Abstract
Multiple mechanisms for feedback control of cholesterol synthesis converge on the rate-limiting enzyme in the pathway, 3-hydroxy-3-methylglutaryl coenzyme A reductase. This complex feedback regulatory system is mediated by sterol and nonsterol metabolites of mevalonate, the immediate product of reductase activity. One mechanism for feedback control of reductase involves rapid degradation of the enzyme from membranes of the endoplasmic reticulum (ER). This degradation results from the accumulation of sterols in ER membranes, which triggers binding of reductase to ER membrane proteins called Insig-1 and Insig-2. Insig binding leads to the recruitment of a membrane-associated ubiquitin ligase called gp78 that initiates ubiquitination of reductase. Ubiquitinated reductase then becomes extracted from ER membranes and is delivered to cytosolic 26S proteasomes through an unknown mechanism that is mediated by the gp78-associated ATPase Valosin-containing protein/p97 and appears to be augmented by nonsterol isoprenoids. Here, we will highlight several advances that have led to the current view of mechanisms for sterol-accelerated, ER-associated degradation of reductase. In addition, we will discuss potential mechanisms for other aspects of the pathway such as selection of reductase for gp78-mediated ubiquitination, extraction of the ubiquitinated enzyme from ER membranes, and the contribution of Insig-mediated degradation to overall regulation of reductase in whole animals.
Keywords: cholesterol metabolism, ubiquitination, 26S proteasome, ubiquitin ligase, sterol regulatory element-binding protein, Scap
Introduction
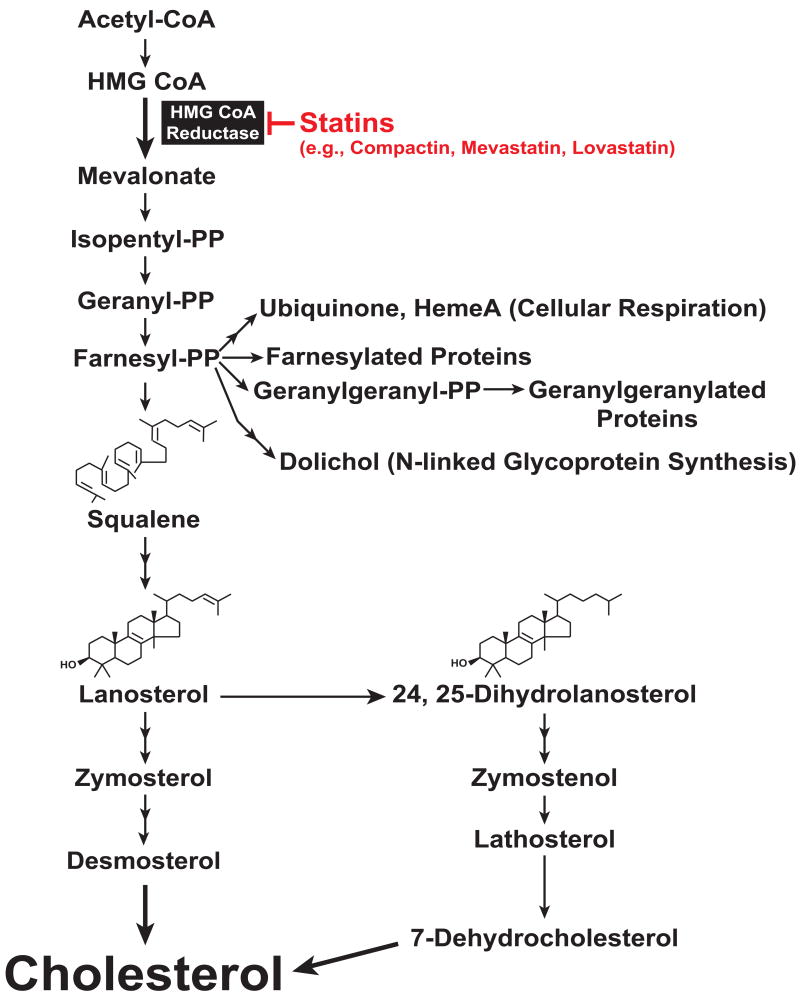
The enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase catalyzes the conversion of HMG CoA to mevalonate, a rate-determining step in the synthesis of not only cholesterol, but also of nonsterol isoprenoids that are essential for normal cell function (Figure 1) (Goldstein and Brown 1990). These molecules include ubiquinone and hemeA, which participate in aerobic cellular respiration, dolichol, which is required for the synthesis of N-linked glycoproteins, and the farnesyl and geranylgeranyl groups that become attached to various cellular proteins, increasing their membrane association. As the rate-limiting enzyme in cholesterol synthesis, reductase is the target of a complex, multivalent feedback regulatory system that is mediated by sterol and nonsterol end-products of mevalonate metabolism (Brown and Goldstein 1980). This complex regulatory system operates at transcriptional and post-transcriptional levels and guards against the overaccumulation of cholesterol while ensuring that essential nonsterol isoprenoids are constantly produced.
Figure 1.
Schematic representation of the mevalonate pathway in animal cells. Statins, competitive inhibitors of HMG CoA reductase, are highlighted in red. The abbreviation “PP” (i.g., isopentyl-PP) designates pyrophosphate.
The complexity of the multivalent control of reductase was first revealed through the use of compactin (also known as ML-236B), a founding member of the statin family of competitive reductase inhibitors that was first isolated from the fungus Penicillium citrinum by Endo and co-workers in the 1970s (Endo et al. 1976a; Endo et al. 1976b). The activity of reductase is largely suppressed when cells are cultured under normal culture conditions (i.e., medium supplemented with fetal calf serum) and, as a result, cholesterol and nonsterol isoprenoids are produced at low rates. This suppression results from the receptor-mediated uptake of cholesterol-rich low-density lipoproteins (LDLs) present in the fetal calf serum of culture medium (Brown and Goldstein 1986). Internalized cholesterol is utilized in the synthesis of cell membranes; excess cholesterol becomes esterified and stored in cytoplasmic lipid droplets as cholesterol esters. The sterol also suppresses reductase activity by inhibiting the enzyme's expression through the multivalent regulatory system. Subjecting cells to cholesterol deprivation through incubation in medium supplemented with lipoprotein-deficient serum plus compactin triggers a massive increase in the amount of reductase protein (Brown et al. 1978). This compensatory increase in reductase results from the combined effect of three regulatory events: enhanced transcription of the reductase gene, enhanced translation of the reductase mRNA, and extended half-life of the reductase protein (Brown and Goldstein 1980). Complete suppression of reductase in compactin-treated cells requires the addition of exogenous mevalonate together with LDL or oxysterols, oxygenated forms of cholesterol that are readily taken up by cells (Goldstein and Brown 1990). Together, these findings formed the basis for the concept that multiple feedback mechanisms mediated by sterol and nonsterol end-products of mevalonate metabolism control the levels and activity of reductase.
Sterol and nonsterol isoprenoids inhibit reductase at different levels. For example, sterols inhibit the activity of sterol regulatory element-binding proteins (SREBPs), a family of membrane-bound transcription factors that enhance the uptake and synthesis of cholesterol by activating transcription of the genes encoding reductase and other cholesterol biosynthetic enzymes as well as the LDL-receptor (Horton et al. 2002). Translation of reductase mRNA is blocked by a nonsterol isoprenoid (Nakanishi et al. 1988). Although the identity of this regulatory product and its mechanism of action is unknown, the reaction may be mediated by the complex 5′-untranslated region of the reductase mRNA (Reynolds et al. 1985). Sterol and nonsterol isoprenoids combine to reduce the half-life of reductase protein in compactin-treated cells from 11-12 h to less than 1 h by accelerating its ER-associated degradation (ERAD) from membranes through a mechanism mediated by the ubiquitin-proteasome system (Inoue et al. 1991; Ravid et al. 2000; Sever et al. 2003b).
The ER-Associated Degradation (ERAD) Pathway
The ER is a major site of protein biogenesis with roughly 30% of all newly synthesized proteins becoming translocated across membranes into the lumen of the organelle (Huh et al. 2003). Soon after their translocation, nascent polypeptides undergo folding and assembly through the assistance of a repertoire of ER-resident molecular chaperones (Buck et al. 2007). Translocated proteins are also subject to co- and post-translational modifications such as N-linked glycosylation and disulfide-bond formation, which promote proper folding (Helenius and Aebi 2004). Proteins that do not fold into their native conformations or fail to become incorporated into oligomeric complexes because of genetic mutation, cellular stress, or translational and transcriptional errors are selectively degraded in the cytosol by the 26S proteasome through a process known as ERAD (Jarosch et al. 2003; Meusser et al. 2005; Vembar and Brodsky 2008). Efficient destruction of defective proteins is essential as they may lead to formation of toxic, insoluble aggregates or compete with functional counterparts for substrate binding and/or complex formation with interacting proteins. Many human diseases and pathologies are linked to known ERAD substrates, which further highlights the importance of the ERAD pathway (Aridor 2007).
The highly conserved ERAD pathway is a multistep process that begins with the recognition of misfolded substrates, which appears to be carried out by a select set of molecular chaperones (Vembar and Brodsky 2008). The variety of ERAD substrates can be enormous; potential substrates can be either completely soluble within the lumen of the ER or integrated in membranes through one or more membrane-spanning segments. Thus, regions of these proteins that are located in the cytosol, within the ER lumen, and embedded in membranes must be stringently screened for misfolding (Carvalho et al. 2006; Denic et al. 2006). In the yeast Saccharomyces cerevisiae, detection of misfolded proteins engages three distinct ERAD pathways, depending upon the location of the misfolded region (Ahner and Brodsky 2004). The ERAD-C pathway becomes engaged when misfolded cytosolic domains are detected, whereas detection of misfolded domains within the ER lumen engages the ERAD-L pathway. Key mediators of the ERAD-C and ERAD-L pathways are cytosolic and ER lumenal heat shock protein homologs (e.g., Hsp70, Hsp40, and Hsp90), which recognize hallmarks of misfolding, such as exposure of hydrophobic amino acid residues that are normally sequestered within the core of the folded protein (Buck et al. 2007). A subset of misfolded glycoproteins present a single glucose moiety on their N-linked glycans, which promotes association with the lectin-like ER lumenal chaperones calnexin and calreticulin for additional rounds of folding cycles (Caramelo and Parodi 2008). Prolonged association with calnexin/calreticulin leads to degradation of these substrates through the ERAD-L pathway. The third ERAD pathway, designated ERAD-M, is engaged through the detection of misfolded regions within the membrane. It has been reasonably postulated that ERAD-M substrates present hydrophilic amino acid residues within the hydrophobic environment of the membrane bilayer (Hampton and Garza 2009). However, the precise mechanism through which these intramembrane lesions are recognized (perhaps through the action of unknown chaperones) and how this recognition engages the ERAD-M pathway is presently unclear. It is important to note that the three ERAD pathways have only been defined in yeast. Although mammalian cells can potentially present a much larger repertoire of misfolded proteins, it seems likely that some aspects of the yeast ERAD pathways, such as chaperone-mediated selection, are applicable to degradation of mammalian substrates. Moreover, these substrates would almost certainly be able to be classified as ERAD-L, ERAD-C, and ERAD-M.
Once selected for ERAD, it is generally accepted that substrates become dislocated from ER membranes into the cytosol where they are fully accessible to proteasomes for degradation. Most ERAD substrates become ubiquitinated, which ensures their efficient delivery to proteasomes, by ubiquitin-conjugating and ligating enzymes that transfer activated ubiquitin from the ubiquitin-activating enzyme (Kostova et al. 2007; Pickart 1997). The specificity of substrate ubiquitination is primarily determined by ubiquitin ligases (Deshaies and Joazeiro 2009). It is assumed that chaperones not only mediate selection of ERAD substrates, but that they also mediate substrate selection by facilitating interactions with ubiquitin ligases through the actions of intermediary proteins or substrate selectors (Buck et al. 2007). In addition to this, it is very likely that other mechanisms for selection of ERAD substrates for ubiquitination exist.
The final steps of the ERAD pathway constitute delivery of ubiquitinated substrates to proteasomes through reactions mediated in part by Valosin-containing protein (VCP)/p97, a member of the AAA (ATPases associated with diverse cellular activities)-ATPase superfamily (Vij 2008; Ye et al. 2001). VCP/p97 associates with ubiquitinated proteins through two substrate recruitment factors, Npl4 and Ufd1, which bind polyubiquitin chains. The ATPase activity of VCP/p97 is thought to drive extraction of ERAD substrates from ER membranes into the cytosol. In some cases, extraction is mediated by the 19S regulatory subunit of the proteasome, which also contains AAA-ATPase activity (Wahlman et al. 2007). It is generally accepted that soluble ERAD substrates are transported across membranes into the cytosol through a protein-conducting channel formed by either Sec61, the major component of the translocation channel that imports polypeptides into the ER, or by the Derlin family of polytopic membrane proteins (Lilley and Ploegh 2004; Meusser et al. 2005; Ye et al. 2004). Like their soluble counterparts, membrane-bound ERAD substrates are dislocated into the cytosol prior to proteasomal degradation. This has been demonstrated for substrates that contain one or more membrane-spanning segments (MHC Class I heavy chains and unpaired T-cell receptor subunits and cystic fibrosis transmembrane conductance receptor, Ste6p*, and connexins, respectively) (Huppa and Ploegh 1997; VanSlyke and Musil 2002; Wiertz et al. 1996a; Wiertz et al. 1996b). Whether cytosolic dislocation of membrane-bound ERAD substrates requires a protein-conducting channel formed by Sec61 or Derlins is not known. It should be noted that some membrane-bound ERAD substrates appear to be degraded directly from membranes (Brodsky and Wojcikiewicz 2009; Ikeda et al. 2009). This degradation could be initiated at either end of the misfolded polypeptide or from an internal site following an endoproteolytic cleavage through a mechanism in which degradation and extraction are tightly coupled.
Following extraction, VCP/p97 appears to play another role in ERAD by facilitating delivery of substrates to proteasomes through interactions with a variety of ubiquitin regulatory X (UBX), ubiquitin-associated (UBA), and ubiquitin-like (UBL) domain-containing proteins (Schuberth and Buchberger 2008). These proteins include Ufd2, an E4 enzyme that extends polyubiquitin chains (Koegl et al. 1999), the deubiquitinating enzyme Otu1, and Rad23 and Dsk2, which simultaneously bind to polyubiquitin chains and the proteasome (Raasi and Wolf 2007).
Current view of sterol-accelerated ERAD of HMG CoA reductase
Mammalian HMG CoA reductase consists of 887 or 888 amino acids that can be separated into two domains (Figure 2A). The N-terminal domain of reductase encompasses 339 amino acids; the region is embedded into ER membranes through eight membrane-spanning segments separated by short hydrophilic loops (Roitelman et al. 1992). The 548 amino acid C-terminal domain projects into the cytosol where it exerts all of the enzymatic activity (Gil et al. 1985; Liscum et al. 1985). The membrane domain of reductase is highly conserved across mammalian species (Luskey and Stevens 1985) and the region plays a key role in sterol-accelerated degradation of the enzyme as indicated by two early observations. First, the truncated, cytosolic C-terminal domain of reductase restores cholesterol synthesis when expressed in reductase-deficient Chinese hamster ovary (CHO) cells (Gil et al. 1985). However, this protein is very stable and does not become rapidly degraded in the presence of sterols. The second observation stemmed from studies of a fusion protein between the membrane domain of reductase and soluble β-galactosidase. This reductase membrane domain- β -galactosidase fusion protein exhibits sterol-accelerated degradation that is similar to the wild type, full-length reductase (Skalnik et al. 1988). Considered together, these key observations are consistent with a mechanism whereby the membrane domain of reductase senses levels of membrane-embedded sterols, triggering reactions that render the enzyme susceptible to proteolytic degradation.
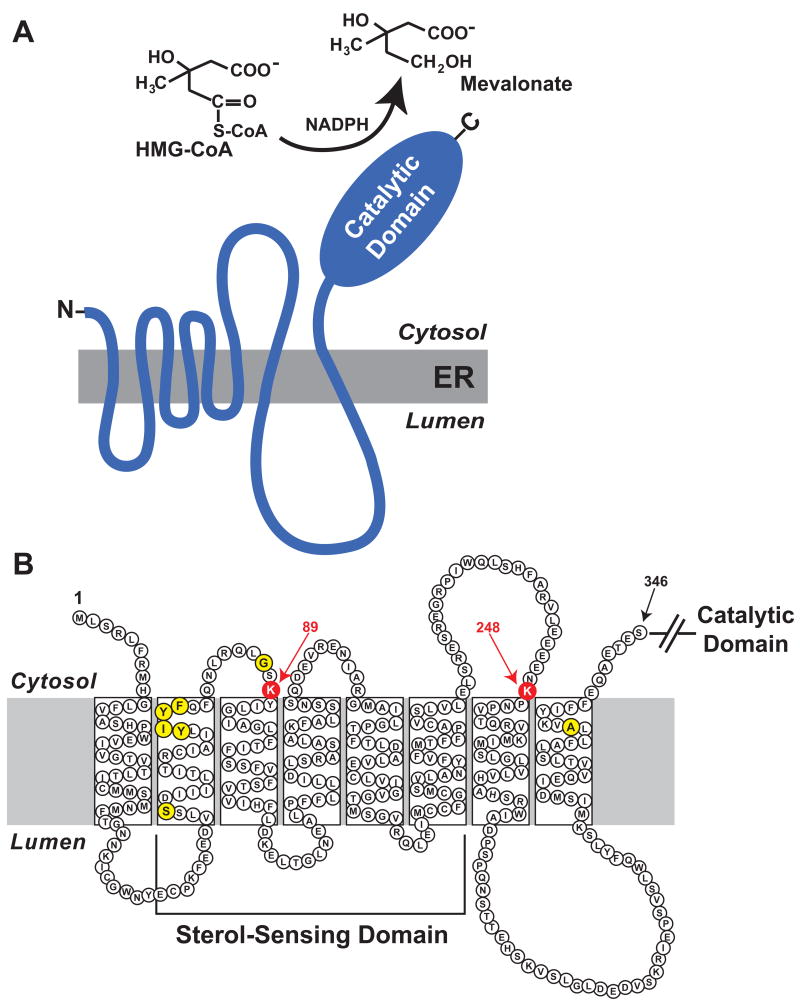
Figure 2.
Domain structure of HMG CoA reductase. (A) As discussed in the text, HMG CoA reductase consists of two distinct domains: a hydrophobic N-terminal domain with eight membrane-spanning segments that plays a key role in sterol-accelerated degradation of the enzyme and a hydrophilic C-terminal domain that directs enzymatic activity. (B) Amino acid sequence and topology of the membrane domain of HMG CoA reductase. The lysine residues that are required for Insig-mediated, sterol-induced ubiquitination of HMG CoA reductase are enlarged, highlighted in red, and denoted by arrows. Sequences required for sterol-regulated binding of HMG CoA reductase to Insigs (YIYF, Ser-60, Gly-87, and Ala-333) are enlarged and highlighted in yellow.
The membrane domain of reductase contains a stretch of ∼ 180 amino acids called the sterol-sensing domain (Figure 2B). This evolutionarily conserved domain comprises five of the eight membrane-spanning segments of reductase and is found in several other polytopic membrane proteins that are postulated to interact with sterols (Kuwabara and Labouesse 2002). These proteins include the sterol-regulated escort protein Scap (Hua et al. 1996), the lipid transport proteins Niemann Pick C1 (NPC1) and NPC1L1 (Altmann et al. 2004; Loftus et al. 1997), the Patched receptor for the cholesterol-modified morphogen Hedgehog (Eaton 2008), and Dispatched, which mediates release of Hedgehog from cells (Burke et al. 1999). The function of the sterol-sensing domain was first demonstrated for Scap, which binds to the SREBP transcription factors in the ER (Hua et al. 1996). In sterol-deprived cells, Scap facilitates transport of SREBPs from the ER to the Golgi where active fragments of the transcription factor are released from membranes by proteolysis (DeBose-Boyd et al. 1999; Goldstein et al. 2006). The processed forms of SREBPs migrate to the nucleus and activate target gene expression, which leads to increased synthesis and uptake of cholesterol and other lipids (Horton et al. 2002). When sterols accumulate in ER membranes, the membrane domain of Scap binds to one of two ER membrane proteins called Insig-1 and Insig-2 (Yabe et al. 2002a; Yang et al. 2002). Insig binding blocks incorporation of Scap-SREBP into COPII-coated vesicles that bud from ER membranes and deliver proteins to the Golgi (Nohturfft et al. 2000; Sun et al. 2007). Sequestration of Scap-SREBP complexes in the ER prevents proteolytic activation of SREBPs; expression of SREBP target genes declines and consequently, cholesterol synthesis and uptake is suppressed.
The topology of Scap in ER membranes is similar to that of reductase. The protein is anchored to membranes through its N-terminal domain, which includes eight membrane-spanning segments (Nohturfft et al. 1998b). The C-terminal domain projects into the cytosol and mediates association with SREBPs (Sakai et al. 1997). The sterol-sensing domain of Scap comprises transmembrane helices 2-6 and exhibits 55% amino acid similarity and 25% identity with the corresponding region of reductase. The importance of the sterol-sensing domain in the regulation of Scap is highlighted by findings that three point mutations (Tyr-298 to Cys, Leu-315 to Phe, and Asp-443 to Asn) within the region abolish sterol-regulated Insig binding, thereby relieving sterol-mediated ER-retention of mutant Scap-SREBP complexes (Hua et al. 1996; Nohturfft et al. 1998a; Nohturfft et al. 1996; Yabe et al. 2002b; Yang et al. 2002).
Insigs also bind to the sterol-sensing domain of reductase in a sterol-regulated fashion (Sever et al. 2003b). This binding is disrupted by mutation of the tetrapeptide sequence YIYF, which is located in the second transmembrane segment of reductase (Figure 2B). Mutation of the YIYF sequence to alanine residues abolishes Insig binding and the mutant enzyme is no longer subjected to sterol-accelerated degradation (Sever et al. 2003a). The first tyrosine of the YIYF tetrapeptide (Tyr-75) is equivalent to Tyr-298 of Scap, which is required for Insig-Scap binding (see above). When overexpressed, the sterol-sensing domain of Scap blocks sterol-accelerated degradation of reductase (Sever et al. 2003b). This effect is ablated by the Tyr-298 to Cys mutation in the Scap sterol-sensing domain, indicating that Scap and reductase binding sites on Insigs overlap. At least three additional amino acids (Ser-60, Gly-87, and Ala-333; see Figure 2B) within the membrane domain of reductase are also required for Insig binding (Lee et al. 2007). Even though Ser-60 and Gly-87 localize to the sterol-sensing domain of reductase, these residues are not present in the corresponding region of the Scap sterol-sensing domain. These observations emphasize the importance of detailed structural analyses of Scap-Insig and reductase-Insig complexes in future studies.
Two major differences exist between the Insig-mediated regulation of Scap and that of reductase. Insig binding to Scap leads to its retention in the ER, whereas Insig binding to reductase causes it to become rapidly ubiquitinated and degraded. This discrepancy can be rationalized when considering the other major difference between Insig-mediated regulation of Scap and reductase: sterol specificity. Cholesterol directly binds to the membrane domain of Scap, triggering a conformational change in the protein that allows for Insig binding (Radhakrishnan et al. 2004). In contrast, cholesterol does not potently induce rapid ubiquitination of reductase, even when added to sterol-deprived membranes in vitro (Song and DeBose-Boyd 2004). Instead, the reaction is potently stimulated by the cholesterol synthesis intermediate 24,25-dihydrolanosterol both in vitro and in intact cells (Song et al. 2005a) (Figure 1). It should be noted that lanosterol, the immediate precursor of 24,25-dihydrolanosterol (see Figure 1), was also found to stimulate ubiquitination of reductase. However, it was subsequently determined that this activity was attributable to small amounts of contaminating 24,25-dihydrolanosterol in the preparations of lanosterol used in the initial studies (Lange et al. 2008). The specificity of reductase ubiquitination is remarkable considering that lanosterol and 24,25-dihydrolanosterol only differ in the degree of side-chain saturation. This suggests that the mechanism through which 24,25-dihydrolanosterol stimulates ubiquitination of reductase likely involves its direct binding to the enzyme. However, attempts to demonstrate direct binding of 24,25-dihydrolanosterol to reductase have so far been unsuccessful. Thus, the possibility that some other protein binds 24,25-dihydrolanosterol and induces reductase to bind Insigs cannot be excluded.
The findings described above not only help to explain how Insigs mediate sterol regulation of Scap and reductase through distinct mechanisms, but they also point to the production of 24,25-dihydrolanosterol as a key focal point in sterol regulation. The demethylation of lanosterol and 24,25-dihydrolanosterol has been implicated as a rate-limiting step in the sterol branch of the mevalonate pathway (Gaylor 2002; Williams et al. 1977). 24,25-Dihydrolanosterol suppresses its own synthesis by reducing flux through the mevalonate pathway via Insig-mediated degradation of reductase. Accumulation of lanosterol and 24,25-dihydrolanosterol is avoided because these sterols do not inhibit ER-to-Golgi transport of Scap-SREBP (Song et al. 2005a). Thus, mRNAs encoding the enzymes that catalyze reactions subsequent to lanosterol synthesis remain elevated and lanosterol and 24,25-dihydrolanosterol are efficiently converted to cholesterol. As cholesterol begins to accumulate, Scap-SREBP transport to the Golgi is blocked, SREBP processing becomes inhibited, and the entire pathway is shut down. The physiologic relevance of 24,25-dihydrolanosterol as a regulator of reductase degradation is highlighted by the finding that oxygen deprivation causes the sterol to accumulate in cells (Nguyen et al. 2007). At the same time, expression of both Insigs is enhanced through the action of the oxygen-sensitive transcription factor hypoxia-inducible factor (HIF)-1α. The accumulation of 24,25-dihydrolanosterol, coupled with HIF-mediated induction of Insigs, leads to rapid degradation of reductase, providing a link between oxygen sensing and cholesterol metabolism.
Targeting HMG CoA Reductase for Proteasomal Degradation: Sterol-Regulated, Insig-Mediated Ubiquitination
An early clue as to the identity of the proteolytic machinery responsible for reductase degradation was provided by the observation that inhibitors of the proteasome block the reaction (Inoue et al. 1991). This led to the finding that proteasome inhibition leads to the accumulation of ubiquitinated forms of reductase on ER membranes (Ravid et al. 2000). A role for the ubiquitin-proteasome pathway is consistent with observations by Hampton and co-workers in S. cerevisiae (Hampton and Bhakta 1997). Hmg2 is one of two reductase isozymes expressed in yeast; the protein is rapidly degraded when flux through the mevalonate pathway is high. Hmg1, the other reductase isozyme, is not subject to regulated degradation.
The genetic analysis of Hmg2 degradation led to the identification of genes encoding several components of the ubiquitin-proteasome pathway as mediators of the reaction (Hampton 1998; Hampton and Garza 2009). These genes are termed HRD genes for HMG CoA reductase degradation and include: Hrd1, a Really Interesting New Gene (RING) finger ubiquitin ligase with multiple membrane-spanning segments followed by a large cytosolic domain; Hrd2, a component of the 26S proteasome; Hrd3, the binding partner of Hrd1 that mediates substrate selection of the enzyme; and Hrd4, the yeast homolog of Npl4, one of at least two ubiquitin-binding substrate selectors for VCP/p97 (Vij 2008). Like mammalian reductase, the membrane domain of Hmg2 is necessary and sufficient for accelerated degradation (Hampton et al. 1996). However, degradation of Hmg2 is stimulated by nonsterol isoprenoids derived from mevalonate, but not by sterols (Garza et al. 2009b; Hampton and Garza 2009). The yeast Insig protein, called Nsg1, does not promote degradation of Hmg2. Instead, Nsg1 stabilizes Hmg2, even in the presence of degradative signals (Flury et al. 2005). Despite these differences, regulated ubiquitination and subsequent ERAD of reductase is a common mechanism both yeast and mammalian systems use to limit synthesis of sterols.
Ubiquitination of mammalian reductase is obligatory for sterol-accelerated degradation of the enzyme and the reaction exhibits an absolute requirement for the action of Insigs. For example, reductase overexpressed in cells by transfection resists both sterol-accelerated ubiquitination and degradation (Sever et al. 2003a). These processes are restored by co-expression of Insig-1 or Insig-2, suggesting saturation of endogenous Insigs by the overexpressed reductase. RNA interference (RNAi)-mediated knockdown of Insig-1 and Insig-2 mRNA or mutation of genes encoding both Insigs abrogates sterol-mediated ubiquitination of reductase and renders the enzyme refractory to accelerated degradation (Lee et al. 2005; Sever et al. 2004; Sever et al. 2003a). Finally, mutation of the YIYF sequence as well as Ser-60, Gly-87, and Ala-333 in reductase abolishes Insig binding and markedly blunts the enzyme's sterol-accelerated ubiquitination (Lee et al. 2007; Sever et al. 2003a). The ubiquitination of reductase is also blocked by conservative substitutions of arginine for two cytosolically exposed lysine residues at positions 89 and 248 in the membrane domain of reductase (Figure 2B). While these mutations prevent reductase degradation, they do not block sterol-induced binding of the enzyme to Insigs. Thus, lysines 89 and 248 in reductase are implicated as sites of Insig-dependent, sterol-induced ubiquitination. The catalytic domain of reductase does not become ubiquitinated as indicated by the observation that mutation of lysines 89 and 248 blocks degradation in the context of the full-length protein. This is consistent with observations that the soluble catalytic domain is dispensable for regulated ubiquitination and degradation.
A subset of Insig molecules is associated with a membrane-bound ubiquitin ligase called gp78 (Song et al. 2005b). The enzyme consists of 643 amino acids and contains an N-terminal domain with 5-7 membrane-spanning segments that mediates association with Insigs. The C-terminal domain of gp78 projects into the cytosol and contains a RING finger domain that is required for ubiquitin ligase activity as well as binding sites for VCP/p97 and the ubiquitin-conjugating enzyme Ubc7 (Kostova et al. 2007). The role for gp78 in sterol-accelerated degradation of reductase is illustrated by several observations. The overexpressed membrane domain of gp78 competes with the full-length enzyme for Insig binding and blocks sterol-accelerated degradation of reductase. Moreover, a mutant form of gp78 harboring inactivating mutations in the RING finger domain exhibits dominant-negative activity towards reductase degradation. Sterols trigger binding of gp78 to reductase, but only when Insigs are co-expressed. The specificity of this interaction is demonstrated by the finding that gp78 does not bind to Scap, regardless of the absence or presence of sterols. Finally, RNAi-mediated knockdown of gp78 blunts sterol-induced ubiquitination and degradation of endogenous reductase. This effect is specific inasmuch as knockdown of mammalian Hrd1, a membrane-bound ubiquitin ligase that resembles gp78 (see below), does not affect reductase ubiquitination. This result is consistent with the failure of Hrd1 to interact with Insig-1 as determined by co-immunoprecipitation experiments.
The pathway for Insig-mediated, sterol-accelerated ERAD of reductase is shown in Figure 3. The process is initiated by the sensing of membrane embedded sterols through direct or indirect interactions with the membrane domain of reductase. This reaction triggers binding of the Insig-gp78 complex to the membrane domain of reductase, resulting in transfer of ubiquitin from the E2 Ubc7 to lysines 89 and 248 in reductase. Ubiquitination marks reductase for recognition by the gp78-associated VCP/p97 that, together with its cofactors, somehow extract ubiquitinated reductase from ER membranes and delivers it to proteasomes for degradation. Although the 20-carbon nonsterol isoprenoid geranylgeraniol (GGOH) augments sterol-accelerated ERAD of reductase, the compound does not appreciably affect ubiquitination of reductase when added to cells (Sever et al. 2003a). In addition, GGOH does not appear to augment in vitro ubiquitination of reductase in a specific manner (unpublished observations). This contrasts the situation in yeast, where it has been recently determined that the phosphorylated derivative of GGOH, GG-pyrophosphate, stimulates ubiquitination of Hmg2p (Garza et al. 2009b). Thus, we postulate that GGOH augments degradation of mammalian reductase by enhancing the extraction of the ubiquitinated enzyme from ER membranes, facilitating its delivery to proteasomes for degradation (Sever et al. 2003a). The possibility exists that GGOH is converted to GG-pyrophosphate and becomes incorporated into a protein that mediates the membrane extraction of ubiquitinated reductase. Geranylgeranylated proteins include the well-known Rab family of proteins that participate in various aspects of vesicular transport (Seabra et al. 2002). Thus, a vesicle-mediated transport event may deliver ubiquitinated reductase from ER membranes to a specific organelle or subdomain of the ER where the protein is subsequently degraded.
Figure 3.
Current model for sterol-accelerated ERAD of HMG CoA reductase. Accumulation of certain sterols (e.g., oxysterols such as 25-hydroxycholesterol and the cholesterol synthesis intermediate 24,25-dihydrolanosterol) stimulates binding of Insigs to the membrane domain of HMG CoA reductase. Some of the Insig molecules are associated with gp78, a membrane-anchored ubiquitin ligase that associates with the ubiquitin conjugating enzyme Ubc7 and the AAA-ATPase VCP/p97. Ubc7 and gp78 combine to initiate the polyubiquitination of two cytosolic lysine residues in the membrane domain of HMG CoA reductase. This ubiquitination triggers extraction of HMG CoA reductase from ER membranes through the action of VCP/p97 and its associated cofactors; this step appears to be enhanced by the 20-carbon nonsterol isoprenoid geranylgeraniol through an undefined mechanism. Once extracted, HMG CoA reductase is delivered to proteasomes for degradation.
Unresolved and Remaining Questions
Despite substantial advances over the past several years in the understanding of sterol-accelerated ERAD of reductase, many questions remain unresolved. For example, how does Insig binding lead to selection of reductase for gp78-mediated ubiquitination? How is ubiquitinated reductase extracted from ER membranes, and do nonsterol isoprenoids augment this reaction? How does sterol-accelerated ERAD contribute to regulation of reductase in the liver, the major site of cholesterol synthesis?
Selection of reductase for gp78-mediated ubiquitination
A model for ERAD of proteins with misfolded lumenal domains (ERAD-L substrates) is beginning to emerge from studies conducted in yeast (Carvalho et al. 2006; Denic et al. 2006). The yeast Hrd1 enzyme mediates ubiquitination of ERAD-L and ERAD-M substrates; another membrane-bound ubiquitin ligase, Doa10, mediates ubiquitination of ERAD-C substrates. Hrd1 exists in a large, multiprotein complex containing its co-factor Hrd3, the cytosolic ubiquitin-conjugating enzyme Ubc7 and its membrane anchor Cue1, the polytopic ER membrane protein Der1 and its recruitment factor Usa1, the UBX domain-containing protein Ubx2, which mediates recruitment of the AAA-ATPase cdc48, and the Hsp70 chaperone Kar2 bound to the lectin Yos9 (Figure 4). The mammalian genome encodes for homologs of all members of the yeast Hrd1 complex, except for Cue1. Sucrose gradient centrifugation experiments indicate that mammalian Hrd1 is present in a large, multiprotein complex (Schulze et al. 2005). However, complete delineation of components of the mammalian Hrd1 complex has not been determined.
Figure 4.
The S. cerevisiae Hrd1 ubiquitin ligase complex. Schematic representation of the Hrd1 complex in yeast that includes factors involved in substrate selection (Kar2 and Yos9), ubiquitination (Ubc7 and Cue1), and recruitment of cdc48 (Ubx2) and Der1 (Usa1). Yeast proteins are shown in black and their mammalian homologs are shown in magenta. Hrd1 complex components required for Insig-mediated degradation of HMG CoA reductase in Drosophila S2 cells are denoted by asterisks.
The model for Hrd1-mediated degradation of glycosylated ERAD-L substrates begins with their recognition by the chaperone Kar2, which in turn associates with the ER lumenal lectin Yos9. These substrates are then presented to the Hrd1 complex for dislocation and ubiquitination through a mechanism mediated by interactions between Yos9 and Hrd3, which forms a 1:1 stoichiometric complex with Hrd1 (see Figure 5A). A similar model appears to apply to Hrd1 mediated degradation of ERAD-L substrates in mammalian cells (Christianson et al. 2008). As selectors for ERAD-L substrates, Kar2 and Yos9 sense hallmarks of protein misfolding such as the exposure of hydrophobic amino acid residues or the presence of mono-glucosylated N-linked glycans. Considering this, selection of ERAD-M substrates for Hrd1-mediated ubiquitination/degradation should involve intramembrane protein-protein interactions. By analogy to the model for ERAD-L substrate recognition/selection, it is reasonable to speculate the existence of molecular chaperones that somehow recognize hallmarks of misfolded intramembrane regions such as exposure of hydrophilic amino acid residues within the membrane (Hampton and Garza 2009) and intermediary proteins that bridge the ERAD-M substrate to the Hrd1 complex through Hrd3-mediated interactions.
Figure 5.
Proposed model for Insig-mediated selection of mammalian HMG CoA reductase for ubiquitination/degradation. (A) As discussed in the text, Hrd1-mediated degradation of proteins with misfolded lumenal domain (ERAD-L substrates) in yeast begins with their recognition by the chaperone Kar2, which associates with the lectin-like protein Yos9. These substrates are then transferred to the Hrd1 complex through a mechanism that is mediated by interactions between Yos9 and Hrd3. In subsequent steps, ERAD-L substrates become dislocated into the cytosol, ubiquitinated, and presented to proteasomes for degradation through the actions of other Hrd1 complex components shown in Figure 4. (B) Reconstitution experiments reveal that Drosophila Hrd1 and Sel1 (the Hrd3 homolog) are required for Insig-mediated, sterol-accelerated degradation of mammalian HMG CoA reductase in S2 cells. By analogy to the model present in A, this degradation may involve a mechanism whereby Insigs bridge HMG CoA reductase to the dHrd1 complex through interactions with an unknown intermediary protein(s) that plays a role similar to that of Yos9 in degradation of ERAD-L substrates in yeast.
A role for an intermediary protein and/or chaperone in the degradation of ERAD-M substrates may be suggested by studies of Hmg2, one of few ERAD-M substrates that have been studied in detail. When certain intermediates of mevalonate metabolism accumulate, Hmg2 acquires features of a misfolded protein and becomes degraded through a mechanism mediated by Hrd1 (Gardner and Hampton 1999; Shearer and Hampton 2005). Binding of apparently misfolded Hmg2 to the Hrd1 complex requires the presence of Hrd3, which contains a large lumenal domain, a single membrane-spanning segment, and a short cytosolic tail (Gardner et al. 2001). Two consequences of Hrd3 deletion is a block in degradation of Hmg2 and auto-ubiquitination followed by proteasomal degradation of Hrd1 (Gardner et al. 2001; Gardner et al. 2000). Hrd1 stability and degradation of Hmg2 is restored by expression of the lumenal domain of Hrd3. These processes are dissociable; in Hrd3-deficient yeast, the lumenal domain of Hrd3 lacking the N-terminal half stabilizes Hrd1, but cannot support Hrd1-Hmg2 complex formation and Hmg2 ubiquitination/degradation. These findings suggest that the N-terminal half of the lumenal domain of Hrd3 mediates associations involved in substrate selection (possibly through binding to an intermediary protein or chaperone), whereas the C-terminal half of Hrd3 mediates interactions with Hrd1. Importantly, a recent study suggests that Hrd1 plays a direct role in selection of Hmg2 as an ERAD-M substrate (Sato et al. 2009). Mutation of several hydrophobic amino acid residues in the membrane domain of Hrd1 impairs the ability of the enzyme to initiate ubiquitination of Hmg2, but not of ERAD-L substrates. These results led to the conclusion that the hydrophilic intramembrane residues in Hrd1 engage in noncovalent interactions with hydrophilic residues in the membrane domain of Hmg2 that become exposed in the presence of degradative signals. An important question for future studies is whether an intermediary protein bridges Hmg2 to Hrd1 through Hrd3-mediated interactions.
Sterol-accelerated degradation of mammalian reductase has been recently reconstituted in Drosophila S2 cells, which lack a recognizable INSIG gene and cannot synthesize sterols de novo (Clark and Bloch 1959; Clayton 1964). Although S2 cells express a homolog for reductase, the enzyme is not subjected to sterol-accelerated degradation (Brown et al. 1983; Gertler et al. 1988). Regulated degradation of mammalian reductase in S2 cells precisely mirrors the reaction that occurs in mammalian cells with regard to the absolute requirement for the action of mammalian Insigs, sterol specificity, sensitivity to proteasome inhibition, and augmentation by nonsterol isoprenoids (Nguyen et al. 2009). These findings indicate that Insig-mediated recognition/selection of reductase and subsequent proteasome-mediated degradation of the enzyme occur through a mechanism that is mediated by highly conserved components of the general ERAD pathway.
In yeast, the membrane-bound Hrd1 and Doa10 are the only ubiquitin ligases known to mediate ubiquitination of ERAD substrates (Kostova et al. 2007). It is becoming increasingly evident that the number of ERAD ubiquitin ligases in mammalian cells far exceeds that in yeast. Thus, it is not surprising that in addition to Hrd1 and Teb4 (the Doa10 homolog in mammals), other ubiquitin ligases such as Trc8, CHIP, as well as gp78 have been implicated in the ERAD pathway of mammalian cells. It has been estimated that more than 50 uncharacterized RING finger proteins contain membrane-spanning segments (Kostova et al. 2007); it seems likely that some of these proteins play key roles in ERAD. The ERAD pathway has not been well-studied in Drosophila cells; however, the reconstitution of reductase degradation in S2 cells points to the existence of a Drosophila ubiquitin ligase that can bind Insigs and initiate ubiquitination of mammalian reductase. Indeed, the Drosophila genome contains homologs for Hrd1 (dHrd1), Trc8 (dTrc8), and Teb4 (dTeb4).
The RNAi-mediated knockdown of dHrd1, but not dTrc8 and dTeb4, abolishes sterol-accelerated degradation of mammalian reductase in S2 cells (Nguyen et al. 2009). This finding is significant considering that mammalian Hrd1 and gp78 belong to a subfamily of membrane-bound ubiquitin ligases. Both proteins contain a hydrophobic N-terminal domain with multiple membrane-spanning segments followed by a cytosolic C-terminal domain with a RING finger motif that directs ubiquitin ligase activity (Kostova et al. 2007). Hrd1 and gp78 are organized in ER membranes with similar topologies and their membrane domains share approximately 50% amino acid homology. Importantly, the membrane domains of these enzymes do not bear significant sequence homology with the corresponding regions of other membrane-bound ubiquitin ligases such as Teb4 and Trc8. Thus, gp78 and Hrd1 can be considered as a subfamily of membrane-bound ubiquitin ligases that mediate degradation of reductase in yeast, Drosophila, and mammalian cells.
Reconstitution experiments in S2 cells reveal that a subset of dHrd1 complex components including dSel1 (Hrd3 homolog), dHerp (Usa1 homolog), dUbxd8 (Ubx2 homolog), dNpl4, and dUfd1 are required for regulated degradation of reductase (see Figure 4). These findings are similar to those reported in yeast where a subset of the Hrd1 complex components, namely Hrd1 and Hrd3, are required for ERAD-M substrates such as Hmg2 (Carvalho et al. 2006; Denic et al. 2006). However, the reconstitution experiments reveal that dHerp may play a role in reductase degradation that is distinct from recruitment of Derlins. In mammalian cells, Herp binds to a family of proteins called ubiquilins, which contain an N-terminal UBL domain that binds to proteasomes and a C-terminal UBA domain that binds polyubiquitin chains (Kim et al. 2008). The yeast equivalent of ubiquilin, Dsk2, is known to participate in ERAD by guiding ubiquitinated substrates to the proteasome (Ko et al. 2004; Walters et al. 2004). Whether Herp mediates degradation of reductase in Drosophila and mammalian cells through a mechanism involving ubiquilins remains to be determined.
Based on the current understanding of how ERAD substrates are selected for ubiquitination and degradation, a model for Insig-mediated selection reductase in Drosophila cells is presented in Figure 5B. The key feature of this model is the recruitment of reductase to the dHrd1 complex through interactions mediated by dSel1 (the Hrd3 homolog), Insig, and an as-of-yet to be identified intermediary protein(s). The proposed intermediary protein(s) is presumed to bridge reductase to the dHrd1 complex through interactions with both Insig and dSel1; a similar mechanism applies to Yos9-mediated degradation of ERAD-L substrates (see Figure 5A). In the model of Figure 5B, Insig plays the role of the intramembrane chaperone that initiates recognition of reductase as ERAD-M substrate. Considering that Insig-mediated degradation of reductase in S2 cells is incredibly specific, it seems very likely that the proposed intermediary protein(s) are conserved components of the general ERAD pathway involved in selection of a subset of dHrd1 substrates. However, the possibility exists that dSel1 plays an indirect role in reductase degradation by stabilizing dHrd1. Thus, exciting avenues for future work will be to define the mechanism for Insig-dependent, Hrd1-mediated degradation of reductase in S2 cells and identify the proposed intermediary protein that mediates the reaction.
Studies of the yeast ERAD pathway have clearly established that the Hrd1 and Doa10 complexes contains an array of conserved factors involved in processes ranging from the selection and recruitment of substrates to the extraction of ubiquitinated substrates from membranes and their delivery to proteasomes (Figure 4) (Vembar and Brodsky 2008). Although not completely defined at the molecular level, complexes of mammalian ERAD ubiquitin ligases are likely to exist. In fact, mammalian Hrd1 associates with gp78 and appears to mediate its proteasomal degradation (Shmueli et al. 2009; Ye et al. 2005). However, it is presently unknown whether gp78 and Hrd1 combine to mediate degradation of ERAD substrates or whether they share common subunits. The complete definition of mammalian Hrd1 and gp78 complexes and determining the role of these proteins in Insig-mediated, sterol-accelerated ERAD of reductase are obvious areas of future investigation. The original experiments demonstrating sterol-regulated binding of reductase-Insig to gp78 relied on co-immunoprecipitation of the proteins from detergent lysates of cells (Song et al. 2005b). It is presently unknown whether binding between reductase-Insig and gp78 is mediated by direct interactions between gp78 and Insig. Thus, the possibility of the existence of an intermediary protein that bridges this interaction cannot be excluded, further emphasizing the importance of defining the gp78 ubiquitin ligase complex.
It should be noted that an ER membrane protein called SPFH2 can be recovered in a multiprotein complex that includes gp78, VCP/p97, and inositol trisphosphate receptors (IP3Rs), polytopic membrane proteins known to undergo regulated ERAD (Brodsky and Wojcikiewicz 2009). RNAi-mediated knockdown of SPFH2 prevents ubiquitination and subsequent degradation of IP3Rs; degradation of other ERAD substrates is also blunted in SPFH2-knockdown cells. Whether SPFH2 plays a role as an intermediary protein in the ERAD of IP3Rs and other substrates mediated by gp78 or other ERAD ubiquitin ligase remains to be determined.
Mechanism for extraction of ubiquitinated reductase from ER membranes
The striking feature of reductase degradation is that the catalytic and membrane domains are degraded together as a unit. That is to say, the soluble catalytic domain is not released from membranes during the degradation process (Gil et al. 1985). Ubiquitination is obligatory for reductase degradation, occurring on two cytosolic lysine residues in the membrane domain. As mentioned earlier, mutation of these lysines abolishes detectable ubiquitination of reductase, even in the context of the full-length protein. Moreover, the membrane domain of reductase confers sterol-accelerated degradation when fused to a normally stable soluble protein such as β-galactosidase, GFP, or luciferase ((Skalnik et al. 1988) and our unpublished observations). Thus, the question arises as to how ubiquitination of the membrane domain renders the entire reductase protein susceptible to proteasomal degradation. Ubiquitinated reductase could be degraded by proteasomes directly from ER membranes or completely extracted from the membrane into the cytosol prior to proteolysis. Evidence in favor of the latter scenario is beginning to accumulate. Recent studies have shown that in both yeast and mammalian systems, full-length reductase becomes dislocated from ER membranes into the cytosol (Garza et al. 2009a; Leichner et al. 2009). However, these observations raise additional questions as to the mechanism for cytosolic dislocation of reductase. Does the reaction require a protein-conducting channel? If so, does Sec61, one of the Derlin proteins, or a novel protein form this channel? It has been proposed that the multiple membrane-spanning segments of ERAD ubiquitin ligases form the channel through which ERAD substrates are dislocated (Nakatsukasa and Brodsky 2008; Nakatsukasa et al. 2008). Finally, an intriguing hypothesis has been put forth in which ERAD substrates become dislocated from ER membranes into the cytosol through lipid droplets. Lipid droplets are cytosolic organelles traditionally regarded as storage depots for neutral lipids such as triglycerides and cholesterol esters. A role for lipid droplets in ERAD is suggested by 1) the accumulation of the ERAD substrate apolipoprotein B-100 on lipid droplets when proteasome activity is blocked (Ohsaki et al. 2006) and 2) identification of several chaperones as well as VCP/p97 and its membrane receptor Ubxd8 as lipid droplet-associated proteins (Bartz et al. 2007; Liu et al. 2004). Other open questions regarding dislocation of reductase pertain to how the solubility of its membrane-spanning segments is maintained during dislocation, how the enzyme is delivered to proteasomes following dislocation, and whether dislocation involves a vesicular budding or fusion event that is mediated by a geranylgeranylated protein. Answers to these questions may be provided through rigorous examination of reductase degradation in vitro.
Contribution of sterol-accelerated degradation to overall regulation of reductase in vivo
Prior to the availability of anti-reductase antibodies and cDNA probes, indirect methods such as measurement of enzymatic activity were used to study the regulation of reductase in livers of whole animals. These key studies demonstrated that a multivalent feedback regulatory system similar to that described in cultured cells operates in the liver to control the levels and activity of reductase (Endo et al. 1979; Kita et al. 1980; Singer et al. 1984). The significance of this regulatory system is highlighted by the effectiveness of statins, which potently inhibit reductase, in lowering plasma LDL-cholesterol and reducing the incidence of coronary heart disease in humans (Heart Protection Study Collaborative 2002; Scandinavian Simvastatin Study 1994). However, statin-mediated inhibition of reductase disrupts feedback regulation of the enzyme and induces a compensatory increase in the levels of reductase. Notably, this compensatory increase has been observed in the livers of patients undergoing statin therapy (Reihner et al. 1990). The accumulation of reductase becomes progressively harder to inhibit, evoking the need for higher doses of statins to maintain their LDL-cholesterol lowering effects. Studies of genetically altered mice suggest that disruption of Insig-mediated degradation accounts, in part, for the compensatory increase in reductase that accompanies statin treatment. In livers of mice deficient in Insig-1 and Insig-2, reductase protein accumulates to a level >100-fold than that in wild type animals (Engelking et al. 2005). This accumulation is presumably attributable to the contribution of both transcriptional and post-transcriptional regulation of reductase. However, it should be noted that reductase protein accumulates disproportionately to its mRNA in the absence of Insigs. A remarkably similar increase in the amount of reductase protein occurs in Insig-deficient CHO cells (Lee et al. 2005), indicating that the response is a common feature of many cell types. Studies focused on Insig-mediated degradation are required to precisely determine the contribution of protein stability to the overall regulation of reductase and its impact on cholesterol metabolism at the level of the whole animal.
The further elucidation of mechanisms for sterol-accelerated degradation of reductase will have important implications for both basic science and clinical medicine. From the basic science perspective, reductase represents an ideal model for the ERAD of polytopic membrane proteins. Degradation of reductase is a highly specific reaction that only occurs in Insig-expressing cells that have been subjected to sterol treatment. The sterol-dependence of reductase degradation is a valuable control that guards against artifactual degradation that may occur once various steps of the reaction are reconstituted in vitro. From a clinical point of view, detailed understanding of reductase degradation may lead to the development of drugs that accelerate this process and counteract the accumulation of the enzyme that occurs during statin treatment. These new drugs may provide an important alternative or adjuvant to statin therapy. Finally, understanding mechanisms for reductase degradation may provide valuable insight into the ERAD of other clinically relevant polytopic membrane proteins such as mutant forms of the cystic fibrosis transmembrane conductance receptor (cytstic fibrosis, (Ward et al. 1995)), connexins-32 (X-linked Charot-Marie-Tooth disease, (VanSlyke et al. 2000)), and polycystin-2 (autosomal dominant polycystic kidney disease, (Liang et al. 2008)).
Acknowledgments
We thank Drs. Jin Ye and Isamu Hartman for critical reading of this manuscript.
Footnotes
Declaration of Interest: Work in the DeBose-Boyd laboratory is supported by grants from the National Institute of Health (HL20948) and the Perot Family Foundation. R.D.-B. is an Early Career Scientist of the Howard Hughes Medical Institute, an Established Investigator of the American Heart Association, and a W.M. Keck Foundation Distinguished Young Scholar in Medical Research. The authors declare no conflict of interest.
References
- Ahner A, Brodsky JL. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Altmann SW, Davis, Zhu LJ, Yao X, Hoos LM, et al. Niemann-Pick C1 Like 1 Protein Is Critical for Intestinal Cholesterol Absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- Aridor M. Visiting the ER: The endoplasmic reticulum as a target for therapeutics in traffic related diseases. Advanced Drug Delivery Reviews. 2007;59:759–781. doi: 10.1016/j.addr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation 2. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Wojcikiewicz RJH. Substrate-specific mediators of ER associated degradation (ERAD) Current Opinion in Cell Biology. 2009;21:516–521. doi: 10.1016/j.ceb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Havel CM, Watson JA. Isoprene synthesis in isolated embryonic Drosophila cells. II. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity 47. J Biol Chem. 1983;258:8512–8518. [PubMed] [Google Scholar]
- Brown MS, Faust JR, Goldstein JL. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. Journal of Biological Chemistry. 1978;253:1121–1128. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. Journal of Lipid Research. 1980;21:505–517. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Buck TM, Wright CM, Brodsky JL. The activities and function of molecular chaperones in the endoplasmic reticulum. Seminars in Cell & Developmental Biology. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, et al. Dispatched, a Novel Sterol-Sensing Domain Protein Dedicated to the Release of Cholesterol-Modified Hedgehog from Signaling Cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct Ubiquitin-Ligase Complexes Define Convergent Pathways for the Degradation of ER Proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD 3. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Bloch K. The Absence of Sterol Synthesis in Insects. Journal of Biological Chemistry. 1959;234:2578–2582. [PubMed] [Google Scholar]
- Clayton RB. The Utilization of Sterols by Insects 1. J Lipid Res. 1964;15:3–19. [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, et al. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A Luminal Surveillance Complex that Selects Misfolded Glycoproteins for ER-Associated Degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP. RING Domain E3 Ubiquitin Ligases. Annual Review of Biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity 9. FEBS Lett. 1976a;72:323–326. doi: 10.1016/0014-5793(76)80996-9. [DOI] [PubMed] [Google Scholar]
- Endo A, Kuroda M, Tsujita Y. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogenesis produced by Penicillium citrinium. J Antibiot (Tokyo) 1976b;29:1346–1348. doi: 10.7164/antibiotics.29.1346. [DOI] [PubMed] [Google Scholar]
- Endo A, Tsujita Y, Kuroda M, Tanzawa K. Effects of ML-236B on cholesterol metabolism in mice and rats: lack of hypocholesterolemic activity in normal animals. Biochim Biophys Acta. 1979;575:266–276. [PubMed] [Google Scholar]
- Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, et al. Schoenheimer effect explained - feedback regulation of cholesterol synthesis in mice mediated by Insig proteins 1. J Clin Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury I, Garza R, Shearer A, Rosen J, Cronin S, et al. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. The EMBO Journal. 2005;24:3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. The EMBO Journal. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. The Journal of Cell Biology. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. Journal of Biological Chemistry. 2009a;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Tran PN, Hampton RY. Geranylgeranyl pyrophosphate (GGPP) is a potent regulator of HRD-dependent HMG-CoA reductase degradation in yeast. Journal of Biological Chemistry. 2009b doi: 10.1074/jbc.M109.023994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylor JL. Membrane-Bound Enzymes of Cholesterol Synthesis from Lanosterol. Biochemical and Biophysical Research Communications. 2002;292:1139–1146. doi: 10.1006/bbrc.2001.2008. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Chiu CY, Richter-Mann L, Chin DJ. Developmental and metabolic regulation of the Drosophila melanogaster 3-hydroxy-3-methylglutaryl coenzyme A reductase. Molecular and Cellular Biology. 1988;8:2713–2721. doi: 10.1128/mcb.8.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G, Faust JR, Chin DJ, Goldstein JL, Brown MS. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41:249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols 1. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Hampton RY. Genetic analysis of hydroxymethylglutaryl-coenzyme A reductase regulated degradation. Curr Opin Lipidol. 1998;9:93–97. doi: 10.1097/00041433-199804000-00003. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci U S A. 1997;94:12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Koning A, Wright R, Rine J. In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc Natl Acad Sci U S A. 1996;93:828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heart Protection Study Collaborative, G. MRC/BHF Heart Protection Study of cholesterol lowering with simvastatin in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 2002;360:7–22. doi: 10.1016/S0140-6736(02)09327-3. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-Linked Glycans in the Endoplasmic Reticulum. Annual Review of Biochemistry. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. In vitro translation and assembly of a complete T cell receptor-CD3 complex. J Exp Med. 1997;186:393–403. doi: 10.1084/jem.186.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Demartino GN, Brown MS, Lee JN, Goldstein JL, et al. Regulated endoplasmic reticulum-associated degradation of a polytopic protein: p97 RECRUITS PROTEASOMES TO Insig-1 BEFORE EXTRACTION FROM MEMBRANES. J Biol Chem. 2009;284:34889–34900. doi: 10.1074/jbc.M109.044875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Bar-Nun S, Roitelman J, Simoni RD. Inhibition of degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in vivo by cysteine protease inhibitors. Journal of Biological Chemistry. 1991;266:13311–13317. [PubMed] [Google Scholar]
- Jarosch E, Lenk U, Sommer T. Endoplasmic reticulum-associated protein degradation 15. Int Rev Cytol. 2003;223:39–81. doi: 10.1016/s0074-7696(05)23002-4. [DOI] [PubMed] [Google Scholar]
- Kim TY, Kim E, Yoon SK, Yoon JB. Herp enhances ER-associated protein degradation by recruiting ubiquilins 1. Biochem Biophys Res Commun. 2008;369:741–746. doi: 10.1016/j.bbrc.2008.02.086. [DOI] [PubMed] [Google Scholar]
- Kita T, Brown MS, Goldstein JL. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. J Clin Invest. 1980;66:1094–1100. doi: 10.1172/JCI109938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains 1. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, et al. A Novel Ubiquitination Factor, E4, Is Involved in Multiubiquitin Chain Assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Seminars in Cell & Developmental Biology. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- Lange Y, Ory DS, Ye J, Lanier MH, Hsu FF, et al. Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methylglutaryl-CoA reductase 2. J Biol Chem. 2008;283:1445–1455. doi: 10.1074/jbc.M706967200. [DOI] [PubMed] [Google Scholar]
- Lee PC, Nguyen AD, DeBose-Boyd RA. Mutations within the membrane domain of HMG-CoA reductase confer resistance to sterol-accelerated degradation 1. J Lipid Res. 2007;48:318–327. doi: 10.1194/jlr.M600476-JLR200. [DOI] [PubMed] [Google Scholar]
- Lee PC, Sever N, DeBose-Boyd RA. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2 2. J Biol Chem. 2005;280:25242–25249. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- Leichner GS, Avner R, Harats D, Roitelman J. Dislocation of HMG-CoA reductase and Insig-1, two polytopic endoplasmic reticulum proteins, en route to proteasomal degradation. Mol Biol Cell. 2009;20:3330–3341. doi: 10.1091/mbc.E08-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Li Q, Tang Y, Kokame K, Kikuchi T, et al. Polycystin-2 is regulated by endoplasmic reticulum-associated degradation. Human Molecular Genetics. 2008;17:1109–1119. doi: 10.1093/hmg/ddm383. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Liscum L, Finer-Moore J, Stroud RM, Luskey KL, Brown MS, et al. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. Journal of Biological Chemistry. 1985;260:522–530. [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, et al. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic 1. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Luskey KL, Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. Journal of Biological Chemistry. 1985;260:10271–10277. [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Goldstein JL, Brown MS. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. Journal of Biological Chemistry. 1988;263:8929–8937. [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. The Recognition and Retrotranslocation of Misfolded Proteins from the Endoplasmic Reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-Associated Degradation of a Misfolded Polytopic Membrane Protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AD, Lee SH, DeBose-Boyd RA. Insig-mediated, sterol-accelerated degradation of the membrane domain of hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase in insect cells. Journal of Biological Chemistry. 2009;284:26778–26788. doi: 10.1074/jbc.M109.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AD, McDonald JG, Bruick RK, DeBose-Boyd RA. Hypoxia stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase through accumulation of lanosterol and hypoxia-inducible factor-mediated induction of insigs 1. J Biol Chem. 2007;282:27436–27446. doi: 10.1074/jbc.M704976200. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Brown MS, Goldstein JL. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc Natl Acad Sci U S A. 1998a;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Brown MS, Goldstein JL. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. Journal of Biological Chemistry. 1998b;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Hua X, Brown MS, Goldstein JL. Recurrent G-to-A substitution in a single codon of SREBP cleavage-activating protein causes sterol resistance in three mutant Chinese hamster ovary cell lines. Proc Natl Acad Sci U S A. 1996;93:13709–13714. doi: 10.1073/pnas.93.24.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B 1. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. The FASEB Journal. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH. Ubiquitin receptors and ERAD: A network of pathways to the proteasome. Seminars in Cell & Developmental Biology. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct Binding of Cholesterol to the Purified Membrane Region of SCAP; Mechanism for a Sterol-Sensing Domain 1. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Journal of Biological Chemistry. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- Reihner E, Rudling M, Stahlberg D, Berglund L, Ewerth S, et al. Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N Engl J Med. 1990;323:224–228. doi: 10.1056/NEJM199007263230403. [DOI] [PubMed] [Google Scholar]
- Reynolds GA, Goldstein JL, Brown MS. Multiple mRNAs for 3-hydroxy-3-methylglutaryl coenzyme A reductase determined by multiple transcription initiation sites and intron splicing sites in the 5′-untranslated region. Journal of Biological Chemistry. 1985;260:10369–10377. [PubMed] [Google Scholar]
- Roitelman J, Olender EH, Bar-Nun S, Dunn WA, Jr, Simoni RD. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. The Journal of Cell Biology. 1992;117:959–973. doi: 10.1083/jcb.117.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, et al. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. Journal of Biological Chemistry. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandinavian Simvastatin Study, G. Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S) Lancet. 1994;344:1383–1389. [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cellular and Molecular Life Sciences (CMLS) 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Standera S, Buerger E, Kikkert M, Van Voorden S, et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway 9. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Seabra MC, Mules EH, Hume AN. Rab GTPases, intracellular traffic and disease. Trends Mol Med. 2002;8:23–30. doi: 10.1016/s1471-4914(01)02227-4. [DOI] [PubMed] [Google Scholar]
- Sever N, Lee PCW, Song BL, Rawson RB, DeBose-Boyd RA. Isolation of Mutant Cells Lacking Insig-1 through Selection with SR-12813, an Agent That Stimulates Degradation of 3-Hydroxy-3-methylglutaryl-Coenzyme A Reductase. Journal of Biological Chemistry. 2004;279:43136–43147. doi: 10.1074/jbc.M406406200. [DOI] [PubMed] [Google Scholar]
- Sever N, Song BL, Yabe D, Goldstein JL, Brown MS, et al. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. Journal of Biological Chemistry. 2003a;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003b;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- Shearer AG, Hampton RY. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein 12. EMBO J. 2005;24:149–159. doi: 10.1038/sj.emboj.7600498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun. 2009;390:758–762. doi: 10.1016/j.bbrc.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer II, Kawka DW, Kazazis DM, Alberts AW, Chen JS, et al. Hydroxymethylglutaryl-coenzyme A reductase-containing hepatocytes are distributed periportally in normal and mevinolin-treated rat livers. Proc Natl Acad Sci U S A. 1984;81:5556–5560. doi: 10.1073/pnas.81.17.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalnik DG, Narita H, Kent C, Simoni RD. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto beta-galactosidase. Journal of Biological Chemistry. 1988;263:6836–6841. [PubMed] [Google Scholar]
- Song BL, DeBose-Boyd RA. Ubiquitination of 3-Hydroxy-3-methylglutaryl-CoA Reductase in Permeabilized Cells Mediated by Cytosolic E1 and a Putative Membrane-bound Ubiquitin Ligase 1. J Biol Chem. 2004;279:28798–28806. doi: 10.1074/jbc.M402442200. [DOI] [PubMed] [Google Scholar]
- Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metabolism. 2005a;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase 1. Mol Cell. 2005b;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Sun LP, Seemann J, Goldstein JL, Brown MS. From the Cover: Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proceedings of the National Academy of Sciences. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSlyke JK, Deschenes SM, Musil LS. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Molecular Biology of the Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSlyke JK, Musil LS. Dislocation and degradation from the ER are regulated by cytosolic stress. The Journal of Cell Biology. 2002;157:381–394. doi: 10.1083/jcb.200111045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N. AAA ATPase p97/VCP: cellular functions, disease and therapeutic potential. J Cell Mol Med. 2008;12:2511–2518. doi: 10.1111/j.1582-4934.2008.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, et al. Real-Time Fluorescence Detection of ERAD Substrate Retrotranslocation in a Mammalian In Vitro System. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective 1. Biochim Biophys Acta. 2004;1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, et al. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996a;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, et al. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996b;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Williams MT, Gaylor JL, Morris HP. Investigation of the rate-determining microsomal reaction of cholesterol biosynthesis from lanosterol in Morris hepatomas and liver 1. Cancer Res. 1977;37:1377–1383. [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci U S A. 2002a;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Xia ZP, Adams CM, Rawson RB. Three mutations in sterol-sensing domain of SCAP block interaction with insig and render SREBP cleavage insensitive to sterols. Proc Natl Acad Sci U S A. 2002b;99:16672–16677. doi: 10.1073/pnas.262669399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol 25. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, Van Voorden S, Wiertz E, et al. Inaugural Article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane 1. Proc Natl Acad Sci U S A. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol 6. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]