Abstract
Rickettsia prowazekii is an obligate intracellular pathogen that possesses a small genome and a highly refined repertoire of biochemical pathways compared to those of free-living bacteria. Here we describe a novel biochemical pathway that relies on rickettsial transport of host cytosolic dihydroxyacetone phosphate (DHAP) and its subsequent conversion to sn-glycerol-3-phosphate (G3P) for synthesis of phospholipids. This rickettsial pathway compensates for the evolutionary loss of rickettsial glycolysis/gluconeogenesis, the typical endogenous source of G3P. One of the components of this pathway is R. prowazekii open reading frame RP442, which is annotated GpsA, a G3P dehydrogenase (G3PDH). Purified recombinant rickettsial GpsA was shown to specifically catalyze the conversion of DHAP to G3P in vitro. The products of the GpsA assay were monitored spectrophotometrically, and the identity of the reaction product was verified by paper chromatography. In addition, heterologous expression of the R. prowazekii gpsA gene functioned to complement an Escherichia coli gpsA mutant. Furthermore, gpsA mRNA was detected in R. prowazekii purified from hen egg yolk sacs, and G3PDH activity was assayable in R. prowazekii lysed-cell extracts. Together, these data strongly suggested that R. prowazekii encodes and synthesizes a functional GpsA enzyme, yet R. prowazekii is unable to synthesize DHAP as a substrate for the GpsA enzymatic reaction. On the basis of the fact that intracellular organisms often avail themselves of resources in the host cell cytosol via the activity of novel carrier-mediated transport systems, we reasoned that R. prowazekii transports DHAP to supply substrate for GpsA. In support of this hypothesis, we show that purified R. prowazekii transported and incorporated DHAP into phospholipids, thus implicating a role for GpsA in vivo as part of a novel rickettsial G3P acquisition pathway for phospholipid biosynthesis.
Rickettsia prowazekii is an obligate intracellular pathogen, a select agent, and the etiologic cause of epidemic typhus fever in humans. R. prowazekii is transmitted by the human body louse, Pediculus humanus, and is prevalent in areas of low socioeconomic conditions where hygiene and sanitation are lacking (3, 12, 35). Known reservoirs include humans with recrudescent Brill-Zinsser disease and flying squirrels (17, 29). If they are misdiagnosed and/or improperly treated, typhus infections result in substantial morbidity and mortality (36).
Upon entering a eukaryotic host cell, R. prowazekii quickly escapes the endosomal vesicle and replicates directly within the cytoplasm (21, 45, 48). As an obligate intracellular pathogen, R. prowazekii has evolved to exploit the nutrient-rich cytoplasmic growth niche by transporting the end products of various host cell metabolic pathways (7, 10, 39, 42, 50, 52). As a presumed consequence of its reliance on the transport of host cell metabolites, R. prowazekii has lost many of its biosynthetic pathway genes through the process of reductive evolution (2, 4-6). In short, essential metabolites that cannot be synthesized by the rickettsiae de novo are transported from the eukaryotic host cell cytosol. The ongoing loss of R. prowazekii biochemical pathways from the genome is evidenced by the identification of pseudogenes, biochemical pathway remnants that have acquired mutations and that encode nonfunctional proteins (2, 4, 5, 22).
Interestingly, there exists another class of rickettsial biochemical pathway remnants that may be mistaken as pseudogenes. We have termed members of this class “functional orphaned enzymes” because the purified enzyme, unlike the product of a pseudogene, exhibits its annotated biochemical activity. However, the other enzymes in the corresponding pathway are absent from the R. prowazekii genome; thus, there is no endogenous enzymatic synthesis of substrate for the functional orphaned enzyme. We hypothesize that rickettsial functional orphaned enzymes are fed their substrate from the host cell cytosol via the activity of novel rickettsial transport systems. Together, functional orphaned enzymes and their cognate transport systems serve as streamlined biosynthetic pathways for required metabolites.
Here we present the characterization of a R. prowazekii functional orphaned enzyme, an sn-glycerol-3-phosphate (G3P) dehydrogenase (G3PDH) enzyme, RP442 (or GpsA), that catalyzes the conversion of dihydroxyacetone phosphate (DHAP) to G3P for phospholipid biosynthesis. GpsA appears to be a pathway remnant because R. prowazekii lacks glycolytic and gluconeogenic pathways and thus does not possess the capacity to synthesize DHAP (6, 30, 56). Evidence is presented to suggest that GpsA is a functional enzyme and that R. prowazekii transports and incorporates DHAP into phospholipid, indicating that DHAP transport and GpsA are part of a novel G3P acquisition pathway. This is the first report of DHAP transport by a bacterium.
MATERIALS AND METHODS
Chemicals, media, and radiolabeled triose phosphate synthesis.
Unless otherwise indicated, chemicals and enzymes were from Sigma. Molecular biology enzymes and thermostable DNA polymerase were from New England Biolabs. Escherichia coli was typically grown at 37°C with aeration (200 rpm) in LB medium containing 100 μg ml−1 of ampicillin (LBAp100). Solid medium contained 1.5% agar (LBAp100 agar). Oligonucleotide primers were synthesized by Integrated DNA Technologies. [γ-32P]ATP and [32P]orthophosphate were from PerkinElmer.
[32P]DHAP and [32P]G3P were synthesized in a coupled enzymatic reaction (0.5 ml) containing 1.1 mM dihydroxyacetone or glycerol, 100 mM Tris-HCl, pH 7.5, 1.1 mM KCl, 1.08 mM MgCl2, 1 mM ATP, 1.05 mM phosphoenolpyruvate, 1.1 mM NADH, 5 U glycerol kinase, 5 U pyruvate kinase, 5 U lactate dehydrogenase, and 0.4 mCi ml−1 [γ-32P]ATP (26). The reaction mixtures were incubated at room temperature for 30 min, and the product yield was determined by measuring the extent of NADH oxidation at an optical density of 340 nm (OD340; extinction coefficient, 6.2 mM−1 cm−1). Unincorporated [γ-32P]ATP was removed by adsorption to 100 mg of Norit A decolorizing carbon suspended in 100 mM Tris-HCl (pH 7.4). Enzymes were removed by ultrafiltration at 4°C using an Amicon YM10 cellulose filter, according to the manufacturer's directions. The quality of each purified radiolabeled substrate was verified by two different chromatographic methods. Products were resolved on polyethyleneimine cellulose using a solvent system of 1 N acetic acid and 4 M lithium chloride (85:15, vol/vol) (34) and on Whatman 3MM filter paper using a solvent system of picric acid-saturated t-butanol-t-butanol-water (10:70:20) (adapted from a previous report [25]). All reagents for paper chromatography were equilibrated at 30°C. Thin-layer chromatography plates/paper were visualized by phosphorimaging (Cyclone; PerkinElmer).
Purification of recombinant R. prowazekii GpsA protein and G3PDH activity assays.
Following standard molecular biology protocols (11), the R. prowazekii gpsA gene (RP442) was amplified via thermocycling using purified R. prowazekii (Madrid E strain) chromosomal DNA and forward and reverse primers (5-GGA ATT CCA TAT GAA CAA ATT TAA GAA TAT TGC AGT TTA TG-3 [the NdeI site is underlined] and 5-CGC GGA TCCTCA AAT TAA AAT GAG TGA AGC TAC TTC-3 [the BamHI site is underlined], respectively). The size of the resulting amplicon was confirmed by agarose gel electrophoresis, purified (Qiagen PCR purification kit), digested with NdeI and BamHI, purified, and ligated (T4 DNA ligase) into similarly digested pET15b expression vector (Novagen) to incorporate an amino-terminal hexahistidine tag (pET15b-N-His6-GpsA). The ligation products were transformed into an electrocompetent DH5α strain of E. coli, and transformants were selected on LBAp100 agar. A single colony transformant was purified and used to prepare plasmid to verify the presence of the insert by restriction endonuclease digestion and sequencing.
Sequence-verified pET15b-N-His6-GpsA plasmid was introduced into an electrocompetent BL21(DE3) strain of E. coli, and a single colony transformant was used for protein purification, as follows. An overnight culture grown in LBAp100 was diluted 1:100 into 200 ml of fresh LBAp100 and grown to an OD600 of 0.5, and recombinant N-His6-GpsA protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (1 mM final concentration). Overnight incubation was continued at room temperature with aeration. Induced cultures were collected by centrifugation (7,741 × g, 4°C, 10 min), suspended in 20 ml binding buffer (10 mM imidazole, 20 mM Tris-HCl, and 500 mM NaCl, pH 8.0, containing 1 mM phenylmethylsulfonyl fluoride), lysed in a French pressure cell (four passes at 20,000 lb/in2), and cleared by centrifugation (7,741 × g, 4°C, 10 min). The supernatant fraction was subjected to ultracentrifugation (490,000 × g, 4°C, 60 min). The N-His6-GpsA protein was purified from the soluble fraction using a Bio-Rad Profinia protein purification system using Ni2+-nitrilotriacetic acid and standard desalting columns, according to the manufacturer's directions, with the following changes. Wash buffer 1 contained 10 mM imidazole, 20 mM Tris-HCl, and 500 mM NaCl, pH 8.0; wash buffer 2 contained 30 mM imidazole, 20 mM Tris-HCl, and 500 mM NaCl, pH 8.0; elution buffer contained 300 mM imidazole, 20 mM Tris-HCl, and 500 mM NaCl, pH 8.0; and purified protein was desalted into buffer A (135 mM NaCl, 2.7 mM KCl, 5.4 mM Na2HPO4, 1.8 mM KH2PO4, 250 mM potassium glutamate, 2 mM MgCl2, pH 7.4) and stored at 4°C. Protein purification was verified by electrophoresis using a 4 to 15% Tris-HCl, SDS-polyacrylamide gel (Bio-Rad) and visualized by staining with Imperial protein stain (Bio-Rad). Western blotting was carried out as described previously (8) using primary mouse anti-His6 monoclonal antibody (1:1,000; Novagen) and secondary goat anti-mouse antibody (1:5,000; GE Lifesciences/Amersham), and the products were visualized using enhanced chemiluminescence (GE Lifesciences/Amersham) on a ChemiDocXRS apparatus (Bio-Rad). The identity of the purified protein was verified by mass spectrometry at the University of South Alabama MCI core facility.
The G3PDH activity of GpsA was assayed by paper chromatography. The reaction mixtures (20 μl) were incubated in modified buffer A (containing 25 mM potassium glutamate) containing the substrate and enzyme concentrations reported in the legend to Fig. 1. The product of the reaction was confirmed to be [32P]G3P by paper chromatography, as described above. Spectrophotometric assays measured the linear oxidation rate of either NADPH or NADH at OD340 in 1-ml reaction mixtures that contained modified buffer A and the substrate and enzyme concentrations reported in the legend to Fig. 1. Pyridine nucleotide baseline levels were established, and reactions were initiated by the addition of enzyme to the cuvette. All kinetic data were determined at room temperature in a continuous assay using the initial reaction rate. The ability of N-His6-GpsA to catalyze the reverse reaction (i.e., conversion of G3P to DHAP) was tested using reaction mixtures containing 10 μg ml−1 N-His6-GpsA, 100 mM G3P, and 0.5 mM NADP in modified buffer A.
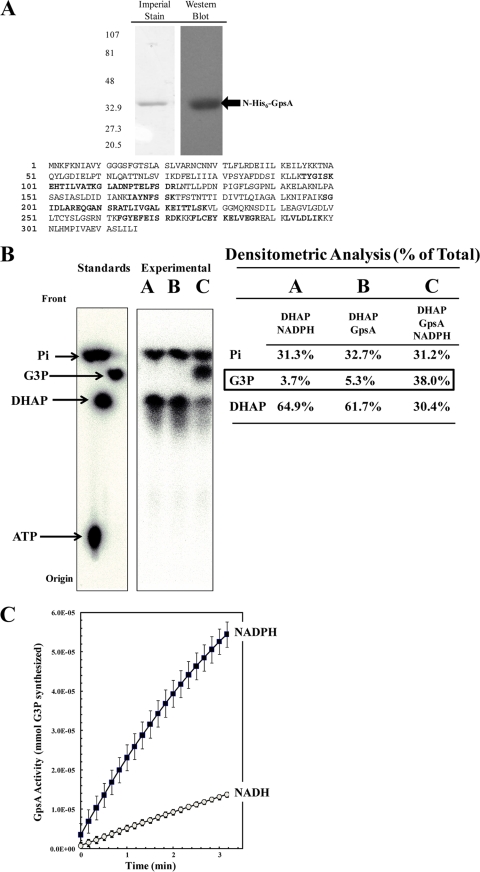
FIG. 1.
Recombinant N-His6-GpsA protein is an active G3P dehydrogenase in vitro. R. prowazekii gpsA was cloned into pET15b, heterologously expressed in E. coli BL21(DE3), and purified with N-His6-GpsA. (A) Purified protein was resolved on a 4 to 15% Tris-HCl SDS-polyacrylamide gel and visualized by Imperial protein staining and Western blot analysis using anti-His6 monoclonal antibody (sizes [in kDa] are indicated on the left). The mass spectrometry analysis coverage map shows matched peptides in bold font (30% coverage). (B) N-His6-GpsA-catalyzed conversion of [32P]DHAP to [32P]G3P was verified by paper chromatography. [32P]ATP, [32P]DHAP, [32P]G3P, and [32P]orthophosphate were used as standards. The reaction mixtures were incubated for 60 min at room temperature prior to chromatographic analysis. Lane A, a negative-control reaction mixture containing 6 μM [32P]DHAP and 100 μM NADPH only; lane B, a negative-control reaction mixture containing [32P]DHAP and 2.5 μg ml−1 N-His6-GpsA protein only; lane C, the N-His6-GpsA-catalyzed conversion of [32P]DHAP to [32P]G3P absolutely required the presence of NADPH. The density of each compound on the chromatograph is expressed as a percentage of the total lane density (summarized in tabular form to the right of each spot on the chromatograph). (C) N-His6-GpsA-catalyzed conversion of DHAP to G3P was measured spectrophotometrically at OD340 by following the oxidation of NADPH or NADH over time. Error bars represent standard deviations. The reaction mixtures contained 1.0 μg ml−1 N-His6-GpsA, 500 μM DHAP, and 100 μM NAD(P)H in modified buffer A; and the reactions were carried out at room temperature.
Complementation assays.
The R. prowazekii gpsA gene was subcloned from the pET15b vector into the arabinose-inducible pBAD24 expression vector (24). Complementation was tested using E. coli gpsA mutant strain BB20-14, whose growth requires either G3P or glycerol (13). BB20-14 was grown at 37°C with aeration in modified M56LP medium, which consisted of 100 mM Tris-HCl (pH 7.4), 10 mM KCl, 15 mM (NH4)2SO4, 10 mM MgCl2, 0.3 mM phosphate (K+ salt, pH 7.4), 0.2% Casamino Acids (CAA), 0.2% glucose, and 0.1% glycerol; and electrocompetent cells were prepared by standard methods. The pBAD24 vector control and pBAD24-gpsA were introduced into BB20-14, and transformants were selected on modified M56LP medium agar plates without CAA and with 100 μg ml−1 ampicillin and 1.5% Noble agar. Transformants were scored on modified M56LP agar plates (without CAA) supplemented with different carbon sources, as described in the legend to Fig. 2.
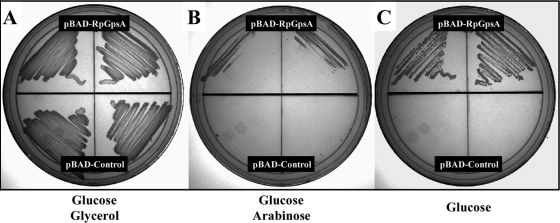
FIG. 2.
Heterologous expression of the R. prowazekii gpsA gene complements an E. coli gpsA auxotrophic mutant. E. coli gpsA mutant strain BB20-14 was transformed with pBAD24-RpGpsA expressing the R. prowazekii gpsA gene (inoculated in duplicate on the top half of each plate) or the pBAD24 vector control (inoculated in duplicate on the bottom half of each plate). (A) M56LP agar medium containing ampicillin (100 μg ml−1), glucose (0.2%), and glycerol (0.2%) (positive control); (B) M56LP agar medium containing ampicillin (100 μg ml−1), glucose (0.2%), and arabinose (0.2%); (C) M56LP agar medium containing ampicillin (100 μg ml−1) and glucose (0.2%).
Propagation and purification of R. prowazekii.
All experiments described in this study were performed using the Madrid E strain of R. prowazekii (hen egg yolk sac passage no. 282), purified as described by Bovarnick and Snyder (16) and modified by Winkler (47). Purified R. prowazekii suspended in a solution of 220 mM sucrose, 12 mM potassium phosphate, 4.9 mM potassium glutamate, and 10 mM magnesium chloride, pH 7.0 (SPGMg2+), was collected by centrifugation (9,000 × g, 4°C, 10 min), suspended in buffer B (SGMg2+ with 1 mM potassium phosphate), and stored on ice until it was assayed. Rickettsial preparation quality was assessed using a modified hemolysis assay which estimates the numbers of metabolically active rickettsiae (44). Additionally, rickettsial preparations were assayed for transport of two known rickettsial substrates: ATP (100 μM) with and without the mitochondrion-specific ATP transport inhibitor atractyloside (1 mM), in order to assess the levels of mitochondrial contamination (50, 54), and lysine (5 μM) with and without the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) at 10 μM as a control for experiments testing energy-dependent transport (57) (by the transport assay described below). All rickettsial preparations contained less than 5% mitochondrial contamination, based on the levels of atractyloside-inhibited ATP uptake. Finally, randomly selected rickettsial preparations were assayed for cytochrome c oxidase activity at different steps of the purification for both infected and mock-infected yolk sacs as a further control for mitochondrial contamination (40, 50).
gpsA mRNA detection by qRT-PCR and detection of G3PDH activity in R. prowazekii lysed-cell extracts.
Purification of total R. prowazekii RNA and quantitative reverse transcriptase (qRT) PCR (qRT-PCR) protocols were performed as described previously (9, 10) using the Express Two-Step qRT-PCR system from Invitrogen, according to the manufacturer's directions. The reverse transcription reaction mixtures contained 500 ng of total R. prowazekii RNA, and the resulting cDNA was serially diluted to generate an internal standard curve for each qPCR, as described previously (9, 10), using forward and reverse primers, each at a 128 nM final concentration. The gpsA mRNA was detected using a forward primer (5′-AGT AGC GAC AAA AGG CTT GG-3′) and a reverse primer (5′-CCG AAG CAG GCA AAT TTT TAA-3′) at an annealing temperature of 54°C. All other reaction conditions and detection of the tlc1 mRNA were carried out as described previously (9, 10).
G3PDH activity was assessed in R. prowazekii lysed-cell extracts as follows. Hen egg yolk sac-purified rickettsiae were concentrated to a final volume of 1 ml in modified buffer A (with Roche complete protease inhibitor cocktail at the manufacturer's suggested working concentration) and lysed by ballistic shearing using a Mini-Beadbeater apparatus (BioSpec Products), to deliver a 20-s pulse (maximum intensity setting), followed by incubation on ice for 2 min (repeated six times) using 0.1 mm zirconium beads. The lysate was cleared by centrifugation (10,000 × g, 4°C, 10 min), and the concentration of total protein was determined at an OD280 on a Nanodrop 2000 apparatus (Thermo Scientific). G3PDH assays were carried out as described above for the purified recombinant N-His6-GpsA protein with the rickettsial lysed-cell extract and substrate concentrations reported in the legend to Fig. 3.
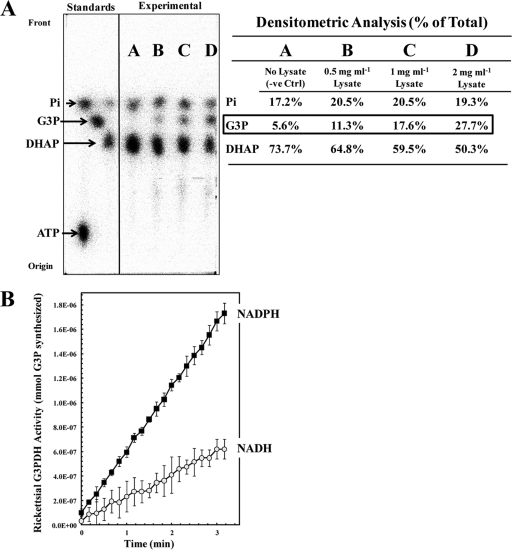
FIG. 3.
G3PDH activity is detectable in R. prowazekii lysed-cell extracts. Hen egg yolk sac-purified R. prowazekii cells were lysed by ballistic shearing, and the lysed-cell extracts were assayed for G3PDH activity. (A) Rickettsial lysed-cell extracts catalyzed conversion of [32P]DHAP to [32P]G3P, as determined by paper chromatography. [32P]ATP, [32P]DHAP, [32P]G3P, and [32P]orthophosphate were used as standards. Lane A, a negative-control (-ve Ctrl) reaction mixture containing 75 μM [32P]DHAP and 100 μM NADPH only; lane B, a reaction mixture containing [32P]DHAP, NADPH, and 0.5 mg ml−1 of rickettsial lysed-cell extract; lane C, a reaction mixture containing [32P]DHAP, NADPH, and 1 mg ml−1 of rickettsial lysed-cell extract; lane D, a reaction mixture containing [32P]DHAP, NADPH, and 2 mg ml−1 of rickettsial lysed-cell extract. The density of each compound on the chromatograph is expressed as a percentage of the total lane density (summarized in tabular form to the right of the chromatograph). (B) Rickettsial lysed-cell extract-catalyzed conversion of DHAP to G3P was measured spectrophotometrically at an OD340 by following the oxidation of NADPH or NADH. Error bars represent standard deviations. The reaction mixtures contained 1.4 mg ml−1 of rickettsial lysed-cell extract, 500 μM DHAP, and 100 μM NAD(P)H in modified buffer A; and the reactions were carried out at room temperature.
Substrate transport assays.
The uptake of radiolabeled substrates by purified R. prowazekii (i.e., transport assays) was measured using the filtration and wash method described previously (50). Briefly, the original suspension of yolk sac-purified rickettsiae was concentrated 10-fold, and assays were initiated by diluting back to the original concentration in buffer B containing 10 μM [32P]DHAP at 34°C. At the time points indicated in Fig. 4, 100-μl samples were removed, filtered through a Millipore 0.45-μm-pore-size HAWP02500 nitrocellulose filter, and washed with 8 ml of buffer B; and the filters were analyzed by liquid scintillation spectrometry (LKB Wallac). To examine the energy dependence of transport, rickettsiae were incubated in the presence of either 1 mM KCN or 10 μM CCCP for 10 min at 34°C, prior to the addition of substrate to initiate the uptake assay. Purified rickettsiae were also tested for transport of [32P]orthophosphate (1 mM, 0.5 μCi ml−1) as a control for the breakdown of [32P]DHAP. All transport data were normalized to the total amount of R. prowazekii protein, as determined using the Bio-Rad DCProtein assay.
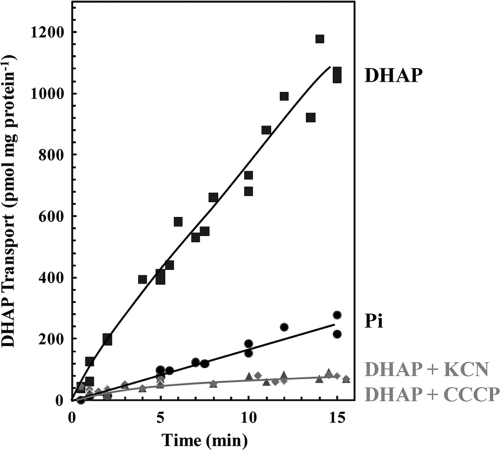
FIG. 4.
R. prowazekii transports DHAP in an energy-dependent manner. Purified R. prowazekii in buffer B was assayed for uptake of 10 μM [32P]DHAP or 1 mM [32P]orthophosphate at 34°C in at least three independent rickettsial preparations. Each data point represents the average of triplicates at the given time point for one rickettsial preparation. Data are expressed as transport activity per mg of total R. prowazekii protein. Energy-poisoned rickettsiae were preincubated for 10 min with 10 μM CCCP (diamonds) or 1 mM potassium cyanide (triangles), prior to the addition of [32P]DHAP to initiate the uptake assay.
Analysis of R. prowazekii phospholipid biosynthesis.
To determine if rickettsiae can incorporate exogenous DHAP into phospholipids, the original yolk sac-purified rickettsial suspension was concentrated 20-fold in SPGMg2+ containing 1 mM ATP, 0.1 mM NADH, 0.05 mM NADPH, 1 mM sodium bicarbonate, 0.4 mM acetyl coenzyme A, 0.01 mM biotin, 40 mM KCl, 0.05 thiamine pyrophosphate, 1 mM dithiothreitol, and 10 μM [32P]DHAP or 1 mM [32P]orthophosphate (0.5 μCi ml−1) and incubated for 1 h at 34°C. Total lipids were extracted using the modified method of Bligh and Dyer (14, 27) and resolved by thin-layer chromatography on a Silica Gel G plate using a solvent system of chloroform-methanol-28% ammonia (65:25:5, vol/vol/vol) (38) with phospholipid, [32P]G3P, and [32P]DHAP standards. Lipids were visualized by exposing the plate to iodine vapor, and the radioactivity incorporated into organic soluble compounds was analyzed by phosphorimaging.
Estimation of rickettsial intracellular DHAP concentration.
The transport assays described above were modified as follows. Purified rickettsiae in buffer B were incubated with 10 μM [32P]DHAP for 10 min, and 800 μl of cells was filtered (200 μl cells filter−1) and washed with 50 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.4). Washed filters were immediately extracted into 3 ml of HCl-acidified 70% ethanol (EtOH; pH 3.0), and the total radioactivity was determined by Cerenkov counting (37). In all experiments, at least 80% of the total radioactivity was recovered from the filter. EtOH was removed by evaporation, and the extracts were suspended at 1/20 the original volume in 70% EtOH (pH 3.0) and analyzed by paper chromatography. The radioactivity incorporated into ethanol-soluble compounds was visualized by phosphorimaging. Densitometry analysis of the paper chromatography was used to determine the percentage of [32P]DHAP in the total EtOH-soluble material in a given lane on the chromatograph, and this percentage was converted to the molar amount of DHAP, on the basis of the known specific activity of the [32P]DHAP added to the transport assay mixture. Rickettsial intracellular water space measurements were estimated for a portion of the purified intact rickettsiae as described previously (49) and used along with the determined molar amount of DHAP to calculate the intrarickettsial DHAP concentration after 10 min of transport.
RESULTS
Recombinant R. prowazekii GpsA protein is an active G3P dehydrogenase in vitro, and its expression functionally complements an E. coli gpsA mutation.
The R. prowazekii RP442 open reading frame is annotated GpsA, a NAD(P)H-dependent G3PDH that catalyzes the conversion of DHAP to G3P. Bioinformatic analysis shows that GpsA is highly conserved among all sequenced Rickettsia species genomes, and comparison to known functional G3PDH enzymes from other bacterial species suggests that RP442 could be a functional rickettsial G3PDH.
The usual metabolic role of GpsA in bacteria is to shuttle triose phosphate from glycolysis into the phospholipid biosynthesis pathway. However, as mentioned above, R. prowazekii lacks glycolytic/gluconeogenic pathways to supply an endogenous source of DHAP as substrate for GpsA (6, 30, 56). Thus, is rickettsial gpsA a pseudogene in the process of being lost from the genome, or has it retained a functional role in rickettsial physiology? Our first step in addressing this question was to purify recombinant N-His6-GpsA protein by heterologous expression in E. coli and assay it for G3PDH activity in vitro. Purified N-His6-GpsA was resolved by SDS-PAGE and analyzed by total protein staining and Western blotting with an antibody to the N-His6-tag, which verified the presence of a single protein that migrated with the expected molecular mass of 36.6 kDa (including the N-His6-tag) (Fig. 1 A). Mass spectrometry confirmed the identity of the protein as R. prowazekii GpsA.
Figure 1B shows that N-His6-GpsA catalyzed the NADPH-dependent reduction of [32P]DHAP, whereby the reaction product was confirmed to be [32P]G3P by paper chromatography. The minor amount of contaminating [32P]orthophosphate in the [32P]DHAP substrate conveniently served as an internal loading control for the chromatography. Kinetic analysis of N-His6-GpsA activity by spectrophotometry revealed it to prefer NADPH over NADH as the cofactor (Fig. 1C), and we determined an apparent Km of 528 ± 52 μM for DHAP and a Vmax of 0.039 ± 0.001 mmol mg−1 min−1 in the presence of NADPH. The apparent Km of NADPH was 48 ± 4 μM. The kinetic profile of purified recombinant rickettsial GpsA is similar to that described for the E. coli and Bacillus subtilis GpsA homologues (20, 28). We also determined that the conversion of DHAP to G3P was the favored reaction direction (data not shown).
For further verification of the G3PDH activity of the R. prowazekii GpsA protein, a complementation assay using E. coli gpsA mutant strain BB20-14 (13) was performed. BB20-14 was transformed with a pBAD24 vector control and the R. prowazekii GpsA-encoding pBAD24-gpsA plasmids and tested for growth in the absence of glycerol in the culture medium. Interestingly, Fig. 2 shows that the pBAD24-gpsA construct expressing the R. prowazekii GpsA protein supported E. coli growth in the absence of glycerol in the uninduced state, suggesting that only very low levels of rickettsial GpsA were required for complementation. Furthermore, the addition of 0.2% arabinose to induce rickettsial gpsA expression resulted in mildly impaired growth. BB20-14 transformed with the pBAD24 control plasmid grew only on medium containing both glucose and glycerol. Together, these data suggest that the R. prowazekii GpsA protein is an active G3PDH enzyme.
Detection of gpsA mRNA and G3PDH activity in purified isolated R. prowazekii.
Having confirmed the in vitro activity of recombinant N-His6-GpsA, we next determined if R. prowazekii actually synthesizes functional GpsA protein. We determined that the levels of gpsA mRNA levels in R. prowazekii isolated from embryonated hen egg yolk sacs were comparable to those of the well-characterized gene tlc1, whose activity is assayable in yolk sac-purified rickettsiae (50), indicating that the gpsA gene is transcribed (data not shown). We next assayed R. prowazekii lysed-cell extracts for the presence of G3PDH activity. Figure 3 A shows that the rickettsial extracts catalyzed the NADPH-dependent conversion of [32P]DHAP to [32P]G3P and that the activity increased with increasing amounts of added extract. Results from spectrophotometric-based G3PDH kinetic assays showed comparable kinetics between rickettsial lysed-cell extracts and purified recombinant N-His6-GpsA protein and the expected preference for NADPH over NADH (Fig. 3B). The G3PDH present in the rickettsial extract possessed an apparent Km of 489 ± 53 μM for DHAP as a substrate, which was within the range of that determined for recombinant purified rickettsial GpsA in vitro. In addition to the supporting kinetic data, it is highly likely that the G3PDH activity is rickettsial in origin and not host cell contamination because (i) it is well-known that the mitochondrial G3PDH is a flavin-linked enzyme not pyridine dependent like the GpsA activity described here and (ii) the cytosolic G3PDH of Gallus gallus domesticus has been shown to be an NAD+-linked enzyme (46) which should be removed by the extensive washing involved in the rickettsial preparation.
DHAP is transported by purified intact R. prowazekii and incorporated into phospholipid.
There are many examples of R. prowazekii de novo biosynthetic pathways that have been replaced by novel carrier-mediated transport systems that acquire the desired metabolite directly from the host cell (41, 42, 50-53). Thus, we hypothesized that R. prowazekii transports DHAP from the host cell to supply GpsA with substrate. Indeed, Fig. 4 shows that purified intact R. prowazekii demonstrated the capacity to transport [32P]DHAP (Fig. 4, squares) from the external environment. Control assays showed that the level of [32P]orthophosphate transport was very low (Fig. 4, circles), confirming that the observed uptake of [32P]DHAP is not spuriously due to its breakdown in the uptake assay medium.
To examine the metabolism of the transported [32P]DHAP inside the rickettsial cytosol, washed filters from the uptake assays were ethanol extracted and analyzed by paper chromatography. On the basis of data from three biological replicates, it was determined that ∼20% of the radioactivity remained associated with the filter (ethanol insoluble) and that 66% ± 8% of the total ethanol-soluble radioactivity corresponded to DHAP. The other major ethanol-soluble radioactive compounds migrated similarly to the standards for orthophosphate (12% ± 4%), ATP (11% ± 2%), and G3P (9% ± 2%). We used the transport and paper chromatography data along with a measurement of the rickettsial cytosolic water volume to estimate that DHAP was concentrated 2.2-fold after 10 min of transport. Figure 4 shows that incubation of the rickettsiae with energy poisons such as KCN (Fig. 4, triangles) and CCCP (Fig. 4, diamonds) prior to the addition of substrate abolished DHAP uptake, as would be expected of an energy-coupled transport system.
Our observations that R. prowazekii GpsA is a functional G3P dehydrogenase and that purified intact R. prowazekii transports DHAP suggest that this pathway is a possible source of G3P for phospholipid biosynthesis. On the basis of the findings of chromatographic analysis of three biological replicates, it was determined that [32P]DHAP incubated with intact rickettsiae was incorporated into organic soluble compounds that displayed migration patterns comparable to those of known phospholipid standards. The major species of phospholipid appear to be phosphatidylethanolamine (64% ± 5% of the total), lysophosphadylethanolamine (21% ± 4%), and phosphatidylglycerol (11% ± 2%), as would be expected from previous studies of the R. prowazekii phospholipid profile (43, 55). As a control, we verified that purified rickettsiae do not incorporate [32P]inorganic phosphate into phospholipid, suggesting that that our observations were not due to the spurious breakdown of [32P]DHAP in the assay medium (data not shown). The incorporation of [32P]DHAP into rickettsial phospholipid likely required its conversion to G3P and is indirect evidence for a physiological role for GpsA.
DISCUSSION
We have presented biochemical evidence demonstrating that the R. prowazekii GpsA is a functional G3P dehydrogenase that works in concert with a novel bacterial DHAP transport system to supply rickettsiae with G3P for phospholipid biosynthesis. The present study is the first to describe the transport of DHAP by a bacterium and to validate an enzymatic activity for the orphaned rickettsial G3P dehydrogenase enzyme.
Transport of DHAP is a rare phenomenon in nature. Until our study, transport of DHAP had been reported only for organelles such as plant plastids and trypanosome glycosomes, with triose phosphate transporters from plastidic inner membranes being the best characterized (15, 23, 31). The known plastidic DHAP transporters share no significant homology with any rickettsial open reading frame when they were examined by BLAST analysis (1). The fact that rickettsiae demonstrated the ability to concentrate DHAP against a gradient and the fact that transport activity was strongly inhibited in the presence of metabolic uncouplers are strongly indicative of an energy-coupled transport system rather than facilitated diffusion. We are currently working to identify the rickettsial DHAP transport system to verify that it belongs to a new family of bacterial triose phosphate transporters.
Presumably, reductive evolution of the ancestral rickettsial glycolytic/gluconeogenic pathway coincided with the acquisition of a DHAP transport system that imposed a selection pressure to maintain gpsA and, thus, a source of G3P for phospholipid biosynthesis. The complexity of this intriguing evolutionary picture is amplified by the R. prowazekii RP054 open reading frame annotated as a homologue of the bacterial G3P/inorganic phosphate obligate exchange antiporter, GlpT (6). Is RP054 really a DHAP transporter, or do rickettsiae also transport G3P and thus possess dual metabolite acquisition pathways? Unpublished work from our laboratory has shown that purified intact rickettsiae are able to transport and incorporate G3P into phospholipid at rates comparable to those reported here for DHAP (K. M. Frohlich and J. P. Audia, unpublished data). Unfortunately, our efforts to clone and assay R. prowazekii GlpT by heterologous expression in E. coli have been hindered by the toxicity associated with overexpression, despite the use of different expression systems, such as pET and pBAD, to tune GlpT expression levels. We are currently exploring alternative model microbial expression systems and the possibility of eventually generating and testing a knockout of the R. prowazekii glpT gene, acknowledging the genetic intractability of rickettsia, to explore these interesting physiological questions.
The observation of dual acquisition pathways for G3P in purified R. prowazekii raises interesting questions regarding the evolutionary pressures that have selected for maintenance of both pathways. Perhaps rickettsial transport of G3P and DHAP is an example of metabolic redundancy where one of the two systems is dispensable. Alternatively, rickettsiae may be in fierce competition with host cell enzymes for limiting amounts of G3P and DHAP in the host cell cytosol and, thus, the two rickettsial transport systems work in parallel because the activity of either on its own is insufficient to sustain growth. Another variable to consider is the effect of rickettsial growth on the host cell's triose phosphate pools. At late stages of infection, when the host cell is under a tremendous burden, the large number of rickettsiae must compete for substrate not only with host enzymes but also with each other. It has been shown in eukaryotic cells that the pools of free metabolic intermediates such as DHAP are subject to dramatic shifts in response to stress (32, 33). Thus, the availability of DHAP and G3P may fluctuate as the rickettsial burden increases and the two systems provide the rickettsiae with metabolic flexibility to maximize utilization of host cell resources. Similar points have been raised to explain the existence of the ATP/ADP translocase and tricarboxylic acid/oxidative-phosphorylation systems as dual pathways to supply ATP as an energy source (18, 19, 50). In the end, our work suggests that the identification of functional orphaned enzymes may serve as a useful indicator of the existence of other novel R. prowazekii transport systems that exploit the host cell cytosolic niche.
Acknowledgments
We especially thank Herb Winkler for many stimulating discussions and for critically reading the manuscript. Thanks go to Christopher McAtee for technical assistance in the recombinant GpsA activity assays as well as Aimee Tucker and Lewis Pannell for assistance with the mass spectroscopy. Rickettsial RNA isolation and qRT-PCR were performed by Mary Patton. We also thank Michael Housley and Robin Daugherty for technical assistance. Additional thanks go to David Wood and John Foster for critical reading of the manuscript.
This work was supported by Public Health Service grant R21 AI-069210 from the National Institute of Allergy and Infectious Diseases to J.P.A.
The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIAID or NIH.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, J. O., and S. G. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, J. O., and S. G. E. Andersson. 2000. A century of typhus, lice and Rickettsia. Res. Microbiol. 151:143-150. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, J. O., and S. G. E. Andersson. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178-1191. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, S. G. E., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. [DOI] [PubMed] [Google Scholar]
- 6.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowdki, A. K. Naslund, A.-S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson, W. H., and H. H. Winkler. 1989. Permeability of Rickettsia prowazekii to NAD. J. Bacteriol. 171:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Audia, J. P., and J. W. Foster. 2003. Acid shock accumulation of sigma S in Salmonella enterica involves increased translation, not regulated degradation. J. Mol. Microbiol. Biotechnol. 5:17-28. [DOI] [PubMed] [Google Scholar]
- 9.Audia, J. P., M. C. Patton, and H. H. Winkler. 2008. DNA microarray analysis of the heat shock transcriptome of the obligate intracytoplasmic pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 74:7809-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Audia, J. P., and H. H. Winkler. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 188:6261-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology, vol. 1, 2, and 3. John Wiley & Sons, Inc., New York, NY.
- 12.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell, R. M. 1974. Mutants of Escherichia coli defective in membrane phospholipid synthesis: macromolecular synthesis in an sn-glycerol 3-phosphate acyltransferase Km mutant. J. Bacteriol. 117:1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 15.Borchert, S., J. Harborth, D. Schunemann, P. Hoferichter, and H. W. Heldt. 1993. Studies of the enzymic capacities and transport properties of pea root plastids. Plant Physiol. 101:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bovarnick, M. R., and J. C. Synder. 1949. Respiration of typhus rickettsiae. J. Exp. Med. 89:561-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozeman, F. M., S. A. Masiello, M. S. Williams, and B. L. Elisberg. 1975. Epidemic typhus rickettsiae isolated from flying squirrels. Nature 255:545-547. [DOI] [PubMed] [Google Scholar]
- 18.Cai, J., H. Pang, D. O. Wood, and H. H. Winkler. 1995. The citrate synthase-encoding gene of Rickettsia prowazekii is controlled by two promoters. Gene 163:115-119. [DOI] [PubMed] [Google Scholar]
- 19.Cai, J., and H. H. Winkler. 1996. Transcriptional regulation in the obligate intracytoplasmic bacterium Rickettsia prowazekii. J. Bacteriol. 178:5543-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark, D., V. Lightner, R. Edgar, P. Modrich, J. E. Cronan, Jr., and R. M. Bell. 1980. Regulation of phospholipid biosynthesis in Escherichia coli. Cloning of the structural gene for the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J. Biol. Chem. 255:714-717. [PubMed] [Google Scholar]
- 21.Cowan, G. 2000. Rickettsial diseases: the typhus group of fevers—a review. Postgrad. Med. J. 76:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driskell, L. O., A. M. Tucker, H. H. Winkler, and D. O. Wood. 2005. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J. Bacteriol. 187:5719-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairlamb, A. H., and F. R. Opperdoes. 1986. Carbohydrate metabolism in African trypanosomes, with special reference to the glycosome, p. 183-224. In M. J. Morgan (ed.), Carbohydrate metabolism in cultured cells. Plenum Publishing Corporation, New York, NY.
- 24.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajra, A. K., and B. W. Agranoff. 1968. Acyl dihydroxyacetone phosphate. Characterization of a 32P-labeled lipid from guinea pig liver mitochondria. J. Biol. Chem. 243:1617-1622. [PubMed] [Google Scholar]
- 26.Hajra, A. K., and C. Burke. 1978. Biosynthesis of phosphatidic acid in rat brain via acyl dihydroxyacetone phosphate. J. Neurochem. 31:125-134. [DOI] [PubMed] [Google Scholar]
- 27.Kolarovic, L., and N. C. Fournier. 1986. A comparison of extraction methods for the isolation of phospholipids from biological sources. Anal. Biochem. 156:244. [DOI] [PubMed] [Google Scholar]
- 28.Morbidoni, H. R., D. de Mendoza, and J. E. Cronan, Jr. 1995. Synthesis of sn-glycerol 3-phosphate, a key precursor of membrane lipids, in Bacillus subtilis. J. Bacteriol. 177:5899-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, E. S., G. Baehr, G. Shwartzman, R. Mandelbaum, N. Rosenthal, J. C. Doane, L. B. Weiss, S. Cohen, and J. C. Snyder. 1950. Brill's disease. I. Clinical and laboratory diagnosis. JAMA 142:1059-1066. [Google Scholar]
- 30.Phibbs, P. V., Jr., and H. H. Winkler. 1981. Regulatory properties of partially purified enzymes of the tricarboxylic acid cycle of Rickettsia prowazekii, p. 421-430. In W. Burgdorfer and R. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY.
- 31.Quick, W. P., and H. E. Neuhaus. 1996. Evidence for two types of phosphate translocators in sweet-pepper (Capsicum annum L.) fruit chromoplasts. Biochem. J. 320(Pt 1):7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralser, M., M. M. Wamelink, A. Kowald, B. Gerisch, G. Heeren, E. A. Struys, E. Klipp, C. Jakobs, M. Breitenbach, H. Lehrach, and S. Krobitsch. 2007. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralser, M., M. M. Wamelink, S. Latkolik, E. E. Jansen, H. Lehrach, and C. Jakobs. 2009. Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat. Biotechnol. 27:604-605. [DOI] [PubMed] [Google Scholar]
- 34.Randerath, K., and E. Randerath. 1967. Thin-layer separation methods for nucleic acid derivatives. Methods Enzymol. 12:323-347. [Google Scholar]
- 35.Raoult, D., J. B. Ndihokubwayo, H. Tissot-Dupont, V. Roux, B. Faugere, R. Ebegbinni, and R. J. Birtles. 1998. Outbreak of epidemic typhus associated with trench fever in Burundi. Lancet 352:353-358. [DOI] [PubMed] [Google Scholar]
- 36.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Rowe, N. J., J. Tunstall, L. Galbraith, and S. G. Wilkinson. 2000. Lipid composition and taxonomy of Pseudomonas echinoides: transfer to the genus Sphingomonas. Microbiology 146(Pt 11):3007-3012. [DOI] [PubMed] [Google Scholar]
- 39.Smith, D. K., and H. H. Winkler. 1977. Characterization of a lysine-specific active transport system in Rickettsia prowazekii. J. Bacteriol. 129:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, L. 1955. Spectrophotometric assay of cytochrome c oxidase, p. 427-434. In D. Glich (ed.), Methods in biochemical analysis, vol. 2. Interscience Publishers, Inc., New York, NY. [DOI] [PubMed] [Google Scholar]
- 41.Speed, R. R., and H. H. Winkler. 1990. Acquisition of polyamines by the obligate intracytoplasmic bacterium, Rickettsia prowazekii. J. Bacteriol. 172:5690-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tucker, A. M., H. H. Winkler, L. O. Driskell, and D. O. Wood. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzianabos, T., C. W. Moss, and J. E. McDade. 1981. Fatty acid composition of rickettsiae. J. Clin. Microbiol. 13:603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker, T. S., and H. H. Winkler. 1979. Rickettsial hemolysis: rapid method for enumeration of metabolically active typhus rickettsiae. J. Clin. Microbiol. 9:645-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss, E. 1982. The biology of rickettsiae. Annu. Rev. Microbiol. 36:345-370. [DOI] [PubMed] [Google Scholar]
- 46.White, H. B., III, and N. O. Kaplan. 1969. Purification and properties of two types of diphosphopyridine nucleotide-linked glycerol 3-phosphate dehydrogenases from chicken breast muscle and chicken liver. J. Biol. Chem. 244:6031-6039. [PubMed] [Google Scholar]
- 47.Winkler, H. H. 1974. Inhibitory and restorative effects of adenine nucleotides on rickettsial adsorption and hemolysis. Infect. Immun. 9:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler, H. H. 1990. Rickettsia species (as organisms). Annu. Rev. Microbiol. 44:131-153. [DOI] [PubMed] [Google Scholar]
- 49.Winkler, H. H. 1976. Rickettsial cell water and membrane permeability determined by a micro space technique. Appl. Environ. Microbiol. 31:146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler, H. H. 1976. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 251:389-396. [PubMed] [Google Scholar]
- 51.Winkler, H. H., R. Daugherty, and F. Hu. 1999. Rickettsia prowazekii transports UMP and GMP, but not CMP, as building blocks for RNA synthesis. J. Bacteriol. 181:3238-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler, H. H., and R. M. Daugherty. 1986. Acquisition of glucose by Rickettsia prowazekii through the nucleotide intermediate uridine 5′-diphosphoglucose. J. Bacteriol. 167:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winkler, H. H., and R. M. Daugherty. 1984. Proline transport and metabolism in Rickettsia prowazekii. J. Bacteriol. 158:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler, H. H., and A. L. Lehninger. 1968. The atractyloside-sensitive nucleotide binding site in a membrane preparation from rat liver mitochondria. J. Biol. Chem. 243:3000-3008. [PubMed] [Google Scholar]
- 55.Winkler, H. H., and E. T. Miller. 1978. Phospholipid composition of Rickettsia prowazekii grown in chicken embryo yolk sacs. J. Bacteriol. 136:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wisseman, C. L., E. B. Jackson, and F. E. Hahn. 1951. Metabolic studies of rickettsia. I. The effects of antimicrobial substances and enzyme inhibitors on the oxidation of glutamate by purified rickettsiae. J. Immunol. 67:123-136. [PubMed] [Google Scholar]
- 57.Zahorchak, R. J., and H. H. Winkler. 1983. Transmembrane electrical potential in Rickettsia prowazekii and its relationship to lysine transport. J. Bacteriol. 153:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]