Abstract
ppGpp regulates gene expression in a variety of bacteria and in plants. We proposed previously that ppGpp or its precursor, pppGpp [referred to collectively as (p)ppGpp], or both might regulate the activity of the enzyme polynucleotide phosphorylase in Streptomyces species. We have examined the effects of (p)ppGpp on the polymerization and phosphorolysis activities of PNPase from Streptomyces coelicolor, Streptomyces antibioticus, and Escherichia coli. We have shown that (p)ppGpp inhibits the activities of both Streptomyces PNPases but not the E. coli enzyme. The inhibition kinetics for polymerization using the Streptomyces enzymes are of the mixed noncompetitive type, suggesting that (p)ppGpp binds to a region other than the active site of the enzyme. ppGpp also inhibited the phosphorolysis of a model RNA substrate derived from the rpsO-pnp operon of S. coelicolor. We have shown further that the chemical stability of mRNA increases during the stationary phase in S. coelicolor and that induction of a plasmid-borne copy of relA in a relA-null mutant increases the chemical stability of bulk mRNA as well. We speculate that the observed inhibition in vitro may reflect a role of ppGpp in the regulation of antibiotic production in vivo.
The alarmone, ppGpp (GDP; 3′-diphosphate) regulates gene expression in bacteria. In the classical stringent response to amino acid starvation in Escherichia coli, ppGpp is synthesized on idling ribosomes by the product of the relA gene, a (p)ppGpp synthetase (6). ppGpp then inhibits the synthesis of ribosomal and transfer RNAs, decreasing the levels of ribosome and tRNA synthesis when rates of protein synthesis decrease (reviewed in references 3, 6, 31, and 35). ppGpp can also activate transcription in E. coli. E. coli mutants lacking ppGpp are auxotrophic for a number of amino acids, and ppGpp has been shown to activate transcription of the operons for those amino acids (3, 6, 31, 35). Recent studies indicate that ppGpp interacts with RNA polymerase in concert with the effector protein DksA, and this interaction results in promoter-specific inhibition of transcription, in the case of stable RNA promoters, or the activation of transcription in the case of the promoters for amino acid biosynthesis (11, 12, 18). It has also been argued that ppGpp regulates growth rate in E. coli (2, 19), although there is other evidence suggesting that it is not essential to this process (15). Microarray studies indicate that the expression of several hundred genes is affected by changes in ppGpp levels in E. coli (13). In other systems, ppGpp is involved in sporulation, stress survival, and virulence (1, 14, 17).
The stringent response also occurs in the soil-dwelling actinomycete, Streptomyces (36). Streptomyces species contain homologs of relA (8, 22, 28), and ppGpp levels increase in response to amino acid starvation in Streptomyces, as in E. coli (36). Of particular interest is the observation that ppGpp serves as both a negative and a positive regulator of antibiotic production in Streptomyces species. Thus, relA-null mutants of Streptomyces clavuligerus overproduce clavulanic acid and cephalomycin C, indicating negative regulation by ppGpp (16). In contrast, ppGpp has been shown to be required for antibiotic production in Streptomyces griseus (streptomycin [30]), Streptomyces coelicolor (actinorhodin and calcium-dependent antibiotic [7, 21]), and Streptomyces antibioticus (actinomycin [22]). In the case of S. coelicolor, there is strong evidence indicating that ppGpp activates the transcription of genes that regulate antibiotic production. Thus, a microarray analysis demonstrated that genes for production of the calcium-dependent antibiotic (CDA) and actinorhodin were induced after ppGpp synthesis (20). Induction of the CDA and actinorhodin clusters was accompanied by an increase in transcription of the pathway-specific regulators for those clusters, cdaR and actII-ORF4, respectively (20). It is noteworthy that induction of the genes for undecylprodigiosin, another of the antibiotics produced by S. coelicolor, was not observed in that study.
An early attempt to identify the relA gene in S. antibioticus identified another gene instead, namely, the gene for the 3′-5′-exoribonuclease, polynucleotide phosphorylase (PNPase) (23). This situation occurred because S. antibioticus PNPase also possesses pppGpp synthetase activity (24). pppGpp (GTP; 3′-diphosphate) is the precursor of ppGpp, and it was initially thought that PNPase might represent an alternative enzyme for ppGpp synthesis in Streptomyces. More recent studies indicate that PNPase does not modulate ppGpp levels in vivo, and we speculated that the pppGpp synthetase activity might reflect possible regulation of PNPase activity by (p)ppGpp (5). PNPase plays an important role in RNA degradation in E. coli and Streptomyces and serves both as an exonuclease and as an RNA 3′-polyribonucleotide polymerase [the functional analog of poly(A) polymerase] in both species. Mohanty and Kushner have shown that PNPase is responsible for the G, C, and U residues that occur at low frequency in the poly(A) tails of E. coli RNAs (29). In Streptomyces, the data suggest that PNPase is the sole RNA 3′-polyribonucleotide polymerase present and that it is responsible for the synthesis of the heteropolymeric tails associated with RNAs in those organisms (4, 34). Thus, both the degradative activity (phosphorolysis) and the synthetic activity (polymerization) of PNPase may be important in Streptomyces. We thus endeavored to examine the possible role of (p)ppGpp in the regulation of PNPase activity and show here that ppGpp effectively inhibits both the polymerization and the phosphorolysis activities of the enzyme. We also provide evidence that ppGpp increases the chemical half-lives of bulk mRNA in S. coelicolor.
MATERIALS AND METHODS
Preparation of (p)ppGpp, substrates, and PNPases.
pppGpp and ppGpp were synthesized in vitro from GTP and ATP by using crude extracts of E. coli strain CF3120, which expresses an inducible form of relA, generously supplied by Michael Cashel. (p)ppGpp was purified from reaction mixtures by ion-exchange chromatography on Q Sepharose Fast Flow in the presence of 7 M urea. For some experiments, ppGpp was obtained from Trilink Biotechnologies. [3H]poly(A) was synthesized from [3H]ADP as described previously (25). Untagged polynucleotide phosphorylases from S. antibioticus, S. coelicolor, and E. coli were expressed and purified as described previously (25). The 5601 transcript, derived from the rpsO-pnp operon of S. coelicolor, was synthesized by using [32P]CTP as described previously (9).
Conditions for PNPase reactions.
Polymerization reactions were performed as described previously (25) in 15-μl reaction mixtures, incubated for 15 min at 37°C. Mixtures contained 5 mM ADP, 0.45 μCi of [3H]ADP (Perkin-Elmer, 33.9 Ci/mmol), and ca. 0.28 μM PNPase. Kinetic assays for polymerization were performed using a stock solution of 125 mM [3H]ADP with a specific activity of 6 μCi/μmol, which was diluted to prepare substrate to obtain initial velocities. Preliminary assays (not shown) showed that product formation proceeded linearly at the protein concentrations and for the incubation times used in the experiments reported here. Phosphorolysis reactions were performed as described previously (25) in 15-μl reaction mixtures, incubated for 10 min at 37°C. Mixtures contained ca. 30,000 cpm of [3H]poly(A), prepared as described previously (25); 5 mM potassium phosphate; and ca. 14 nM PNPase. Phosphorolysis of the 5601 transcript was performed as described previously (9) in 15-μl reaction mixtures containing 0.43 μM 5601 RNA (ca. 30,000 cpm), 5 mM potassium phosphate and 28 nM PNPase, which were incubated for 10 min at 37°C. Some reactions contained 1 mM ppGpp. Reactions were stopped by adding Sequencing Stop Solution (Promega) containing formamide, and samples were heated for 5 min at 75°C. Phosphorolysis products were separated on 7 M urea-5% polyacrylamide gels and visualized by autoradiography.
Measurement of bulk mRNA half-lives.
S. coelicolor M600 (a relA+ parental strain), M653 (a relA strain containing the relA gene under the control of the inducible tipA promoter) (21), and M570 (a relA strain) (8) were generously provided by Andrew Hesketh of the John Innes Centre, Norwich, England. M570 containing pIJ8600 was constructed in our laboratory and was designated JSE571. pIJ8600 is the cloning vector used for overexpression of relA in M653 (21).
All Streptomyces strains were grown in minimal medium (26) containing carboxymethyl cellulose rather than polyethylene glycol as a dispersant. To measure mRNA half-lives liquid cultures were grown at 30°C for the appropriate lengths of time when 15 ml was removed to a 50-ml screw cap tube. [3H]uridine (37.5 μCi [DuPont NEN], 31.9 Ci/mmol) was added, and the samples were incubated for 10 min at 30°C. Actinomycin D (Sigma) was then added to these samples to a final concentration of 75 μg/ml. Incubation was continued with the removal of 1-ml samples to 0.2 ml of 50% trichloroacetic acid (TCA) at 0, 5, 10, 15, 20 and 60 min, after actinomycin addition. TCA samples were collected as described previously, and the cpm in mRNA were calculated by subtracting the cpm values obtained at 60 min, representing stable RNAs, from the values obtained at the earlier time points (5). Half-lives were determined by regression analysis of the decay data. S. coelicolor M600 and M570 were grown to A450 values of 0.4 to 0.5 (ca. 7 h postinoculation) for the measurement of half-lives during exponential phase and to A450 values of ≥1.2 (26 h postinoculation) to measure half-lives in stationary phase. Strains M653 and JSE571 were grown to A450 values of ≥1.2 (26 h postinoculation) when half-lives were measured as described above. Thiostrepton was then added to separate 15-ml samples of each culture to a final concentration of 25 μg/ml, and the samples were incubated for 60 min at 30°C to induce ppGpp production in M653 (21). The half-lives were then measured as described above.
The half-lives of bulk mRNA were also measured in two E. coli strains, CF1648 (relA+) and CF1652 (ΔrelA::kan), generously donated by Michael Cashel (38). The strains are isogenic apart from the relA mutation. The strains were grown in 10 to 12 ml of M9 medium (33) containing 0.2% each of glucose and Casamino Acids. Exponential-phase measurements were performed at A600 values of 0.2 to 0.3, and stationary-phase measurements were performed at A600 values of ∼1.0. To measure half-lives, 25 μCi of [3H]uridine was added to 10-ml cultures, and the samples were incubated for 2 min at 37°C. Rifampin and nalidixic acid (Sigma) were then added to these cultures to final concentrations of 500 and 20 μg/ml, respectively, and the cultures were incubated for two additional minutes. Incubation was continued with the removal of duplicate 0.4-ml samples to 0.1 ml of 50% TCA at 0, 2, 4, 6, 8, and 30 min after rifampin addition. The half-lives were measured as described for the Streptomyces assays with the 30-min cpm values representing stable RNAs.
RESULTS AND DISCUSSION
Examination of the effects of (p)ppGpp on the polymerization reactions catalyzed by PNPase.
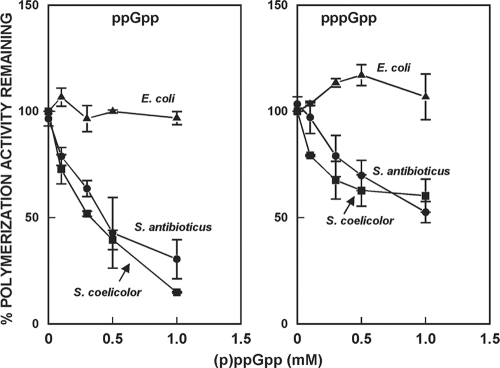
The exact intracellular concentrations of ppGpp during normal growth have not been determined in Streptomyces; however, Riesenberg et al. attempted to correlate intracellular concentrations of ppGpp with ppGpp levels presented as pmol/mg of dry weight. These authors estimated that an intracellular ppGpp concentration of 0.5 mM corresponded to ca. 1,500 pmol/mg of dry weight (32). ppGpp levels of 200 to 500 pmol/mg of mycelial dry weight have been measured in S. antibioticus, S. coelicolor, and other streptomycetes (7, 20, 22, 30), corresponding to intracellular concentrations of ca. 0.07 to 0.17 mM, based on the estimates of Riesenberg et al. Local concentrations within the mycelium might well be higher. In E. coli, it is known that millimolar concentrations of ppGpp accumulate during the stringent response and that (p)ppGpp concentrations of 0.1 to 0.5 mM inhibit exopolyphosphatase from E. coli (27). Thus, we utilized concentrations up to 1 mM in the studies reported here. The effects of (p)ppGpp on polymerization of [3H]ADP are shown in Fig. 1. It can be seen that ppGpp effectively inhibited the polymerization reactions catalyzed by the streptomycete enzymes but did not inhibit the E. coli enzyme. ppGpp at 1 mM inhibited polymerization by the Streptomyces enzymes ca. 90%. pppGpp also inhibited the streptomycete PNPases but to a lesser extent than did ppGpp. We consistently observed a slight stimulation of the E. coli PNPase by pppGpp as shown in Fig. 1. The mechanism of this stimulation is not currently known. No inhibition of polymerization by either compound was observed at the concentrations tested.
FIG. 1.
Inhibition of the polymerization activity of PNPase by (p)ppGpp. The data in the figure show the percentage of the polymerization activity observed in the absence of (p)ppGpp that remained when the reactions were performed in the presence of the indicated concentrations of (p)ppGpp and are the averages of duplicates from two separate experiments ± the standard error of the mean (SEM).
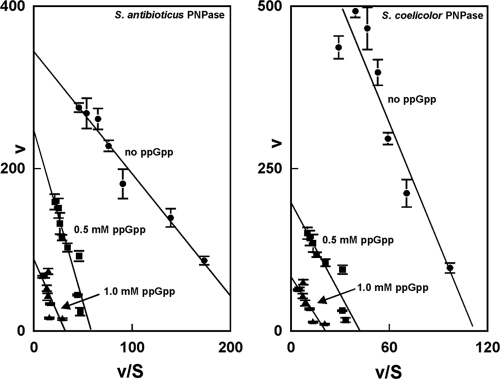
To examine this phenomenon further, we performed a kinetic analysis by varying the concentration of ADP in reaction mixtures containing either S. antibioticus or S. coelicolor PNPases. The results of this analysis are shown in Fig. 2 and Table 1. Figure 2 shows Eadie-Hofstee plots of the kinetic data, and it can been seen that both the maximum velocity and the Km values of the reaction were affected by 0.5 and 1 mM ppGpp, indicating that ppGpp is a mixed or true noncompetitive inhibitor of the polymerization reaction. This conclusion is confirmed by the data of Table 1, which show a change in Km and kcat in the presence of ppGpp. ppGpp affects kcat to a greater degree than it does Km and, interestingly, the Km for ADP decreased somewhat at 1 mM ppGpp in the case of S. antibioticus PNPase and at both ppGpp concentrations in the case of the S. coelicolor enzyme. kcat/Km decreased by ∼8-fold in the case of S. antibioticus PNPase and >5-fold in the case of the S. coelicolor enzyme.
FIG. 2.
Eadie-Hofstee plots of kinetic data for S. antibioticus and S. coelicolor PNPases in the polymerization reaction. Kinetic assays were performed as described previously (25) and in Materials and Methods. Velocities are expressed as pmol min−1, and substrate concentrations are indicated in mM. The data in the figure are averages of two sets of duplicates ± the SEM. The kinetic parameters were calculated from the regression statistics corresponding to the slopes and intercepts of the plots.
TABLE 1.
Kinetic parameters for the polymerization reactions in the presence or absence of ppGppa
| ppGpp concn (mM) | Mean ± SEM |
kcat/Km (l mol−1 min−l, 103) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
Km (mM) |
kcat (min−1) |
||||||||
| S. antibioticus | S. coelicolor | E. coli | S. antibioticus | S. coelicolor | E. coli | S. antibioticus | S. coelicolor | E. coli | |
| 0 | 1.46 ± 0.12 | 6.14 ± 0.43 | 14.6 ± 1.2 | 80.5 ± 2.9 | 492 ± 43.4 | 731 ± 224 | 55.1 | 80.1 | 50.1 |
| 0.5 | 4.24 ± 0.77 | 4.68 ± 0.32 | 12.6 ± 2.0 | 59.1 ± 6.5 | 141 ± 14.1 | 672 ± 179 | 13.9 | 30.1 | 53.3 |
| 1.0 | 2.78 ± 0.26 | 3.96 ± 0.68 | - | 21.2 ± 4.2 | 59.3 ± 6.7 | - | 7.6 | 15.0 | |
The Km and kcat data are averages of two sets of duplicates.
For comparison with the results described above, we also measured the kinetic constants for polymerization using E. coli PNPase. The results of this analysis are also shown in Table 1. Under our assay conditions, the kcat value for E. coli polymerase was greater than that observed for either of the Streptomyces enzymes, but the Km value was somewhat higher as well. As expected, 0.5 mM ppGpp had essentially no effect on the kinetic constants for the E. coli enzyme.
Effects of (p)ppGpp on phosphorolysis.
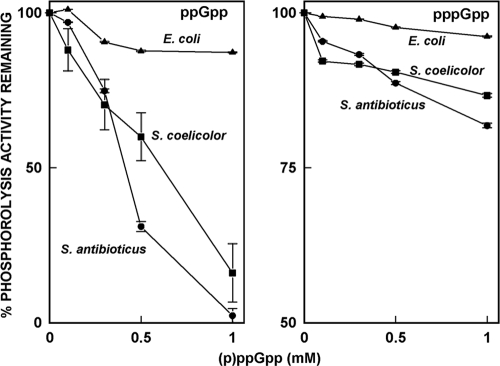
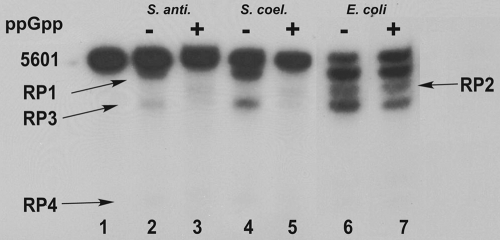
We examined the effects of (p)ppGpp on phosphorolysis using [3H]poly(A), synthesized as described previously (25). The results of the analysis are shown in Fig. 3. In this figure, the 100% value is the level of phosphorolysis observed in the absence of ppGpp and lower percentages reflect the inhibition of phosphorolysis by ppGpp. Thus, for example, the phosphorolysis activity of the S. antibioticus PNPase measured in the presence of 0.5 mM ppGpp was only ca. 30% of that observed in the absence of ppGpp. Again, the activities of the Streptomyces enzymes were inhibited by ppGpp; 1 mM ppGpp essentially abolished phosphorolysis of the poly(A) substrate. pppGpp was much less effective than ppGpp with the Streptomyces enzymes and did not substantially affect the activity of E. coli PNPase. ppGpp produced only about a 10% inhibition of the E. coli enzyme, even at a concentration of 1 mM. To obtain additional information on the effects of ppGpp on the phosphorolysis reactions, we examined the phosphorolysis of a model RNA substrate, the 5601 transcript, derived from the rpsO-pnp operon of S. coelicolor and used by us in other studies (9). The effects of 1 mM ppGpp on phosphorolysis of the substrate by the three PNPases are shown in the autoradiogram presented in Fig. 4. Again, ppGpp had no effect on the phosphorolysis of this substrate catalyzed by the E. coli PNPase (lanes 6 and 7). All of the products normally observed upon phosphorolysis of the 5601 transcript were observed in the presence or absence of ppGpp. In contrast, 1 mM ppGpp completely abolished the production of RP1, RP3, and RP4 by PNPases from S. coelicolor (lanes 2 and 3) and S. antibioticus (lanes 4 and 5). These results indicate that ppGpp inhibits the phosphorolysis of not only a model substrate, poly(A), but also a substrate corresponding to a naturally occurring transcript in S. coelicolor.
FIG. 3.
Inhibition of the phosphorolysis activity of PNPase by (p)ppGpp. In this figure, the 100% value is the level of phosphorolysis observed in the absence of ppGpp for each enzyme, and lower percentages reflect the inhibition of phosphorolysis by ppGpp. The data in the figure are the averages of duplicates from two separate experiments ± the SEM.
FIG. 4.
Gel electrophoresis of PNPase digests of the 5601 transcript. 32P-labeled 5601 RNA was synthesized as described previously (9). Gels were subjected to autoradiography. Lane 1, undigested 5601 transcript; lanes 2 and 3, digestion with S. antibioticus PNPase; lanes 4 and 5, digestion with S. coelicolor PNPase; lanes 6 and 7, digestion with E. coli PNPase. The reaction mixtures corresponding to lanes 3, 5, and 7 contained 1 mM ppGpp. The product designations at the left and right of the figure relate the bands produced to the 3′-endpoints mapped previously by cDNA cloning and sequencing (9).
Effects of ppGpp on the stability of bulk mRNA in S. coelicolor.
How might the observation of ppGpp inhibition of PNPase reflect a regulatory role for ppGpp and PNPase in Streptomyces? As indicated above, ppGpp positively regulates antibiotic production in several Streptomyces species, including the paradigm for streptomycete studies, S. coelicolor (7, 20, 22, 30). ppGpp levels rise just prior to the onset of antibiotic production in these organisms and, although the levels decline subsequently, they do not fall to zero. ppGpp levels are maintained at ca. 25% of peak levels for hours to days following the initial rise in concentration (22, 36). Again, as indicated above, PNPase functions as both an exonuclease and as an RNA 3′-polynucleotide polymerase in Streptomyces (4, 34). Both functions appear to be important in Streptomyces since: (i) Streptomyces spp. do not contain an RNase R homolog (39), so PNPase is the major if not the only 3′-5′-exonuclease present in Streptomyces; (ii) Streptomyces spp. do not contain a dedicated poly(A) polymerase, so PNPase must synthesize the 3′ tails that facilitate the degradation of streptomycete RNAs (4, 34); and (iii) PNPase is an essential enzyme in Streptomyces (4). Thus, PNPase almost certainly functions as a critical determinant of transcript stability in Streptomyces. As demonstrated here, ppGpp inhibits PNPase activity in Streptomyces. The inhibition of both the polymerization and the phosphorolysis activities of PNPase would be expected to increase transcript stability. It is conceivable, therefore, that ppGpp, which persists at significant levels in Streptomyces after antibiotic synthesis is initiated, could increase the stability of transcripts for proteins required for antibiotic production, by inhibiting the activity of PNPase. This strategy, along with the persistence of low levels of protein and RNA synthesis, would ensure that sufficient amounts of the necessary proteins remain available to support antibiotic production.
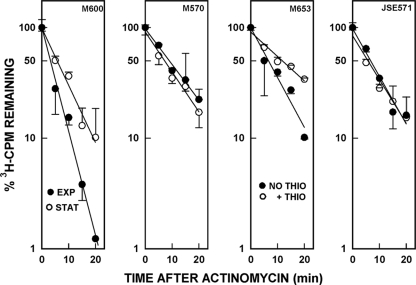
To test this hypothesis, we have determined the chemical half-lives of bulk mRNA in S. coelicolor under several sets of conditions. In the initial set of experiments, S. coelicolor M600 and M570 were grown to mid-exponential phase (A450 of ∼0.5, ca. 7 h postinoculation), a portion of the cultures was removed, RNAs were radioactively labeled, and RNA synthesis was then inhibited with actinomycin as described in Materials and Methods. A second portion of the cultures was removed 26 h postinoculation in the stationary phase, when the production of undecylprodigiosin and actinorhodin had begun in M600. The chemical half-lives of the bulk mRNA were determined as described previously (5) and in Materials and Methods. It is apparent from Fig. 5 and Table 2 that the half-life of bulk mRNA increased by a factor of 1.8 in stationary phase in M600, from 3.2 to 5.7 min. No such increase was observed with M570, the S. coelicolor relA-null mutant. This is exactly the outcome that is predicted if ppGpp inhibits mRNA decay during the stationary phase.
FIG. 5.
Decay curves of bulk mRNAs in various S. coelicolor strains. Cultures were grown and treated as described in Materials and Methods, and actinomycin D was used to inhibit transcription. Measurements were made during exponential (EXP) and stationary (STAT) phases for M600 and M570 and during stationary phase for M653 and JSE571. Samples of the latter two strains were also treated with thiostrepton to induce expression of the tipA promoter. The cpm values obtained at each time point were plotted as a percentage of the cpm present at time zero, set arbitrarily as 100. The data in the figure are the averages of duplicate measurements from two independent experiments ± the SEM.
TABLE 2.
Chemical half-lives of bulk mRNA in various S. coelicolor and E. coli strains
| Strain | Conditions or description | Mean half-life (min) ± SEMa |
|---|---|---|
| S. coelicolor | ||
| M600 | Parental strain, exponential phase | 3.2 ± 0.2 |
| Parental strain, stationary phase | 5.7 ± 0.6 | |
| M570 | relA-null mutant, exponential phase | 8.9 ± 0.8 |
| relA-null mutant, stationary phase | 7.2 ± 0.7 | |
| M653 | Strain M570 with inducible relA with no thiostrepton | 6.6 ± 0.9 |
| Strain M570 with inducible relA plus thiostrepton | 11.8 ± 1.6 | |
| JSE571 | With pIJ8600, no thiostrepton | 6.8 ± 0.8 |
| With pIJ8600 plus thiostrepton | 5.7 ± 0.9 | |
| E. coli | ||
| CF1648 | relA+; exponential phase | 2.6 ± 0.3 |
| relA+; stationary phase | 2.2 ± 0.3 | |
| CF1652 | relA-null mutant, exponential phase | 2.3 ± 0.2 |
| relA-null mutant, stationary phase | 3.0 ± 0.2 |
Half-lives were calculated from the data shown in Fig. 5 or from similar plots in the case of the E. coli measurements.
To determine whether ppGpp rather than some other agent was likely to be responsible for the observations just described, we examined the effects of induction of the relA gene on the stability of bulk mRNA. We utilized S. coelicolor M653, containing the relA gene under the control of the thiostrepton-inducible tipA promoter (21). As a control, M570 containing pIJ8600 (JSE571), the vector utilized to clone the relA gene, was used. M653 and JSE571 were grown to stationary phase, and a portion of each culture was treated with [3H]uridine and actinomycin as described in Materials and Methods. A separate portion of each culture was brought to 25 μg/ml with thiostrepton to induce relA expression (21). The half-lives of bulk mRNA were measured, and the results are shown in Fig. 5 and Table 2. The half-life of bulk mRNA prior to induction of relA in M653 was ca. 6.6 min, whereas the half-life increased to ca. 11.8 min after induction, again a 1.8-fold increase. Thiostrepton treatment produced no increase in half-life in JSE571, the strain containing the cloning vector only. These data strongly suggest that induction of relA in S. coelicolor M653 and the subsequent production of ppGpp by RelA leads to an increase in the stability of bulk mRNA. The observation that the half-life of bulk mRNA in exponential phase in M600 was shorter than in any of the strains in which relA was disrupted suggests that in addition to its role in ppGpp synthesis, relA may be involved in other physiological processes that affect mRNA stability. Effects of relA disruption on the growth and morphology of S. coelicolor have been reported (7).
We also examined the half-lives of bulk mRNA in two strains of E. coli to provide a comparison with the data described above for S. coelicolor. As shown in Table 2, the chemical half-lives were about 2 min during the exponential phase of growth in the parental relA+ strain and in the relA-null mutant. The half-life observed in stationary phase for the parental strain was also approximately 2 min, while a slight increase (∼1.3-fold) was observed in the relA-null mutant. If this increase has physiological significance, it is presumably not due to an effect of ppGpp.
In summary, we have shown that (p)ppGpp inhibits both polymerization and phosphorolysis by Streptomyces PNPases but not by E. coli PNPase. The observation that ppGpp exhibits mixed noncompetitive inhibition of polymerization suggests that the inhibitor binds to a region of the Streptomyces PNPases other than the active site (10). Analysis of the crystal structure of S. antibioticus PNPase suggests a candidate region for ppGpp binding. In particular, helices α3 and α4 of the PNPase, along with two of the β strands, form a solvent-filled pocket which does not bind tungstate (37). Thus, these structural elements are potential components of the site for (p)ppGpp binding. It should be possible to identify the binding site using analogs of ppGpp that can be cross-linked to the protein.
Our data also suggest that ppGpp stabilizes bulk mRNA during stationary phase in S. coelicolor and in strains in which ppGpp synthesis is ectopically induced. Although we did not measure ppGpp levels in the experiments reported here, we note that no stabilization of mRNA was observed when exponential- and stationary-phase cultures of S. coelicolor M570 (relA) were compared or when thiostrepton-treated and untreated cultures of JSE571 were compared (Fig. 5 and Table 2). It should be noted further that although the half-life of bulk mRNA was only increased by a factor of 1.8 in the experiments described here, ppGpp might well exert a greater effect on the stability of particular mRNAs. Although it is possible to speculate as to the identities of mRNAs whose stability might be affected by ppGpp levels, the most effective way to examine this question would be to perform a global analysis of mRNA half-lives under various physiological conditions in S. coelicolor. Experiments are in progress to examine further the roles of ppGpp and PNPase as determinants of mRNA stability during the stationary phase in this organism.
Acknowledgments
This study was supported by grant MCB 0817177 from the National Science Foundation.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Abranches, J., A. R. Martinez, J. K. Kajfasz, V. Chavez, D. A. Garsin, and J. A. Lemos. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191:2248-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baracchini, E., and H. Bremer. 1988. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J. Biol. Chem. 263:2597-2602. [PubMed] [Google Scholar]
- 3.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45-54. [DOI] [PubMed] [Google Scholar]
- 4.Bralley, P., B. Gust, S. A. Chang, K. F. Chater, and G. H. Jones. 2006. RNA 3′-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and PNPase. Microbiology 152:627-636. [DOI] [PubMed] [Google Scholar]
- 5.Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149:2173-2182. [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 7.Chakraburtty, R., and M. J. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraburtty, R., J. White, E. Takano, and M. J. Bibb. 1996. Cloning and characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2). Mol. Microbiol. 19:357-368. [DOI] [PubMed] [Google Scholar]
- 9.Chang, S. A., M. Cozad, G. A. Mackie, and G. H. Jones. 2008. Kinetics of polynucleotide phosphorylase: comparison of enzymes from Streptomyces and Escherichia coli and effects of nucleoside diphosphates. J. Bacteriol. 190:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland, R. A. 2000. Enzymes: a practical introduction to structure, mechanism, and data analysis, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 11.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo, A., H. Nicoloff, S. E. Barchinger, A. B. Banta, R. L. Gourse, and S. E. Ades. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67:619-632. [DOI] [PubMed] [Google Scholar]
- 13.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eymann, C., G. Mittenhuber, and M. Hecker. 2001. The stringent response, sigmaH-dependent gene expression and sporulation in Bacillus subtilis. Mol. Gen. Genet. 264:913-923. [DOI] [PubMed] [Google Scholar]
- 15.Gaal, T., and R. L. Gourse. 1990. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:5533-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Escribano, J. P., J. F. Martin, A. Hesketh, M. J. Bibb, and P. Liras. 2008. Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154:744-755. [DOI] [PubMed] [Google Scholar]
- 17.Gong, L., K. Takayama, and S. Kjelleberg. 2002. Role of spoT-dependent ppGpp accumulation in the survival of light-exposed starved bacteria. Microbiology 148:559-570. [DOI] [PubMed] [Google Scholar]
- 18.Gourse, R. L., W. Ross, and S. T. Rutherford. 2006. General pathway for turning on promoters transcribed by RNA polymerases containing alternative sigma factors. J. Bacteriol. 188:4589-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez, V. J., and H. Bremer. 1990. Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J. Biol. Chem. 265:11605-11614. [PubMed] [Google Scholar]
- 20.Hesketh, A., W. J. Chen, J. Ryding, S. Chang, and M. Bibb. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesketh, A., J. Sun, and M. J. Bibb. 2001. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-orf4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 39:136-141. [DOI] [PubMed] [Google Scholar]
- 22.Hoyt, S., and G. H. Jones. 1999. relA is required for actinomycin production in Streptomyces antibioticus. J. Bacteriol. 181:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, G. H. 1994. Purification and properties of ATP:GTP 3′-pyrophosphotransferase (guanosine pentaphosphate synthetase) from Streptomyces antibioticus. J. Bacteriol. 176:1475-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, G. H., and M. J. Bibb. 1996. Guanosine pentaphosphate synthetase from Streptomyces antibioticus is also a polynucleotide phosphorylase. J. Bacteriol. 178:4281-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, G. H., M. F. Symmons, J. S. Hankins, and G. A. Mackie. 2003. Overexpression and purification of untagged polynucleotide phosphorylases. Protein Expr. Purif. 32:202-209. [DOI] [PubMed] [Google Scholar]
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 27.Kuroda, A., H. Murphy, M. Cashel, and A. Kornberg. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240-21243. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Costa, O. H., P. Arias, N. M. Romero, V. Parro, R. P. Mellado, and F. Malpartida. 1996. A relA/spoT homologous gene from Streptomyces coelicolor A3(2) controls antibiotic biosynthetic genes. J. Biol. Chem. 271:10627-10634. [DOI] [PubMed] [Google Scholar]
- 29.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochi, K. 1987. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 169:3608-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35-51. [DOI] [PubMed] [Google Scholar]
- 32.Riesenberg, D., F. Bergter, and C. Kari. 1984. Effect of serine hydroxamate and methyl alpha-d-glucopyranoside treatment on nucleoside polyphosphate pools, RNA and protein accumulation in Streptomyces hygroscopicus. J. Gen. Microbiol. 130:2549-2558. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sohlberg, B., J. Huang, and S. N. Cohen. 2003. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase can polyadenylate RNA. J. Bacteriol. 185:7273-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivatsan, A., and J. D. Wang. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11:100-105. [DOI] [PubMed] [Google Scholar]
- 36.Strauch, E., E. Takano, H. A. Bayliss, and M. J. Bibb. 1991. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5:289-298. [DOI] [PubMed] [Google Scholar]
- 37.Symmons, M., G. H. Jones, and B. Luisi. 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity and regulation. Structure 8:1215-1226. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 39.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]