophobic in nature (Fig. 3). It is likely that this surface patch is important for the association of CPT-II with the inner mitochondrial membrane [3, 4, 6]. Moreover, this surface patch is located near the entrance to the active site tunnel of the enzyme, suggesting that long-chain acylcarnitines that have been transported across the inner membrane could be shuttled directly into the active site of CPT-II (Fig. 3). In this regard, it might be speculated that this insert could also mediate direct interactions between CPT-II and the carnitine-acylcarnitine translocase (CACT) in the inner membrane of the mitochondria [3, 4], which could coordinate acylcarnitine transport and its back conversion to acyl-CoA.
Fig. 3.
A hydrophobic surface for the unique insert of CPT-II. It is likely that this surface interacts with the mitochondrial inner membrane, which could allow direct transport of acylcarnitine into the active site of CPT-II, indicated by the arrow. Produced with Ribbons [24].
The presence of this hydrophobic surface patch may explain how the BAM detergent can disrupt the oligomer of CPT-II and promote its crystallization. This insert also helps to stabilize the longer N-terminal segment of CPT-II (Figs. 2A–C), as some of these residues are held in place between the two helices and the small β-sheet of this insert (Fig. 3). The short α1 helix in the N-terminal segment, observed in the structure of CrAT, is absent in CPT-II, as is the case with CrOT (Figs. 2A–C).
A possible binding site for long-chain acyl groups
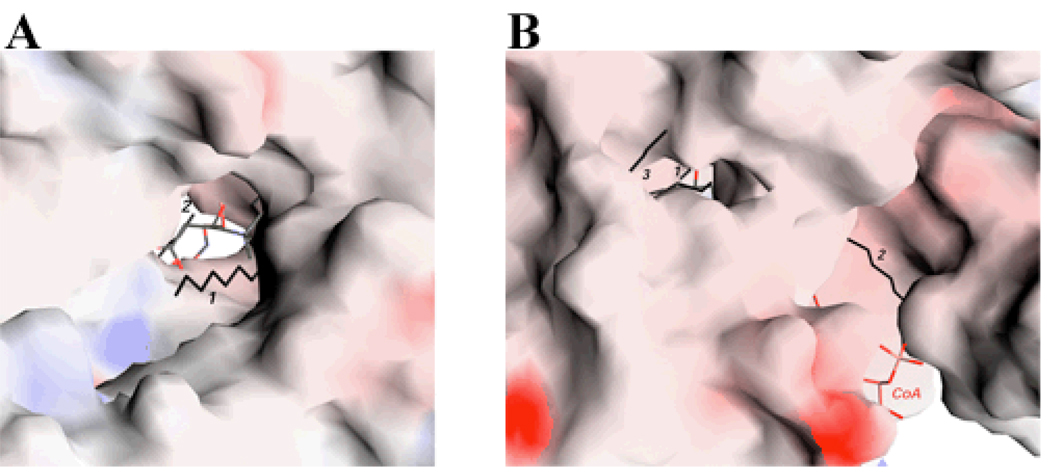
Our crystallographic analyses revealed three pieces of extended electron density in the CPT-II molecule, which we have interpreted as aliphatic chains of 12 carbons (Fig. 2A), very likely from the dodecyl group of the BAM detergent. The possibility that these entities have copurified from E. coli cells can probably be excluded based on the fact that the purified enzyme is catalytically active. Two of these molecules are located in the active site tunnel (Fig. 4A), and are expected to block access of the carnitine and CoA substrates to the enzyme (Figs. 2A, 4A). In fact, our kinetic assays showed that the detergent is an inhibitor of CPT-II activity, with 90% inhibition at 1.3 mM concentration (BAM does not form micelles).
Fig. 4.
The active site tunnels in CPT-II. (A). Molecular surface of CPT-II looking down the tunnel for binding carnitine and CoA. Carnitine is shown in green, CoA in brown, and the three aliphatic chains in black and labeled. (B). A narrow tunnel for binding the third aliphatic group. The tunnel is connected to that for binding carnitine and CoA (both visible in the figure). Produced with Grasp [25].
Interestingly, the third aliphatic chain is located in another, narrower tunnel that is connected to the active site of the enzyme (Fig. 4B), suggesting that it could be a mimic of the long-chain acyl group in the substrate of CPT-II. This tunnel does not exist in the structures of CrAT and CrOT (see next section). The molecule is situated between strands β14 (part of N domain) and one face of strands β15, β16 and β10 in the C-domain (Fig. 2A). The tip of the acyl group in this binding site would be exposed to the solvent (Fig. 4B). This orientation of the acyl group is almost perpendicular to that observed for the medium-chain acyl group bound to CrOT (Fig. 2C).
Having the structure of CPT-II in complex with the acyl-CoA or acylcarnitine substrate will confirm whether this aliphatic chain is truly a mimic of the acyl group of the substrate. Unfortunately, repeated attempts at replacing these aliphatic chains with acyl-CoA and acylcarnitine, by soaking and co-crystallization experiments, were not successful.
Conformational differences to CrAT and CrOT
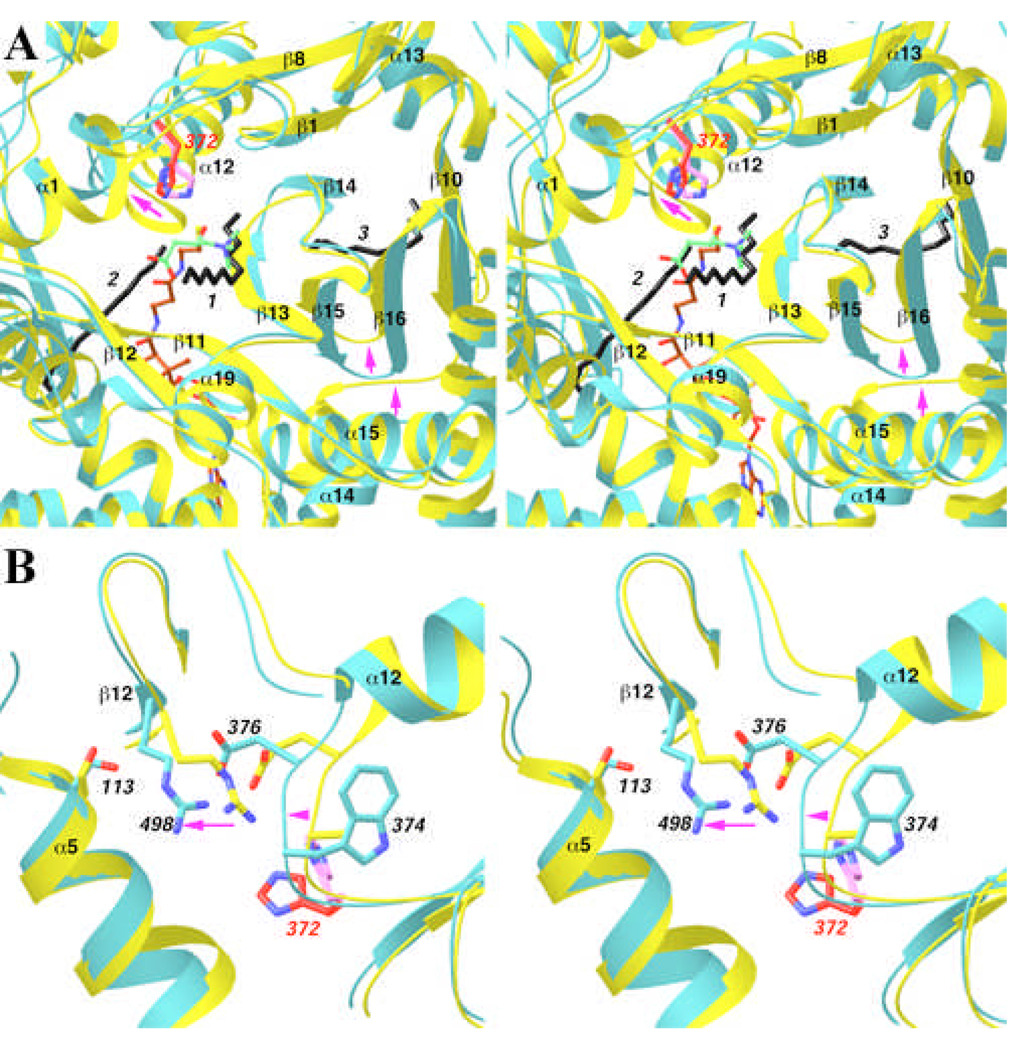
Despite the overall structural homology, there are important conformational differences between CPT-II, CrAT and CrOT. The binding site for the third aliphatic chain does not exist in CrAT and CrOT, due to conformational differences among the structures (Fig. 5A). Most importantly, this aliphatic chain has severe clashes with the tip of the β15–β16 hairpin in CrAT, which shows large conformational differences compared to its equivalent in CPT-II (Fig. 5A). The trigger for this structural difference could be the loop connecting strand β10 and helix α14. It shifts by about 3 Å towards the β15–β16 hairpin in CrAT as compared to CPT-II (Fig. 5A), pushing the hairpin towards the β14 strand and eliminating the tunnel for the aliphatic chain in CrAT.
Fig. 5.
Structural differences between CPT-II and CrAT. (A). Overlap of the structure of CPT-II (in cyan) and CrAT (in yellow) near the binding site for the third aliphatic group. His372 in CPT-II is shown in red, and His343 in CrAT in magenta. Arrows in magenta indicate regions of conformational differences between the two structures. (B). Overlap of the structure of CPT-II (in cyan) and CrAT (in yellow) near the Ser113 residue. Produced with Ribbons [24].
Another difference between CPT-II and CrAT is the conformation of the α12 helix (Fig. 5A). The most pronounced difference is in the position of the N-terminal residues of the helix, near the active site of the enzymes. These residues shift by approximately 3 Å in CPT-II, away from the active site region (Fig. 5A). This change significantly increases the size of the active site tunnel at the interface between the N and C domains (Figs. 2A–C), and the larger tunnel could be important for the binding of the long-chain acyl groups in the substrates of CPT-II.
Finally, the conformation of the side chain of the catalytic His372 residue in CPT-II is different from that of His343 in CrAT (Fig. 5A). This conformation is probably not optimal for catalysis, and a structural change in His372 may be expected upon substrate binding.
Implications for disease causing mutations
Mutations in CPT-II have been linked to many human diseases [3, 4, 6]. Most of these mutations are located away from the active site of the enzyme [11, 13], some of which affect the stability rather than the catalytic activity of the enzyme [3, 4, 6]. The most frequently found mutation in CPT-II is S113L, and it is linked to mild disease symptoms. This residue is conserved in CrAT (Fig. 1), although it does not play an important role in that structure [11]. In the structure of CPT-II, the side chain hydroxyl of Ser113 has a long hydrogen-bond with the guanidinium group of Arg498, which in turn is ion-paired to Asp376 (Fig. 5B). Asp376 is 4 residues from the catalytic His372 residue in the primary sequence, and is conserved among all carnitine acyltransferases as either Asp or Glu (Fig. 1). Therefore, the S113L mutation may disturb this hydrogen-bonding and ion-pair network, and thereby indirectly affect the catalytic efficiency of the His372 residue. In contrast, this network does not exist in the structure of CrAT (Fig. 5B), due to conformational differences between the two structures in this region. The replacement of Ala345 in CrAT by the bulkier Trp374 in CPT-II may have helped trigger this change (Fig. 5B). The main chain of Trp374 shifts by about 1.8 Å to accommodate the much larger Trp side chain. This causes a change in the position of the Arg498 side chain (as well as the shift in the N terminal region of helix α12 as discussed above, Fig. 5A). As a result, the Arg498 side chain is brought into hydrogen-bonding distance of the Ser113 side chain in CPT-II (Fig. 5B).
Another commonly found mutation in CPT-II is P50H, and our structural analysis suggests a distinct mechanism for its detrimental effects. The Pro50 residue is conserved, in sequence and structure, among CPT-II, CrAT, and CrOT (Fig. 1). This residue is 23 Å from the active site, but is located directly below the unique insert in CPT-II (Fig. 3). It might be possible that the mutation interferes indirectly with the association between CPT-II and the mitochondrial membrane, and disturbs the shuttling of the acylcarnitine substrate from the membrane into the active site of CPT-II.
The structure of CPT-II has very recently been reported by another group [23]. Our structural analyses are generally consistent with their findings. Most importantly, the binding site for the third aliphatic chain in our structure is found to be the long-chain acyl group binding site [23].
In summary, we have determined the crystal structure of rat carnitine palmitoyltransferase II (CPT-II) at 1.9 Å resolution. The overall structure shares strong similarity to those of short- and medium-chain carnitine acyltransferases. We observed the binding of three aliphatic chains in the structure, two of which are located in the carnitine and CoA binding sites, respectively. The third aliphatic chain may mimic the long-chain acyl group in the substrate of CPT-II. The binding site for this aliphatic chain does not exist in CrAT and CrOT, due to conformational differences among the enzymes. Moreover, the active site tunnel in CPT-II appears to be larger than that in CrAT and CrOT, which may be important for the binding of long-chain acyl groups as well. A unique insert in CPT-II is positioned on the surface of enzyme, with a highly hydrophobic surface. It is likely that this surface patch mediates the association between CPT-II and the inner membrane of the mitochondria.
Acknowledgements
We thank Randy Abramowitz and John Schwanof for access to the X4C beamline at NSLS. This research was supported in part by a grant from the National Institutes of Health (DK67238) to LT.
REFERENCES
- 1.Bieber LL. Carnitine. Ann. Rev. Biochem. 1988;57:261–283. doi: 10.1146/annurev.bi.57.070188.001401. [DOI] [PubMed] [Google Scholar]
- 2.Foster DW. The role of the carnitine system in human metabolism. Ann. N.Y. Acad. Sci. 2004;1033:1–16. doi: 10.1196/annals.1320.001. [DOI] [PubMed] [Google Scholar]
- 3.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay RR, Gandour RD, van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim. Biophys. Acta. 2001;1546:21–43. doi: 10.1016/s0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- 5.Ramsay RR, Zammit VA. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol. Aspects Med. 2004;25:475–493. doi: 10.1016/j.mam.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol. Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Steiber A, Kerner J, Hoppel CL. Carnitine: a nutritional, biosynthetic, and functional perspective. Mol. Aspects Med. 2004;25:455–473. doi: 10.1016/j.mam.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RC. Carnitine palmitoyltransferase: a viable target for the treatment of NIDDM? Curr. Pharm. Des. 1998;4:1–16. [PubMed] [Google Scholar]
- 9.Lenhard JM, Gottschalk WK. Preclinical developments in type 2 diabetes. Advanced Drug Delivery Reviews. 2002;54:1199–1212. doi: 10.1016/s0169-409x(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 10.Ronnett GV, Kim E-K, Landree LE, Tu Y. Fatty acid metabolism as a target for obesity treatment. Physiol. Behav. 2005;85:25–35. doi: 10.1016/j.physbeh.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Jogl G, Tong L. Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport. Cell. 2003;112:113–122. doi: 10.1016/s0092-8674(02)01228-x. [DOI] [PubMed] [Google Scholar]
- 12.Jogl G, Hsiao Y-S, Tong L. Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity. J. Biol. Chem. 2005;280:738–744. doi: 10.1074/jbc.M409894200. [DOI] [PubMed] [Google Scholar]
- 13.Jogl G, Hsiao Y-S, Tong L. Structure and function of carnitine acyltransferases. Ann. N.Y. Acad. Sci. 2004;1033:17–29. doi: 10.1196/annals.1320.002. [DOI] [PubMed] [Google Scholar]
- 14.Ramsay RR, Naismith JH. A snapshot of carnitine acetyltransferase. Trends Biochem. Sci. 2003;28:343–346. doi: 10.1016/S0968-0004(03)00137-3. [DOI] [PubMed] [Google Scholar]
- 15.Wu D, Govindasamy L, Lian W, Gu Y, Kukar T, Agbandje-McKenna M, McKenna R. Structure of human carnitine acetyltransferase. Molecular basis for fatty acyl transfer. J. Biol. Chem. 2003;278:13159–13165. doi: 10.1074/jbc.M212356200. [DOI] [PubMed] [Google Scholar]
- 16.Hsiao Y-S, Jogl G, Tong L. Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase. J. Biol. Chem. 2004;279:31584–31589. doi: 10.1074/jbc.M403484200. [DOI] [PubMed] [Google Scholar]
- 17.Govindasamy L, Kukar T, Lian W, Pedersen B, Gu Y, Agbandje-McKenna M, Jin S, McKenna R, Wu D. Structural and mutational characterization of L-carnitine binding to human carnitine acetyltransferase. J. Struct. Biol. 2004;146:416–424. doi: 10.1016/j.jsb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Jogl G, Tao X, Xu Y, Tong L. COMO: A program for combined molecular replacement. Acta Cryst. 2001;D57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- 20.Tong L. Combined molecular replacement. Acta Cryst. 1996;A52:782–784. [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Rufer AC, Thoma R, Benz J, Stihle M, Gsell B, de Roo E, Banner DW, Mueller F, Chomienne O, Hennig M. The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment. Structure. 2006;14:713–723. doi: 10.1016/j.str.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Carson M. Ribbon models of macromolecules. J. Mol. Graphics. 1987;5:103–106. [Google Scholar]
- 25.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]