Abstract
Myxococcus xanthus social gliding motility, which is powered by type IV pili, requires the presence of exopolysaccharides (EPS) on the cell surface. The Dif chemosensory system is essential for the regulation of EPS production. It was demonstrated previously that DifA (methyl-accepting chemotaxis protein [MCP]-like), DifC (CheW-like), and DifE (CheA-like) stimulate whereas DifD (CheY-like) and DifG (CheC-like) inhibit EPS production. DifD was found not to function downstream of DifE in EPS regulation, as a difD difE double mutant phenocopied the difE single mutant. It has been proposed that DifA, DifC, and DifE form a ternary signaling complex that positively regulates EPS production through the kinase activity of DifE. DifD was proposed as a phosphate sink of phosphorylated DifE (DifE∼P), while DifG would augment the function of DifD as a phosphatase of phosphorylated DifD (DifD∼P). Here we report in vitro phosphorylation studies with all the Dif chemosensory proteins that were expressed and purified from Escherichia coli. DifE was demonstrated to be an autokinase. Consistent with the formation of a DifA-DifC-DifE complex, DifA and DifC together, but not individually, were found to influence DifE autophosphorylation. DifD, which did not inhibit DifE autophosphorylation directly, was found to accept phosphate from autophosphorylated DifE. While DifD∼P has an unusually long half-life for dephosphorylation in vitro, DifG efficiently dephosphorylated DifD∼P as a phosphatase. These results support a model where DifE complexes with DifA and DifC to regulate EPS production through phosphorylation of a downstream target, while DifD and DifG function synergistically to divert phosphates away from DifE∼P.
The proper regulation of bacterial motility is critical for the survival of bacteria in their natural environment. One such form of regulation is bacterial chemotaxis, which enables organisms to move toward more favorable niches and away from hazardous ones. Chemotaxis regulation in flagellated swimming bacteria has been well studied in model organisms such as Escherichia coli and Bacillus subtilis (2, 36). In general, environmental changes are detected and transduced to the cytoplasmic side of the cell by a transmembrane ternary signaling complex composed of methyl-accepting chemotaxis proteins (MCPs), CheW, and CheA. Typically, MCPs anchor the complex to the membrane through their two transmembrane (TM) domains. Chemical changes in the environment are detected by the periplasmic domain of an MCP, resulting in conformational changes in the conserved cytoplasmic signaling domain. These changes can modulate the activity of the CheA kinase via interactions with CheW in the signaling complex. The response regulator CheY, another essential component of the bacterial chemotaxis pathway, is a substrate of the CheA kinase that accepts a phosphate from autophosphorylated CheA. Phosphorylated CheY (CheY∼P) interacts with the flagellar motor complex to effect bacterial swimming behavior. Although the dephosphorylation of CheY∼P can occur spontaneously, it is accelerated by phosphatases such as CheZ in E. coli and CheC as well as FliY in B. subtilis. The dephosphorylation of CheY∼P is critical for chemotaxis since it is one of the mechanisms for the desensitization of a stimulated chemotaxis pathway. This basic architecture of chemotaxis pathways is generally conserved in all the flagellated swimming bacteria examined to date (31, 36).
Myxococcus xanthus is a gliding Gram-negative bacterium that encodes eight chemosensory systems based on the genome sequence (18, 54). This bacterium, which develops fruiting bodies under nutrient deprivation (25), is motile on surfaces by adventurous (A) and social (S) gliding motility (22). While A motility enables the movement of a cell that is well separated from others, S motility is functional only when cells are in close proximity. S motility is analogous to bacterial twitching in that both are powered by retraction of the type 4 pilus (Tfp) (24, 30, 38). M. xanthus S motility additionally requires exopolysaccharides (EPS) to function (29). For S motility, EPS on one cell is thought to provide the anchor and trigger for the retraction of Tfp from a neighboring cell, thus explaining the proximity requirement. Chemotaxis regulation in M. xanthus has also been investigated extensively (27, 54). One of the surprises from these investigations was that among the eight chemosensory systems, only Frz signal transduction plays a primary role in chemotaxis regulation and mutants in other systems have no or only specific defects in chemotaxis under certain experimental conditions.
The M. xanthus Dif chemosensory system, while also important for tactic responses to certain species of phosphatidylethanolamine (PE) (8), plays a primary role in the regulation of EPS production (7, 53). difA, difC, and difE mutants produce no detectable levels of EPS, whereas difD and difG mutants overproduce EPS (3, 7, 53). Mutations in difD and difG have additive effects on EPS production but failed to suppress mutations in difE (6). Additional analysis, including the use of yeast two- and three-hybrid (Y2H and Y3H, respectively) systems, led to a working model for the regulation of EPS by the Dif system (6, 52). DifA (MCP-like), DifC (CheW-like), and DifE (CheA-like) were projected to form a ternary signaling complex as do the MCPs, CheW, and CheA in bacterial chemotaxis. DifE is proposed to be an autokinase whose activity is modulated by DifA in combination with DifC (6, 52). The output of the signaling complex is the phosphorylation of an unidentified downstream component by DifE. DifD (CheY-like) and DifG (CheC-like), negative regulators of EPS production, are proposed to be ancillary modulators of the output of DifE by partially diverting phosphate from the DifE kinase and thus away from its downstream target(s) (6). That is, DifD may accept phosphate from autophosphorylated DifE (DifE∼P) and DifG may function as a phosphatase to accelerate the autodephosphorylation of phosphorylated DifD (DifD∼P). Phosphorylation and dephosphorylation events, which are obviously critical to this model, had not been examined prior to this present report.
In this study, we used purified Dif proteins expressed in E. coli to examine the autophosphorylation, phosphotransfer, and dephosphorylation properties of the Dif proteins in vitro. Our results provide strong evidence for most of the proposed biochemical and physical interactions among the Dif proteins. Necessary modifications of the model based on the results here are also discussed.
MATERIALS AND METHODS
Strains and growth conditions.
Escherichia coli strains used in this study were XL1-Blue (Stratagene) and Rosetta (Novagen). E. coli cultures were propagated, and protein expression was induced in lysogeny broth (LB) (1% tryptone, 0.5% yeast extract, 1% NaCl) (4). For solid media, 1.5% agar was added. When necessary, liquid and solid media were supplemented with carbenicillin and chloramphenicol at 50 μg/ml and 70 μg/ml, respectively.
Plasmid construction for protein expression.
Plasmids for protein expression were constructed by PCR amplification (Table 1 ) of the appropriate dif genes and cloned into vectors of the pQE-30 series (Qiagen). The recombinant plasmids code for N-terminal hexahistidine (6×His)-tagged fusions. For the initial construction, the E. coli strain XL1-Blue was used as the host. All constructs were verified by DNA sequencing. For every expression construct, two tandem TAA stop codons were used for translational termination. For DifA, a fragment encoding amino acid residues 185 to 461 was cloned into the BamHI-HindIII sites of pQE-30 to create pWB706. For DifD, its full-length coding region (except the start codon) was cloned into the BamHI (blunted with T4 DNA polymerase) and HindIII sites of pQE-32 to create pWB700. Coding regions for difC (without the start codon) and difG (without the start and second codons) were cloned into the BamHI-HindIII sites of pQE-32 to create pWB701 and pWB702, respectively. For DifE, its coding region (excluding the first 5 codons) was cloned into the SphI-HindIII sites of pQE-31 to construct pWB703.
TABLE 1.
Primers used to construct expression plasmidsa
| Plasmid | Primer sequence (5′ to 3′) | Enzyme |
|---|---|---|
| pWB706 (difA) | ATGGATCCGGTGGCGGTGGCTCGTCGATGGAGAAGAT | BamHI |
| GCTAAGCTTATTATGGGCCCAGGCGGAAGCTGCGCA | HindIII | |
| pWB701 (difC) | ATGGATCCGGCACGTCATCTTCCGGGTG | BamHI |
| GCTAAGCTTATTACTTGGAATGGGTGAACAAGG | HindIII | |
| pWB700 (difD) | GCTAAGCGGGTCCTGGTCGT | None |
| GCTAAGCTTATTACGTCTCGCCCAGGACCTTCT | HindIII | |
| pWB703 (difE) | AGACATGCATGCGTCCCGCTACCTCGGGCTCT | SphI |
| GCTAAGCTTATTACGCGGACAGTAACCTCGGCA | HindIII | |
| pWB702 (difG) | ATGGATCCAGCCCCTGCACAGCGACGCG | BamHI |
| GCTAAGCTTATTACACGCCCAGCCGCGCCAGCA | HindIII |
Restriction sites engineered into primers are underlined with the corresponding enzymes indicated on the right.
Protein purification.
Nickel (Ni) affinity chromatography was used to purify DifA, DifC, DifD, and DifG. E. coli XL1-Blue transformed with an appropriate expression construct was grown at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.6 in 1 liter of LB plus carbenicillin. Protein expression was induced by addition of IPTG (isopropyl-β-d-1-thiogalactopyranoside) to a final concentration of 100 μM, followed by incubation at 19°C for 14 to 18 h. Cells were harvested by centrifugation at 5,000 × g for 15 min. Cell pellets were resuspended in 20 ml of binding buffer (20 mM NaH2PO4, 500 mM NaCl, 25 mM imidazole, 10% glycerol, pH 7.4) and lysed by two passes at 18,000 lb/in2 using a French press (Thermo Scientific). Cellular debris was removed first by low-speed centrifugation at 16,000 × g for 30 min, followed by high-speed centrifugation at 100,000 × g for 1 h. The supernatant was filtered through a 0.45-μm filter prior to being loaded on a 5-ml HisTrap FF (GE Healthcare) column equilibrated with binding buffer. Protein purification was performed by fast protein liquid chromatography (FPLC) using an Äkta Prime chromatography system (GE Healthcare). The column was washed until the initial baseline A280 was restored, typically requiring 10 to 15 column volumes of binding buffer. Proteins were eluted from the column using a 25 to 500 mM linear imidazole gradient equivalent to 10 column volumes. Elution fractions with peak A280 readings were collected and analyzed by SDS-PAGE. Fractions judged to contain the target protein at over 90% purity were buffer exchanged into storage buffer (10 mM Tris-Cl [pH 7.5], 25 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM β-mercaptoethanol [β-ME], and 10% glycerol) using either HiPrep 26/10 or HiTrap desalting columns (GE Healthcare). Protein concentrations were measured by the Bio-Rad protein assay using bovine serum albumin (BSA) as the standard or by spectrophotometry using extinction coefficients calculated from the primary sequence using ProtParam on the ExPASy server (17). Protein stocks at 10 μM in storage buffer were prepared and stored at −80°C for future use.
For the purification of DifE, pWB703 was transformed into the E. coli strain Rosetta. Cell lysates with 6×His-DifE were first fractionated by Ni-affinity chromatography as described above with a few modifications. First, chloramphenicol was used for the maintenance of pRARE in the Rosetta strain. Second, protein expression was induced with IPTG at 10 μM for 5 to 6 h at 22.5°C. Third, 10 mM β-ME was included in the binding and elution buffers. Lastly, three protease inhibitors (phenylmethylsulfonyl fluoride [PMSF] at 1 mM, leupeptin at 10 μM, and pepstatin at 10 μM) were added as supplements to the cell suspension prior to lysis. Peak A280 elution fractions for DifE from Ni-affinity chromatography were analyzed by SDS-PAGE and subjected to buffer exchange as described for the other Dif proteins above. These fractions were analyzed for DifE autophosphorylation initially by using conditions adopted from previous studies as described below. Those fractions with the highest specific DifE kinase activity were further fractionated by size exclusion chromatography (SEC) on a Superdex 200 preparation-grade 10/300 column (GE Healthcare). Most DifE eluted in the void volume and lacked kinase activity. DifE that eluted as a second smaller peak after the void volume was active and was used to prepare protein stocks as described above.
Autophosphorylation, phosphotransfer, and dephosphorylation assays.
Initial autophosphorylation assays for DifE were carried out using buffer conditions (50 mM Tris-Cl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 1 mM β-ME, and 10% glycerol) similar to those described for other CheA kinases (1, 11, 16, 20). For optimization of conditions for DifE autophosphorylation, either Tris-based buffers (10 mM Tris-Cl, 50 mM KCl, 5 mM MgCl2, 1 mM β-ME, and 10% glycerol) at pH 7.5, 8.0, 8.5, and 9.0 or HEPES-based buffers (10 mM HEPES, 50 mM KCl, 5 mM MgCl2, 1 mM β-ME, and 10% glycerol) at pH 6.5, 7.0, 7.5, and 8.0 were used with DifE at a final concentration of 1 μM. To examine the effect of KCl, DifE at a 1 μM concentration was incubated in HEPES buffer at pH 8.0 supplemented with various amounts of KCl (0, 5, 10, 25, 50, 75, 100, 150, and 200 mM). All subsequent autophosphorylation, phosphotransfer, and dephosphorylation assays were carried out at room temperature in 1× kinase reaction buffer (10 mM HEPES [pH 8], 25 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM β-ME, and 10% glycerol).
DifE autophosphorylation assays in the presence and absence of other Dif proteins were performed by incubating DifE alone or with the indicated protein at a final concentration of 1 μM. All autophosphorylation reactions were initiated by the addition of 1 μCi of [γ-32P]ATP (3,000 Ci/mmol, 10 mCi/ml). Reactions were stopped at the time indicated by adding 5 μl of 4× SDS-PAGE loading buffer (250 mM Tris-Cl [pH 6.8], 8% SDS, 0.004% bromphenol blue, 40% glycerol, 20% β-ME) containing 200 mM EDTA. Samples were separated by SDS-PAGE, and the gels were dried and exposed to a phosphor screen for 5 to 15 h. The phosphor screen was imaged using a Typhoon Trio (GE Healthcare) scanner. The intensities of radioactive bands from phosphorimaging were analyzed and quantified using ImageQuant TL (GE Healthcare).
For DifE prephosphorylation, DifE at 5 μM was incubated with 15 μCi of [γ-32P]ATP in a total volume of 75 μl for 45 min. [γ-32P]ATP was removed thereafter by desalting using Micro Bio-Spin 6 columns (Bio-Rad) preequilibrated in 1× kinase reaction buffer following the manufacturer's instructions. To examine phosphotransfer, DifE∼P was mixed with DifD in 1× kinase buffer to give final concentrations of approximately 1 μM for each protein and the reaction was allowed to proceed for the indicated times. The reaction setup for the dephosphorylation assays by DifG was identical to that for the above phosphate transfer assay except that DifG was additionally provided in the DifD aliquot at the indicated concentration. Phosphotransfer and dephosphorylation were examined by phosphorimaging as described above.
RESULTS
Purification of DifA, DifC, DifD, and DifG.
A plasmid for the expression of DifD as an N-terminal hexahistidine (6×His)-tagged fusion in E. coli had been constructed previously for the generation of anti-DifD antibodies (6). Although this construct had been confirmed by DNA sequencing, it had always produced two distinct 6×His-tagged proteins when purified by nickel (Ni) affinity chromatography. One had the predicted size for the fusion at 14.5 kDa, and the other was about 1.5 kDa larger (data not shown). Mass spectrometry and other analysis indicated that the larger protein arose due to translational read-through of the DifD stop codon TGA as a sense codon for Trp. In E. coli, TGA is used as the stop codon for ∼29% of open reading frames (ORFs), but TAA is used twice as frequently (∼64%) (34). A new plasmid (pWB700) was constructed for the expression of 6×His-DifD with two tandem TAA stop codons as described in Materials and Methods. When pWB700 was transformed into E. coli XL1-Blue for DifD expression, only one protein at 14.5 kDa was induced. Purification of 6×His-DifD was performed using Ni-affinity chromatography (21) as described in Materials and Methods and analyzed by SDS-PAGE (data not shown).
Similar strategies were employed for the purification of DifA, DifC, and DifG. That is, proteins were expressed as N-terminal 6×His-tagged fusions with tandem TAA codons for translational termination. For DifA, an MCP homolog with two predicted transmembrane regions (51), only the coding region for the conserved C-terminal signaling domain was used for the construction of the expression plasmid pWB706. For DifC and DifG, full-length coding regions were used to construct pWB701 and pWB702, respectively. Transformants of E. coli-XL1-Blue with these three expression constructs were used for the expression and purification of 6×His-tagged DifA, DifC, and DifG as described in Materials and Methods. Each protein eluted as a single peak in a narrow range of the imidazole gradient as monitored by A280 (data not shown). Peak fractions were collected and analyzed by SDS-PAGE (data not shown). The appropriate fractions were pooled, buffer exchanged, and normalized into working stocks as described in Materials and Methods for later use.
Purification and autophosphorylation of DifE.
DifE is a predicted CheA-like protein with an atypical domain structure. Like other CheAs, DifE contains the conserved domains P1 through P5 (5, 14). Unlike other CheAs, DifE has a P′ domain or region sandwiched between P1 and P2. The P′ region is about 200 amino acids long with no significant homology to proteins with known function (52). pWB703 was constructed to express DifE as an N-terminal 6×His-tagged protein (see Materials and Methods). This plasmid was initially transformed into E. coli XL1-Blue for expression and purification. However, induction and purification of this DifE fusion consistently resulted in proteins that appeared truncated, likely at the C terminus (data not shown). Since M. xanthus has a high GC content (∼68.9%) (18) and difE uses many codons that are rare for E. coli (34), the truncations could be due to premature termination at the rare codons in E. coli. As a large multidomain protein, DifE may also be more susceptible to degradation when overexpressed in a heterologous host (32). The E. coli Rosetta strain, which expresses rare E. coli tRNAs and is deficient in both Lon and OmpT proteases (42), was used as the host for DifE expression. Transformants of Rosetta with pWB703 were found to slightly reduce DifE truncation when the fusion protein was induced for expression (data not shown).
The full-length 6×His-DifE produced in Rosetta was heterogeneous even with low levels of induction at low temperatures. Preliminary experiments indicated that induction with 10 μM IPTG at 22.5°C resulted in mostly soluble DifE. However, when purified by Ni-affinity chromatography, DifE eluted as a relatively broad peak over a wide range of the imidazole gradient (data not shown). When the peak fractions were analyzed by SDS-PAGE, all fractions contained significant amounts of full-length DifE protein (Fig. 1 A). These observations indicated sample heterogeneity. To examine which fraction might contain the DifE species with the highest specific activity, fractions were analyzed in autophosphorylation assays with [γ-32P]ATP using conditions similar to those in previous studies of CheA kinases (1, 11, 16, 20). As shown in Fig. 1B, even though all fractions yielded 32P-labeled full-length DifE, fractions 5 to 7 had high levels of DifE with autophosphorylation activity (Fig. 1A). These early fractions (5 to 7) were therefore collected for further purification. Size exclusion chromatography (SEC) was used to further purify DifE in the early fractions from the above Ni-affinity chromatography. There was a large A280 peak corresponding to the void volume of the column followed by a smaller second peak in subsequent elution fractions (data not shown). Although the fractions from both peaks contained DifE that was not as pure as the other Dif proteins, there were marked reductions in the smaller polypeptides in comparison with the starting material (data not shown). Fractions from both peaks were examined for autophosphorylation using [γ-32P]ATP (data not shown). 32P-labeled DifE was observed only in fractions from the smaller peak, and these fractions with active DifE were pooled and used in subsequent experiments.
FIG. 1.
Fractionation of DifE by Ni-affinity chromatography. Equal volumes of selected fractions from Ni-affinity chromatography were subjected to SDS-PAGE and Coomassie blue staining (A) or autophosphorylation assays (B). The fraction numbers indicated at the top of the figure apply to both panels (fractions 4 to 10). The positions of DifE and DifE∼P are indicated on the left. For the phosphorylation studies in panel B, the radioactivity associated with 32P-labeled DifE was analyzed by phosphorimaging as described in Materials and Methods. Indicated at the bottom of the figure is the relative intensity (RI) of each band, which was normalized to the one with the highest radioactivity.
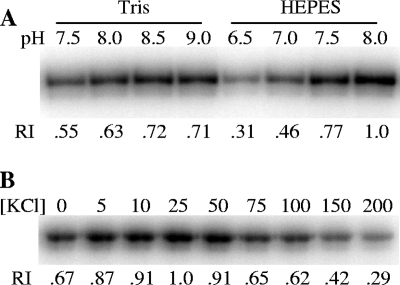
Since the initial conditions for DifE autophosphorylation (Fig. 1B and data not shown) were adopted from previous studies, we attempted further optimization of experimental conditions for the DifE autokinase. We first tested Tris- and HEPES-based buffer systems with pH ranges optimal for each: pH 7.5 to 9 for Tris and pH 6.5 to 8 for HEPES. As shown in Fig. 2 A, more 32P-labeled DifE∼P from [γ-32P]ATP was generated in both buffer systems at higher pH, with HEPES buffer at pH 8 consistently yielding the largest amount of DifE-phosphate (DifE∼P) (Fig. 2A). Since potassium ions had been shown to influence the activity of CheA kinase (43), we next examined the effect of KCl on DifE activity by using HEPES buffer at pH 8. As shown in Fig. 2B, the level of DifE∼P, as measured by phosphorimaging, increased with increasing KCl concentrations of 5 mM up to 25 mM and decreased thereafter. Therefore, HEPES buffer at pH 8 supplemented with 25 mM KCl was used for all subsequent phosphorylation studies. In addition, we determined the time required for DifE to reach a maximal level of phosphorylation under our assay conditions. The results indicated that the level of DifE∼P plateaued between 30 and 60 min of incubation (data not shown). Whenever DifE needed prephosphorylation, it was autophosphorylated for 30 to 45 min in later experiments.
FIG. 2.
Optimization of conditions for DifE autophosphorylation. (A) Optimization of buffering conditions. Either Tris- or HEPES-based buffer at the indicated pH (top of the panel) was used for DifE autophosphorylation. (B) Effect of KCl on DifE autophosphorylation activity. DifE phosphorylation was conducted in HEPES buffer at pH 8 supplemented with KCl at indicated mM concentrations (top of the panel). DifE at 1 μM was used for all phosphorylation reactions. Indicated at the bottom of each panel (A and B) is the relative intensity (RI) of each band, as determined in Fig. 1.
DifA and DifC have inhibitory effects on DifE autophosphorylation in vitro.
MCPs, CheW, and CheA are known to form ternary signaling complexes, and the presence of both MCP and CheW has been demonstrated to affect CheA kinase activity in vitro (10, 13, 15, 23, 44). In the Dif chemosensory system, complex formation by DifA (MCP-like), DifC (CheW-like), and DifE (CheA-like) had been demonstrated by Y2H and Y3H experiments (52). We analyzed whether DifA and DifC could form complexes with DifE to influence its kinase activity by using purified proteins. Inclusion of DifA or DifC separately in phosphorylation reactions with DifE resulted in amounts of DifE∼P similar to those observed for DifE alone (Fig. 3, compare lane 1 with lanes 2 and 3). In the presence of both DifA and DifC, on the other hand, a consistent reduction in the amount of DifE∼P was detected by phosphorimaging (Fig. 3, compare lane 1 with lane 4). These results are consistent with the formation of a ternary signaling complex because an effect on DifE activity was observed only in the presence of both DifA and DifC but not individually.
FIG. 3.
Effects of other Dif proteins on DifE autophosphorylation. Dif proteins at a final concentration of 1 μM each were incubated in 1× kinase reaction buffer with [γ-32P]ATP for 15 min and analyzed as described in Materials and Methods. The presence and absence of a protein in a reaction are indicated by plus and minus signs, respectively. Indicated at the bottom of the figure is the relative intensity (RI) of each 32P-labeled band normalized to that of DifE∼P alone (lane 1). ND, not determined.
DifD accepts phosphate from DifE∼P.
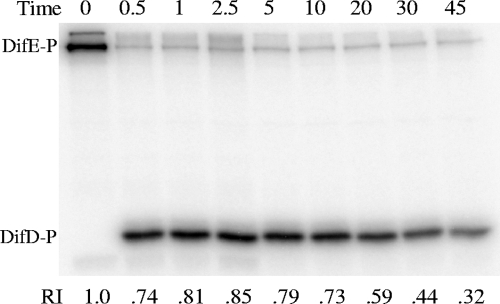
DifD is homologous to the single domain response regulator CheY (51). Typically, CheYs interact with and accept phosphate from autophosphorylated CheA kinases. In the Dif system, DifD negatively regulates EPS production whereas DifE promotes EPS production. While DifD was shown to interact with DifE by Y2H analysis (52), genetic analyses demonstrated that DifD does not function downstream of DifE (6). These results indicated that DifD could negatively regulate DifE, either as an inhibitor of DifE kinase activity or as a phosphate sink of DifE∼P. To examine these possibilities, DifE was incubated with DifD in the presence of [γ-32P]ATP. When analyzed by SDS-PAGE and phosphorimaging, DifD phosphate (DifD∼P) was readily detected but not DifE∼P (Fig. 3, compare lane 1 with lane 5). When DifD was incubated with [γ-32P]ATP by itself, no DifD∼P was detected (data not shown). A plausible explanation was that DifE autophosphorylated but transferred most of the phosphate to DifD during the 15 min of incubation. However, it was theoretically possible that DifD inhibited DifE autophosphorylation while it autophosphorylated using ATP in the presence of DifE. Purified DifE was used to examine if phosphotransfer occurred between DifE∼P and DifD in the absence of ATP. DifE was prephosphorylated, and the free [γ-32P]ATP was removed using a desalting column (see Materials and Methods). DifE∼P was incubated with DifD, and the progress of the reaction was monitored by SDS-PAGE and phosphorimaging. As shown in Fig. 4, after 30 s of incubation, most of the 32P labeling disappeared from DifE and resided on DifD instead (lane 2). After 2.5 min, essentially all the phosphates were transferred from DifE to DifD (lane 4), and there was no further increase in the level of DifD∼P thereafter. Because the only available radioactive phosphate (32P) was from DifE∼P at the onset of the experiment, these observations confirm that DifD is able to accept phosphate from DifE∼P. These results suggest that DifD acts as a phosphate sink to negatively regulate the activity of the DifE kinase rather than functioning as a direct inhibitor of DifE autophosphorylation. It is noted that a small amount of 32P remained associated with DifE throughout the duration of the experiment as observed for other CheA-like kinases (41, 47).
FIG. 4.
Phosphate transfer from DifE∼P to DifD. Purified DifE that was prephosphorylated with [γ-32P]ATP was incubated with DifD at 1 μM concentrations for both Dif proteins. Reactions were stopped at the indicated times in minutes (top) and analyzed as described for Fig. 1. Indicated at the bottom is the relative intensity (RI) of 32P-labeled DifD normalized to that of DifE∼P at time zero.
The half-life of DifD∼P can be estimated from the data in Fig. 4 as well. Since there was no further decrease in the amount of 32P-labeled DifE and no further increase in that of 32P-labeled DifD after 2.5 min, the data points from 5 min on were used to calculate the dephosphorylation half-life of DifD∼P (data not shown). This resulted in an apparent half-life of approximately 30 min under our experimental conditions. In Sinorhizobium meliloti and Rhodobacter sphaeroides, certain CheYs have also been proposed to function as phosphate sinks, similarly to DifD; however, these CheY∼Ps were determined to have half-lives of only 10 to 40 s, or 60 times shorter than that of DifD∼P (35, 39). The half-life of 30 min for DifD∼P is therefore strikingly long in comparison with those of other CheY-like phosphate sinks, suggesting that additional factors might speed up DifD∼P dephosphorylation in vivo.
DifG accelerates dephosphorylation of DifD∼P.
DifG, which is homologous to the CheC protein phosphatase (33, 46), has been proposed to accelerate the dephosphorylation of DifD∼P (6, 52). Y2H analysis had demonstrated interactions between DifG and DifD previously (52). When DifG was included in a phosphorylation reaction with DifE and DifD, there was a marked reduction in the amount of DifD∼P compared to that in the reaction without DifG (Fig. 3, lanes 5 and 7). While consistent with DifG being a phosphatase of DifD∼P, the results could also be explained if DifG inhibits DifE autophosphorylation in the presence of DifD. Prephosphorylated DifE was therefore purified and incubated with DifD in the presence of DifG at various concentrations (Fig. 5). In the absence of DifG, the level of DifD∼P appeared similar to that shown in Fig. 4. However, the presence of DifG in the reaction significantly diminished the amount of DifD∼P. When DifG was present at 1 μM, the same concentration as that of the other two proteins, no DifD∼P was detected at any of the time points examined. Even at 0.01 μM, a concentration 100-fold less than that of DifD, very little DifD∼P was left after 20 min of incubation. These results, especially those with 0.01 μM DifG, indicated that DifG functions as a phosphatase to accelerate the dephosphorylation of DifD∼P.
FIG. 5.
Dephosphorylation of DifD∼P by DifG phosphatase. Reaction conditions were identical to those in Fig. 4 except that DifG was included at the indicated concentrations. Reactions were stopped at the indicated times (in minutes) and analyzed as described for Fig. 1. The equivalent amount of purified DifE∼P in all the reactions was loaded in lane 1 for comparison.
While the results are consistent with DifG acting as a phosphatase of DifD∼P, they do not support the notion of DifG interacting with the DifA-DifC-DifE ternary complex to inhibit signaling activity (6, 7). As shown in Fig. 3, DifG had no discernible effect on DifE autophosphorylation in the presence of DifA and DifC (compare lanes 4 and 8). Neither did DifG by itself have an effect (compare lanes 1 and 6). These results indicated that DifG had no detectable effect on DifE activity in either the absence or the presence of DifA and DifC under our assay conditions. These results with DifG are inconsistent with previous models, and the role of DifG as a direct modulator of the activity of the DifA-DifC-DifE complex will therefore need to be revisited and redefined.
DISCUSSION
The M. xanthus Dif proteins are essential for the regulation of EPS production (7, 53) as well as phosphatidylethanolamine (PE) taxis (9). Previous genetic and Y2H analysis had suggested models for signaling complex formation, protein phosphorylation, and dephosphorylation in the Dif system (6, 9, 52). In this study, we expressed and purified the Dif chemotaxis proteins and studied phosphorylation and dephosphorylation events among them in vitro. Specifically, we demonstrated the autokinase activity of DifE (Fig. 1 and 2) and phosphate transfer from DifE∼P to DifD (Fig. 4). In addition, we provided evidence that DifG functions as a phosphatase to accelerate the dephosphorylation of DifD∼P (Fig. 5). Our results are also consistent with the formation of a DifA-DifC-DifE signaling complex in which the kinase activity of DifE may be modulated by DifA through the mediation of DifC.
Purification of active DifE was unusually challenging and required careful evaluation of protein fractions from tandem chromatography. Although significant amounts of soluble 6×His-tagged DifE could be purified using Ni-affinity chromatography, the protein obtained as such was largely inactive. SEC proved valuable in the separation of active and inactive forms of DifE (see Results; also data not shown). The inactive DifE existed as relatively large aggregates and eluted in the void volume. Even the active DifE in our purification likely existed as aggregates/oligomers because its elution peak corresponds to a molecular mass of about 700 kDa and monomeric DifE is ∼88 kDa (data not shown). There has been no report of difficulties in the purification of CheAs, which usually exist as dimers (2, 5, 36). DifE is highly homologous to other CheAs throughout its primary sequence, except for the P′ region between the conserved P1 and P2 domains (52). It is unclear if P′ contributes to the unexpected aggregation and/or oligomerization of DifE since the deletion of P′ from DifE rendered DifE insoluble when it was expressed in E. coli (data not shown). While the dif genes, including difE, are found in two other sequenced myxobacterial chromosomes (37, 48), those two DifE orthologs have no P′. It is possible that DifE may require interactions with other M. xanthus proteins, possibly through P′, to achieve its native and/or active conformation.
Although there are limitations on how the kinetics of DifE phosphorylation in our study may reflect that of DifE in vivo, our results certified DifE as a bona fide autokinase. Moreover, our observations also suggest the formation of a DifA-DifC-DifE signaling complex. The results in Fig. 3 show that while neither DifA nor DifC individually influenced the kinase activity of DifE, their combination clearly reduced the activity of the DifE autokinase. Previous Y2H and Y3H analysis had shown that DifC directly interacts with both DifA and DifE to bridge the formation of a very stable ternary complex (52). Since there was no direct interaction detected between DifA and DifE (52), these observations together strongly support the formation of a DifA-DifC-DifE signaling complex in which DifA receives signals to modulate DifE activity through DifC.
The inhibition of DifE activity by DifA and DifC was to a certain extent surprising. In previous studies, the combination of MCPs and CheWs was usually found to stimulate the activity of CheA kinases in vitro (10, 13, 15, 23, 44). In the only other known case in M. xanthus, the in vitro autophosphorylation of FrzE required both FrzCD and FrzA; without FrzCD and FrzA, very little if any kinase activity was observed (23). In E. coli, CheA activity is stimulated by the presence of membranes containing MCPs and CheW (10). In addition, the cytoplasmic domains of MCP, either by themselves or fused to a leucine zipper domain, stimulated CheA activity in the presence of CheW (1, 13, 44). In B. subtilis, CheA requires CheW and MCPs to reach maximum kinase activity in vitro (15). M. xanthus DifA is likely unmethylated (49), and its activity should therefore be modulated solely by conformational changes rather than covalent modification. Since the DifA system responds to both positive (in EPS regulation) and negative (in PE taxis) stimulation (9, 50), DifA should exist in at least two signaling conformations. We are unsure whether our experiment indicates that DifA, in its default state, is normally inhibitory of DifE or that the TM truncation has altered DifA from its default stimulatory signaling state to an inhibitory conformation.
The results in this study support the notion that DifD and DifG function synergistically to divert phosphate from the branch of the Dif pathway that regulates EPS production (6). Both DifD and DifG are negative regulators of EPS production in M. xanthus. It had been proposed previously that DifD might function as a phosphate sink of DifE∼P and DifG could be a phosphatase of DifD∼P, but experimental evidence for this model was lacking (6, 52). We have shown in this study that DifD can indeed accept and sequester phosphate from DifE as proposed. However, DifD is apparently a poor phosphate sink on its own because the 30-min half-life of DifD∼P is about 60 times longer than that observed for other known or proposed phosphate sinks (35, 39). The long half-life of DifD∼P is somewhat puzzling by all appearances, because DifD was expected to display faster autodephosphorylation based on the conservation of certain key residues (47). Nevertheless, DifD can function as an effective phosphate sink in the Dif system because we have demonstrated that DifG is a phosphatase that efficiently accelerates the dephosphorylation of DifD∼P (Fig. 5). Kinetically, the activity of DifG (Fig. 5) appears most comparable with that of FliY, which is the stronger of the two phosphatases (FliY and CheC) in B. subtilis (45, 46). The combination of DifD and DifG can therefore effectively divert phosphate from the DifE kinase to negatively regulate EPS production in M. xanthus.
DifD∼P has a significantly longer half-life than do other CheYs that function as phosphate sinks (35, 39). If a CheY with a high autodephosphorylation rate is able to function as an effective sink, why should DifD∼P decay slower? One explanation could be related to the additional role of DifD in PE taxis in M. xanthus, where it is proposed to function downstream of DifE (9). The response of gliding M. xanthus to PE consists of two phases: excitation and adaptation. Excitation refers to the initial suppression of reversal frequency by PE, and adaptation is the eventual return of the reversal frequency to the prestimulation level. Both phases of the PE response in M. xanthus are quite long in comparison to the response times observed in chemotaxis of flagellated bacteria such as E. coli and B. subtilis. For example, the time required for adaptation in E. coli and B. subtilis chemotaxis is usually in the range of seconds instead of minutes (12, 19). In M. xanthus PE taxis, adaptation takes about 120 min (9, 26). This slow adaptation is presumed necessary for M. xanthus as its gliding is about 3 orders of magnitude slower than the swimming of E. coli and B. subtilis (40, 49, 54). Since DifA is unlikely to be methylated (49), the dephosphorylation of DifD∼P may be critical for both desensitization and adaptation. The slow dephosphorylation of DifD∼P may therefore be a prerequisite for the slow adaptation of M. xanthus to chemotactic stimuli mediated by the Dif pathway. In this scenario, DifG would be expected to affect PE taxis, which had been observed previously (9).
Previous genetic analysis concluded that DifG likely regulates EPS production in addition to or independently of its role as a phosphatase of DifD∼P (6). This conclusion was primarily based on the additive effect of difD and difG mutations on EPS production (6). Our previous model had attributed the additional function of DifG in EPS regulation to a potential interaction with the DifA-DifC-DifE signaling complex based on observed interactions of CheC with CheA and McpB in B. subtilis (28). However, we failed to detect interaction of DifG with DifA, DifC, or DifE in our Y2H studies (52). The results here also failed to demonstrate any influence on DifE kinase activity by DifG alone or in combination with DifA and DifC. Most CheCs require CheDs as interacting partners, and these two proteins are usually encoded by two adjacent genes in many organisms (33). It has been shown that B. subtilis CheC reaches full phosphatase activity only in the presence of its cognate CheD (46). In contrast, there are no CheD-like proteins encoded on the M. xanthus chromosome (18). Sequence analysis indeed suggested that DifG belongs to a different class of CheCs that function as dedicated phosphatases (33). As an alternative explanation for the additive effects of difD and difG mutations, we suggest that DifG may interact with one or more components downstream of DifE in EPS regulation. This modification of the model requires further analysis and verification.
Acknowledgments
We are grateful to Carla Finkelstein for her expertise in signal transduction and protein phosphorylation. We also thank Benjamin Orsburn for mass spectrometry analysis.
This work was supported by grant GM071601 and ARRA supplements from the National Institutes of Health to Z.Y.
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9-22. [DOI] [PubMed] [Google Scholar]
- 3.Bellenger, K., X. Ma, W. Shi, and Z. Yang. 2002. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J. Bacteriol. 184:5654-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 6.Black, W. P., Q. Xu, and Z. Yang. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447-456. [DOI] [PubMed] [Google Scholar]
- 7.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonner, P. J., and L. J. Shimkets. 2006. Phospholipid directed motility of surface-motile bacteria. Mol. Microbiol. 61:1101-1109. [DOI] [PubMed] [Google Scholar]
- 9.Bonner, P. J., Q. Xu, W. P. Black, Z. Li, Z. Yang, and L. J. Shimkets. 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol. Microbiol. 57:1499-1508. [DOI] [PubMed] [Google Scholar]
- 10.Borkovich, K. A., and M. I. Simon. 1990. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell 63:1339-1348. [DOI] [PubMed] [Google Scholar]
- 11.Bourret, R. B., J. F. Hess, and M. I. Simon. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. U. S. A. 87:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, D. A., and H. C. Berg. 1974. Temporal stimulation of chemotaxis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 71:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochran, A. G., and P. S. Kim. 1996. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science 271:1113-1116. [DOI] [PubMed] [Google Scholar]
- 14.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrity, L. F., and G. W. Ordal. 1997. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology 143:2945-2951. [DOI] [PubMed] [Google Scholar]
- 16.Garzon, A., and J. S. Parkinson. 1996. Chemotactic signaling by the P1 phosphorylation domain liberated from the CheA histidine kinase of Escherichia coli. J. Bacteriol. 178:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 18.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman, D. J., and G. W. Ordal. 1981. Sensory adaptation and deadaptation by Bacillus subtilis. J. Bacteriol. 147:267-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 21.Hochuli, E., H. Dobeli, and A. Schacher. 1987. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J. Chromatogr. 411:177-184. [DOI] [PubMed] [Google Scholar]
- 22.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 23.Inclan, Y. F., H. C. Vlamakis, and D. R. Zusman. 2007. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol. Microbiol. 65:90-102. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 26.Kearns, D. B., and L. J. Shimkets. 1998. Chemotaxis in a gliding bacterium. Proc. Natl. Acad. Sci. U. S. A. 95:11957-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirby, J. R., J. E. Berleman, S. Muller, D. Li, J. C. Scott, and J. M. Wilson. 2008. Chemosensory signal transduction systems in Myxococcus xanthus, p. 135-147. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 28.Kirby, J. R., C. J. Kristich, M. M. Saulmon, M. A. Zimmer, L. F. Garrity, I. B. Zhulin, and G. W. Ordal. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42:573-585. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 31.Miller, L. D., M. H. Russell, and G. Alexandre. 2009. Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 66:53-75. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, T. B., and J. S. Parkinson. 1994. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91:5485-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muff, T. J., and G. W. Ordal. 2008. The diverse CheC-type phosphatases: chemotaxis and beyond. Mol. Microbiol. 70:1054-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter, S. L., and J. P. Armitage. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J. Mol. Biol. 324:35-45. [DOI] [PubMed] [Google Scholar]
- 36.Rao, C. V., and G. W. Ordal. 2009. The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib. Microbiol. 16:33-64. [DOI] [PubMed] [Google Scholar]
- 37.Ronning, C. M., and W. C. Nierman. 2008. The genomes of Myxococcus xanthus and Stigmatella aurantiaca, p. 285-298. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 38.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327-2335. [DOI] [PubMed] [Google Scholar]
- 40.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, R. C. 1997. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 36:2030-2040. [DOI] [PubMed] [Google Scholar]
- 42.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 43.Surette, M. G., M. Levit, Y. Liu, G. Lukat, E. G. Ninfa, A. Ninfa, and J. B. Stock. 1996. Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J. Biol. Chem. 271:939-945. [DOI] [PubMed] [Google Scholar]
- 44.Surette, M. G., and J. B. Stock. 1996. Role of alpha-helical coiled-coil interactions in receptor dimerization, signaling, and adaptation during bacterial chemotaxis. J. Biol. Chem. 271:17966-17973. [DOI] [PubMed] [Google Scholar]
- 45.Szurmant, H., M. W. Bunn, V. J. Cannistraro, and G. W. Ordal. 2003. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J. Biol. Chem. 278:48611-48616. [DOI] [PubMed] [Google Scholar]
- 46.Szurmant, H., T. J. Muff, and G. W. Ordal. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem. 279:21787-21792. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, S. A., J. A. Brewster, and R. B. Bourret. 2008. Two variable active site residues modulate response regulator phosphoryl group stability. Mol. Microbiol. 69:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, S. H., R. D. Wagner, A. K. Arakaki, J. Skolnick, J. R. Kirby, L. J. Shimkets, R. A. Sanford, and F. E. Loffler. 2008. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS One 3:e2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, Q., W. P. Black, C. L. Cadieux, and Z. Yang. 2008. Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol. Microbiol. 69:714-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, Q., W. P. Black, S. M. Ward, and Z. Yang. 2005. Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera. J. Bacteriol. 187:6410-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123-1130. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Z., and Z. Li. 2005. Demonstration of interactions among Myxococcus xanthus Dif chemotaxis-like proteins by the yeast two-hybrid system. Arch. Microbiol. 183:243-252. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862-872. [DOI] [PubMed] [Google Scholar]