Abstract
Legionella pneumophila is an intracellular pathogen that infects protozoa in aquatic environments and when inhaled by susceptible human hosts replicates in alveolar macrophages and can result in the often fatal pneumonia called Legionnaires' disease. The ability of L. pneumophila to replicate within host cells requires the establishment of a specialized compartment that evades normal phagolysosome fusion called the Legionella-containing vacuole (LCV). Elucidation of the biochemical composition of the LCV and the identification of the regulatory signals sensed during intracellular replication are inherently challenging. l-Arginine is a critical nutrient in the metabolism of both prokaryotic and eukaryotic organisms. We showed that the L. pneumophila arginine repressor homolog, ArgR, is required for maximal intracellular growth in the unicellular host Acanthamoeba castellanii. In this study, we present evidence that the concentration of l-arginine in the LCV is sensed by ArgR to produce an intracellular transcriptional response. We characterized the L. pneumophila ArgR regulon by global gene expression analysis, identified genes highly affected by ArgR, showed that ArgR repression is dependent upon the presence of l-arginine, and demonstrated that ArgR-regulated genes are derepressed during intracellular growth. Additional targets of ArgR that may account for the argR mutant's intracellular multiplication defect are discussed. These results suggest that l-arginine availability functions as a regulatory signal during Legionella intracellular growth.
Legionella pneumophila is a Gram-negative gammaproteobacterial species with a remarkable capacity for robust growth in eukaryotic host cells (24, 35, 58). In both natural and man-made aquatic systems, L. pneumophila replicates within a wide variety of unicellular protozoa (23). Inhalation of aerosolized water contaminated with Legionella species, often from showers or whirlpool baths, can result in infection of human alveolar macrophages (60, 74). In susceptible individuals, this infection can lead to the development of a potentially fatal form of pneumonia called Legionnaires' disease (35, 58, 77).
Replication of Legionella in both unicellular protozoa and human alveolar macrophages occurs through a series of ordered events that begins with phagocytosis (32). Following uptake, L. pneumophila abrogates normal host vesicular trafficking to prevent the phagosome from acidifying and fusing with lysosomes (33, 34). Subsequent steps of phagosome maturation include interactions with host cell organelles such as mitochondria and endoplasmic reticulum, which will ultimately decorate the vacuole (1, 32). At approximately 8 h after uptake, the phagosome has developed into a Legionella-containing vacuole (LCV) that supports robust bacterial replication (32). Between 18 and 24 h postinfection, vacuoles filled with L. pneumophila are released and able to infect the next round of host cells (32, 73). This process requires the Icm/Dot type IVB secretion system (TFBSS), which is essential for the evasion of lysosomal fusion with the Legionella-containing phagosome and prevention of vacuole acidification (66, 70, 72). The Icm/Dot TFBSS is able to translocate >150 protein substrates into the host cytoplasm (12, 16, 20, 21, 61, 63). Icm/Dot-translocated substrates can alter host cell functions in a variety of ways that are predicted to promote bacterial intracellular survival (16, 37, 54, 61).
The ordered nature of the Legionella infection suggests that it is a highly regulated process, likely to require the ongoing detection of, and response to, specific environmental signals (29, 36, 59). Although the two-component systems CpxR/A, LetA/S, and PmrA/B and the global regulator σS have been shown to affect L. pneumophila intracellular multiplication, the signals that they respond to intracellularly are unknown (2, 4, 25, 30, 36, 51). Both LetA/S and σS accumulate in response to ppGpp during growth in rich medium in a process that affects the accumulation of the small regulatory RNAs RsmY and RsmZ (6, 36, 59), but it is not known if this also occurs during intracellular growth. Determining the internal composition of the LCV and identifying signaling molecules required for intracellular growth are inherently challenging.
Recently we reported that the σS-regulated gene lpg0490, a homolog of the arginine repressor, argR, is required for maximal intracellular multiplication of L. pneumophila within the protozoan host Acanthamoeba castellanii but not in the stabilized macrophage cell line THP-1 (36). The ArgR protein has been characterized in other bacteria as a repressor of arginine biosynthetic genes, which are typically distributed throughout the genome and required for the synthesis of the amino acid l-arginine from l-glutamate (19, 46, 52). This set of genes is commonly referred to as the ArgR regulon (46). In multiple bacterial genera, it has been demonstrated that ArgR monomers oligomerize to form homohexamers (28, 39, 62). The ArgR hexamers are allosterically activated by bound l-arginine to form a transcriptional repressor that binds to well-conserved DNA operator sites (28, 39, 78). Thus, in other bacteria ArgR is a direct sensor of l-arginine availability that represses the transcription of its target genes when arginine biosynthesis is not required.
Legionella species are arginine auxotrophs because they lack genes encoding the enzymes that carry out the preliminary steps of the biosynthetic conversion of l-glutamate into l-arginine (26). Legionellae are, however, capable of synthesizing l-arginine from compounds that occur later in this biosynthetic pathway such as l-ornithine and citrulline (26). Amino acid metabolism is of central importance in Legionella biology because amino acids can be utilized as its sole source of carbon and nitrogen and meet most of its energy needs (81).
Based on the function of ArgR in other bacteria and the intracellular replication defect of an L. pneumophila ΔargR mutant, we hypothesized that l-arginine availability is a regulatory signal affecting gene expression in the LCV. In order to understand how arginine availability affects gene expression during intracellular growth, we studied the global gene expression profile of an ΔargR mutant and used it to define the L. pneumophila ArgR regulon. Using a novel dual-fluorescence transcriptional reporter system, the regulation of genes controlled by ArgR and l-arginine was analyzed in chemically defined medium (CDM) and during intracellular growth. This system, in conjunction with quantitative PCR (qPCR) estimates of mRNA abundance from intracellularly growing bacteria, allowed us to demonstrate that several L. pneumophila genes whose transcription is regulated by ArgR and l-arginine availability are derepressed during intracellular growth. These results contribute to understanding how nutrient availability can affect gene expression during L. pneumophila intracellular growth.
MATERIALS AND METHODS
Bacterial strains and mutants.
The bacterial strains used in this study are listed in Table 1. Media and antibiotics were used as previously described (15). The L. pneumophila strains used in this study were JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1 (68); LELA3118, an isogenic dotA (lpg2686) derivative of JR32 (68); KS79, an isogenic ΔcomR (lpg2717) derivative of JR32 that renders the bacteria genetically competent for DNA uptake (20); and GAH280, an isogenic ΔargR (lpg0490) derivative of KS79 (therefore ΔcomR ΔargR) (36). KS79 (ΔcomR) is the isogenic parent of all strains used in this study and is referred to as the wild type (WT) in all experiments. Mutants made in this study were produced by creating allelic exchange fragments using long flanking homology (LFH)-PCR as described previously (36). The oligonucleotides used in mutant formation are lpg0493 P1 (GGCCAGGATGACGTTGCGATTGCC), lpg0493 P2 (CCCGGCCAAGCTAAAAAACACGGATCCCGATGCGTTAATGACTTGCAGC), lpg0493 P3 (GCTCGATGAGTTTTTCTAAGGATCCGCGATTGTTGGTTGTAAAGCAAGG), lpg0493 P4 (GCTGCGTCACATCAATGAATGCATCC), lpg0495 P1 (CCACTAATTGCTCAAAAGCTTGTTGAAATTGC), lpg0495 P2 (CCCGGCCAAGCTAAAAAACACGGATCCCCTGACTGCCATTAATGTCCTGGTC), lpg0495 P3 (GATGCTCGATGAGTTTTTCTAAGGATCCGCAGGAGGATAAAGAAGGATTATTTGATACG), and lpg0495 P4 (GCCTCCTTAACAATAGCCTCATTAGGC).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or feature(s) | Source or reference |
|---|---|---|
| L. pneumophila strains | ||
| JR32 | Am511 salt-sensitive isolate | 68 |
| KS79 | JR32 ΔcomR | 20 |
| LELA3118 | JR32 dotA::Tn903dIIlacZ | 68 |
| GAH199 | KS79 dotA::Tn903dIIlacZ | 36 |
| GAH280 | KS79 ΔargR::Kmr | 36 |
| GAH318 | KS79 Δlpg0493::Kmr | This study |
| GAH319 | KS79 Δlpg0495::Kmr | This study |
| GAH281 | KS79/pXDC31 | 36 |
| GAH282 | GAH199/pXDC31 | 36 |
| GAH286 | GA280/pXDC31 | 36 |
| GAH339 | GAH318/pXDC31 | This study |
| GAH340 | GAH319/pXDC31 | This study |
| GAH348 | KS79/pXDC95 | This study |
| GAH349 | KS79/pGAH140 | This study |
| GAH350 | KS79/pGAH141 | This study |
| GAH354 | GAH280/pXDC95 | This study |
| GAH355 | GAH280/pGAH140 | This study |
| Plasmids | ||
| pMMB207c | pMMB207 mobA | 70 |
| pXDC31 | pMMB207c Ptac-GFP | 36 |
| pXDC94 | pMMB207c Ptac-mCherry-2Ω-GFP | This study |
| pXDC95 | pMMB207c Ptac-mCherry-GFP | This study |
| pGAH140 | pMMB207c Ptac-mCherry-2Ω-P491-GFP | This study |
| pGAH141 | pMMB207c Ptac-mCherry-2Ω-P491ΔO-GFP | This study |
Growth of bacterial strains and preparation of media.
Chemically defined medium (CDM) was prepared as previously described (85); however no l-arginine was added to stock solutions (CDM with no l-arginine). For CDM growth and transcription assays, bacteria were grown to the mid-exponential phase in AYE complex growth medium [N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract], washed three times in CDM with no l-arginine, and resuspended to an optical density (OD) of 0.10 for time zero measurement in CDM containing the desired concentration of l-arginine or its biosynthetic precursors. The standard final concentration of l-arginine or its biosynthetic precursors for experiments conducted in this study was 1.0 mg/ml. Absorbance (optical density at 600 nm [OD600]) and fluorescence measurements were conducted in a TECAN Infinite M200 plate reader in triplicate. Growth experiments were performed three times, and the data shown are from one representative experiment. The data points reflect the average value of three technical replicates. Isolation of exponentially and postexponentially growing L. pneumophila for microarray analysis from complex medium AYE was performed as described previously (36) on triplicate samples.
Plasmid construction.
Plasmids pGAH140 and pGAH141, which contain the promoter fragments P491 and P491ΔO, respectively, were constructed from the restriction digest of PCR fragments bearing the restriction sites XmaI and KpnI cloned into digested pXDC94 vector (for the map, see Fig. 8A). The oligonucleotides used in reporter construction are p0491AFw (TATCCCGGGATAGCAAGACTGTTTGCATTACCAGG), p0491ARv (TATGGTACCATCGTTTTGTGAATATACAGGATCAGG), and p0491CFw (TATCCCGGGATACCATTTTTTCTTGACAGTACAAATCC).
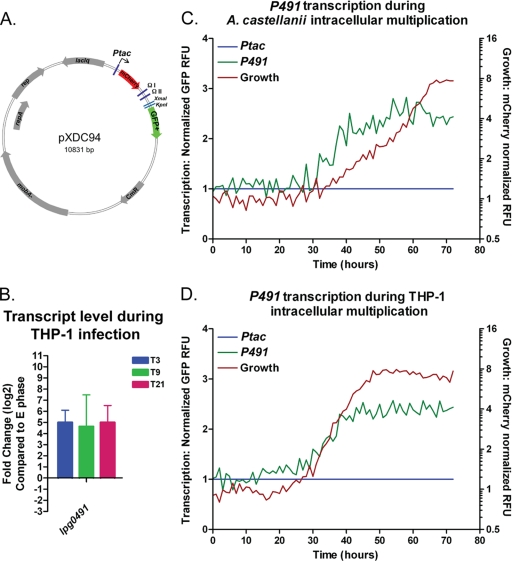
FIG. 8.
Transcription during intracellular growth. Map of pXDC94 transcriptional reporter plasmid (A). Reporter fragments were cloned using XmaI and KpnI enzyme sites (A). Following the infection of THP-1 macrophages with wild-type strain KS79, transcripts of lpg0491 were measure by qRT-PCR at 3, 9, and 21 h postinfection and compared to those of wild-type bacteria during the exponential growth phase (E phase) (B). Infection of A. castellanii and THP-1 macrophage cells in the context of a microplate spectrofluorometer was performed as described previously (36). During growth in both A. castellanii (C) and THP-1 cells (D) cells of the wild-type strain (GAH349) harboring the P491 reporter plasmid (pGAH140) were monitored for intracellular replication (mCherrry-normalized RFU on the secondary y axis; red line) and GFP transcription from P491 reporter (on primary y axis; green line) alongside the wild type (GAH348) harboring pXDC95, from which GFP is Ptac transcribed. Transcription data (primary axis; green [P491] and blue [Ptac] lines) (C and D) was normalized to growth (mCherry; secondary axis red line) and Ptac (GFP) transcription, as described in Materials and Methods.
ClustalW alignments and tree formation.
Amino acid alignments and phylogenetic trees were constructed based on ClustalW pairwise alignment algorithms on a BLOSUM 30 matrix using MacVector software version 7.2.3. The phylogenetic tree of arginine repressor homologs corresponds to the following GenBank accession numbers (in no particular order): Lactococcus lactis subsp. lactis Il1403 (NP_267015.1), Streptococcus pneumoniae TIGR4 (NP_345670.1), Bacillus subtilis (BAA12578.1), Bacillus stearothermophilus (1B4AC), Listeria innocua Clip11262 (NP_470740.1), Staphylococcus aureus subsp. aureus N315 (NP_374634.1), Streptococcus pyogenes M1 group A streptococcus (GAS) (NP_269620.1), Haemophilus influenzae 86-028NP (AAX88207.1), Pasteurella multocida subsp. multocida strain Pm70 (NP_245486.1), Escherichia coli strain K-12 (AAC76269.1), Vibrio cholerae O1 biovar eltor strain N16961 (NP_230085.2), Chlamydophila pneumoniae J138 (NP_300252.1), Salmonella enterica serovar Typhimurium LT2 (P_463323.1), Thermotoga maritima (Q9WW19.1), Mycobacterium leprae TN (NP_302004.1), Mycobacterium tuberculosis CDC1551 (NP_336150.1), and Pseudomonas aeruginosa PAO1 (NP_249584.1).
Microarray procedure.
The L. pneumophila microarray has been previously described (GLP7283) (36). Bacterial growth, RNA isolation, and cDNA preparation were performed as described previously (36). Bacterial genomic DNA (gDNA) was used as the reference channel on each slide to allow comparison of each time point and different samples (79). Five micrograms of genomic DNA was labeled with amino-allyl dUTP by using the Klenow fragment and random primers (Invitrogen) at 37°C for 18 h (22). DNA was subsequently coupled to the succinimidyl ester fluorescent dye Alexa Fluor 546 (for cDNA) or Alexa Fluor 647 (for gDNA) (Invitrogen) following the manufacturer's protocols. Hybridization and data acquisition were performed as previously described (36). Local background was removed from spot signal intensity, and normalization was carried out by calculating the fraction over the total signal intensity in both channels as previously described (22). Signal levels that were lower than background were filtered out. Statistical analysis between test and control conditions was performed using unpaired one-tailed Student's t test. Genes were considered as differentially expressed if they demonstrated a ratio to the control value of log2 ±1.5-fold with a P value of <0.005. Hierarchical clustering diagrams were produced using the TIGR Multiexperiment Viewer (MeV) (69).
Kinetic measurements of growth and transcription.
Kinetic measurements were obtained using a TECAN Infinite M200 fluorescence plate reader as described previously (36). Preparation of growth assays in liquid CDM is described above, and the method used is identical to that for assaying transcription during growth in CDM. A. castellanii was maintained in PYG (peptone-yeast extract-glucose) medium as described previously (29). The protocol for assaying L. pneumophila intracellular multiplication in a microplate spectrofluorometer in A. castellanii at a multiplicity of infection (MOI) of 1.0 and the human macrophage cell line THP-1 at an MOI of 0.10 has been described previously, and the experiments were conducted following previously established parameters (36). Measurement of transcription was performed on strains bearing plasmids derived from pXDC94, which contains a copy of the gene encoding the red fluorescence protein mCherry under Ptac control and a promoterless copy of a gene encoding green fluorescent protein (GFP). Bacterial growth was measured by the production of mCherry from a Ptac promoter induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an excitation of 580 nm and an emission of 620 nm. Transcription was measured by the production of green fluorescent protein (GFP) at an excitation of 485 and an emission of 520. For each strain background or condition tested, transcription from a control plasmid, pXDC95, from which mCherry and GFP are transcribed from Ptac induced with 0.5 mM IPTG, was also analyzed. Transcriptional data analysis during intracellular growth and growth in liquid medium was performed as follows. Relative fluorescence units (RFU) for GFP and mCherry triplicate samples for each time point were averaged, averaged GFP RFU were divided by averaged mCherry RFU, and the value from each time point was normalized (i.e., divided) by the value of Ptac-GFP/Ptac-mCherry obtained from the pXDC95 control for each strain background or condition. This process of data analysis allows measurement of transcription independent of bacterial growth. Kinetic experiments were performed three times, and the data shown are from one representative experiment. Data points reflect the average value of three biological replicates.
Quantitative real-time PCR during infection.
The THP-1 (ATCC TIB-202) cells were maintained in Advanced RPMI (Invitrogen) medium supplemented with 10% (vol/vol) fetal calf serum (Gibco) and 2 mM l-glutamine (Cellgro). A stock culture of these cells was maintained as monocyte-like, nonadherent cells at 37°C in an atmosphere containing 5% (vol/vol) CO2. For macrophage infection, cells were seeded at 2 × 107 cells per well in petri culture dishes and were differentiated by addition of 10−7 M phorbol 12-myristate 13-acetate for 48 h. Before infection, macrophages were treated with antibody raised against the major outer membrane protein (MOMP) of L. pneumophila for 30 min. Bacteria were grown overnight shaking in AYE at 37°C and were then added to the cell monolayer at an MOI of 1 and centrifuged for 5 min at 800 × g to synchronize bacterial uptake. After incubation for 2 h at 37°C, the infected cells were washed three times with phosphate-buffered saline (PBS), pH 7.4, and 18 ml of fresh complete RPMI medium containing 100 μg/ml gentamicin was added to each well. After incubation for 1 h at 37°C, cells were washed three times with PBS and the cells were either harvested at 3 h postinfection (T3) or incubated with 18 ml of fresh complete RPMI medium for a further 6 h (T9) or 18 h (T21). At each time point, cells were scrapped in 2 ml PBS and 100 μl was removed to determine the CFU. Samples were then centrifuged for 5 min at 1,000 × g, and the pellet was lysed in 2 ml TRIzol (Invitrogen) and stored at −80°C. RNA was isolated by using TRIzol reagents as described by manufacturer (Invitrogen). The RNA was subsequently treated with DNase I (Invitrogen) for 1 h at 37°C. The DNase was then inactivated by incubation at 75°C for 5 min and acid phenol-chloroform (Ambion) extracted, and the RNA was precipitated with NaAc-ethanol. The purity and quantity of RNA was assessed by spectrophotometry. cDNA was synthesized in triplicate using Superscript II (Invitrogen) with random hexamers (Invitrogen), according to the manufacturer's instructions. For each sample, a no-reverse transcriptase reaction served as a no-template control (NTC). qPCRs were performed using the Applied Biosystems StepOne Plus 96-well reverse transcription (RT)-PCR system (qRT-PCR) with Power Syber green PCR master mix following the manufacturer's instructions (Applied Biosystems). For each qPCR run, the calculated threshold cycle (CT) was normalized to the CT of the internal control 16S rRNA gene amplified from the corresponding sample and the fold change was calculated as previously described (47). qRT-PCR experiments were performed three times, and the data shown are from one representative experiment. Data points reflect the average value of three biological replicates. The oligonucleotides used in qRT-PCR measurements were lpg0491qF (CAACCAAGCGATAGAAGCTTTAATC) and lpg0491qR (CCTTGTGCCCCATCCATAAG).
RESULTS
Conservation of the L. pneumophila ArgR protein.
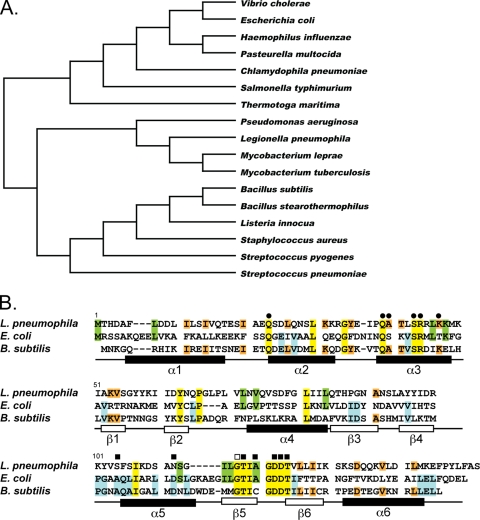
Mutational analysis of putative regulatory genes whose transcription is affected by σS resulted in the discovery that the lpg0490 open reading frame (ORF) is required for efficient intracellular multiplication in the unicellular host A. castellanii but not in the macrophage cell line THP-1 (36). The lpg0490 mutant intracellular growth defect was restored by expressing the gene in trans from an IPTG-inducible promoter, thus demonstrating its significance during A. castellanii infection (36). Based on conservation of DNA-binding and l-arginine-binding domains in the predicted protein sequence, lpg0490 was annotated as the L. pneumophila arginine repressor (18, 36). Arginine repressor proteins, termed ArgR or AhrC based on their similarity to either the E. coli or the B. subtilis proteins, respectively, are common in bacteria (19, 46, 55). To determine the relative similarity of the lpg0490 gene product to the arginine repressors of other bacteria, we used ClustalW to produce a phylogenetic tree of ArgR amino acid sequences from genetically diverse bacterial species (Fig. 1A). The identity of Lpg0490 to the arginine repressors of the bacteria analyzed ranged between 28% and 35%. Overall, the Lpg0490 sequence was slightly more similar to the arginine repressors of both mycobacteria and Gram-positive species such as B. subtilis, (30% identity and 54% similarity) than those from Gram-negative bacteria such as E. coli (28% identity and 52% similarity).
FIG. 1.
Conservation of ArgR. A phylogenetic tree of arginine repressor homologs from diverse bacterial species was produced from a ClustalW alignment (A). GenBank accession numbers for all proteins can be found in Materials and Methods. Shown is the ClustalW alignment of amino acid sequences from E. coli and B. subtilis arginine repressors, ArgR and AhrC, respectively, with the translated sequence from lpg0490 of L. pneumophila (B). Residues conserved among all three bacterial species are highlighted in yellow, those conserved between E. coli and L. pneumophila are in green, those conserved between B. subtilis and L. pneumophila are in orange, and those conserved between E. coli and B. subtilis are in blue (B). Residues required for DNA-binding (•), l-arginine binding (▪), or oligomerization (□) demonstrated by mutational analysis in either E. coli or B. subtilis are indicated above the alignment (B) (8, 41, 76, 82, 83). The general structural features from B. stearothermophilus are shown below the amino acid alignments (B) (62).
The structure and function of arginine repressor proteins have been studied extensively in many bacteria, including E. coli and B. subtilis. Mutational analysis has demonstrated the critical amino acid residues required for ArgR function (41, 52, 82). Alignment of the putative L. pneumophila arginine repressor, Lpg0490, shows that the amino acids required for DNA binding, oligomerization, and l-arginine binding of E. coli and B. subtilis arginine repressors are conserved (Fig. 1B). Based on these observations, lpg0490 will be called the argR gene.
ArgR is not required for growth in media containing l-arginine biosynthetic precursors.
Arginine auxotrophy is common among all of the strains of L. pneumophila that have been tested (26), including Philadelphia-1, the strain used in this study. Although the chemical composition of the LCV is unknown, L. pneumophila's ability to grow intracellularly, despite its arginine auxotrophy (26), suggests that sufficient l-arginine is available in the LCV to support growth. Alternatively, L. pneumophila may be able to utilize the terminal arginine biosynthetic precursors l-ornithine and citrulline during growth (26). It is not known if the ability to utilize l-arginine precursors is required for intracellular replication. If mutation of argR resulted in an inability to grow in l-arginine or its biosynthetic precursors, this might explain the reported intracellular growth defect (36).
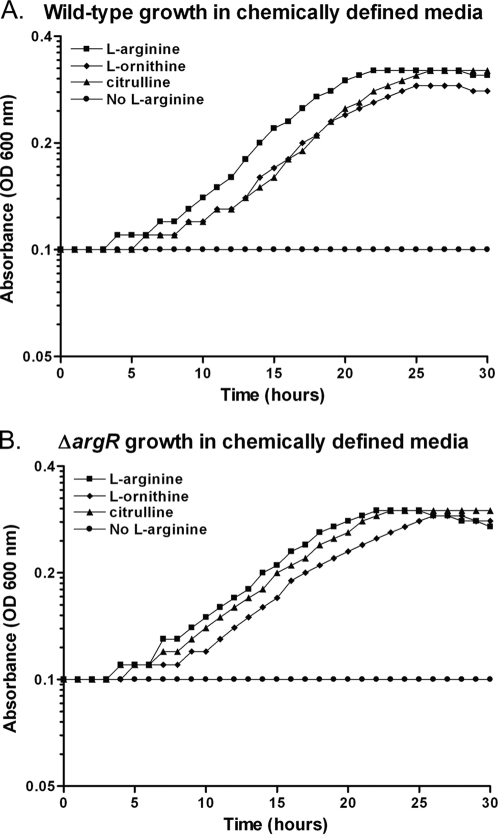
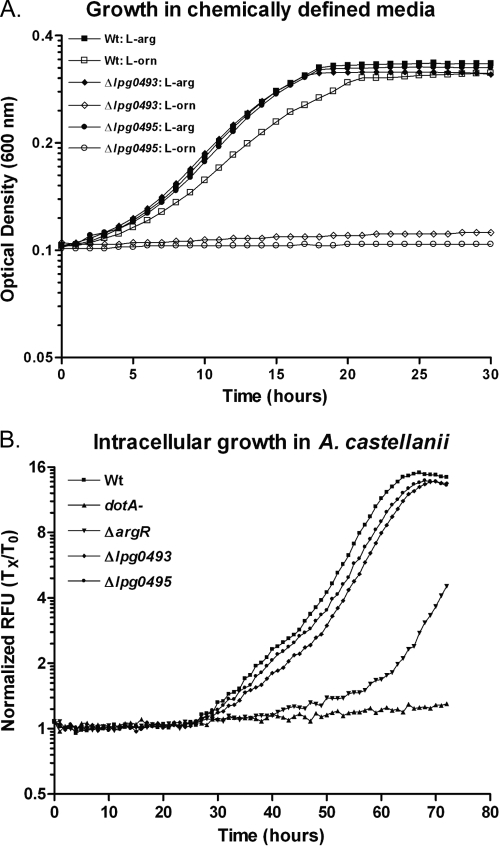
To test the ability of a ΔargR mutant to grow in media containing l-arginine or its biosynthetic precursors, chemically defined medium lacking l-arginine (CDM-A) was prepared (64). L. pneumophila strains were grown in CDM-A or supplemented with l-arginine (1.0 mg/ml), l-ornithine (1.0 mg/ml), or citrulline (1.0 mg/ml). As expected, wild-type Legionella (KS79) is unable to grow in the absence of l-arginine, but the addition of l-arginine or its precursors restored growth (Fig. 2A). The ΔargR mutant GAH280 grew similarly to the wild type in CDM-A containing either l-arginine or its biosynthetic precursors (Fig. 2B). Therefore, the intracellular growth defect of the ΔargR mutant does not result from an inability to utilize l-ornithine or l-citrulline as l-arginine precursors.
FIG. 2.
Growth of wild-type and ΔargR mutant strains in CDM. Bacterial growth was monitored in a TECAN Infinite M200 plate reader at an absorbance of 600 nm. At time zero, bacteria were washed and diluted back to an OD of 0.10 in chemically defined medium (CDM) prepared either with l-arginine (1.0 mg/ml) (▪), without l-arginine (no l-arginine) (•), without l-arginine but including l-ornithine (1.0 mg/ml) (⧫), or without l-arginine but including citrulline (1.0 mg/ml) (▴). Absorbance (OD600) was monitored for 30 h under the four CDM growth conditions at 37°C for both the ΔargR mutant (GAH280) (B) and its isogenic parent (KS79), the wild type (A).
Global gene expression analysis of an L. pneumophila ΔargR mutant.
Deletion of argR results in a significant defect in L. pneumophila intracellular multiplication (36). In other bacterial species, ArgR responds rapidly to changes in the intracellular concentration of l-arginine by binding DNA as a transcriptional regulator (78). Using global gene expression analysis, we sought to identify L. pneumophila genes regulated by ArgR that contribute to the observed intracellular growth defect as well as those that could be used as reporters of the ArgR response to l-arginine.
RNA was extracted from a ΔargR mutant (GAH280) and its isogenic, wild-type (WT) parent strain (KS79) during exponential and postexponential growth in rich media. The transcriptional profiles were compared by microarray analysis as previously described (36). For the purpose of this study, significantly affected genes were defined as having a log2 ratio of mutant to wild-type ± 1.5 (3-fold) and with a P value of ≤0.005. The steady-state transcript level of 116 genes was significantly affected by the deletion of the argR gene during growth in rich media (Fig. 3A; see Table S1 in the supplemental material). The transcript levels of 12 genes were affected during the exponential phase, 11 of which were increased in the mutant. An increase in transcript level in the mutant background compared to the wild type suggests that the ArgR protein acts negatively on the transcription of these 11 genes. The transcript levels of 111 genes were affected during the postexponential phase, 51 were increased in the mutant compared to the wild type, while the transcripts of another 60 genes were reduced in the mutant background. Reductions in gene transcripts in the mutant compared to the level in the wild type suggest that the ArgR protein acts positively on these genes. Seven of the 11 genes negatively affected by argR during the exponential phase are also negatively affected during the postexponential phase, representing the only set of genes that is similarly regulated during both phases of growth (Fig. 3B, red vertical bar). There are no genes that are positively affected by ArgR in one growth phase and negatively in the other. The changes in transcript levels of affected genes could reflect either the direct interaction of ArgR with their promoters or could occur indirectly through ArgR-affected regulators. Although additional experiments are required to determine if ArgR is acting directly on specific gene targets, we were able to identify putative ArgR operator sites in the promoter regions of many of affected genes identified from this microarray (see Table S2 in the supplemental material).
FIG. 3.
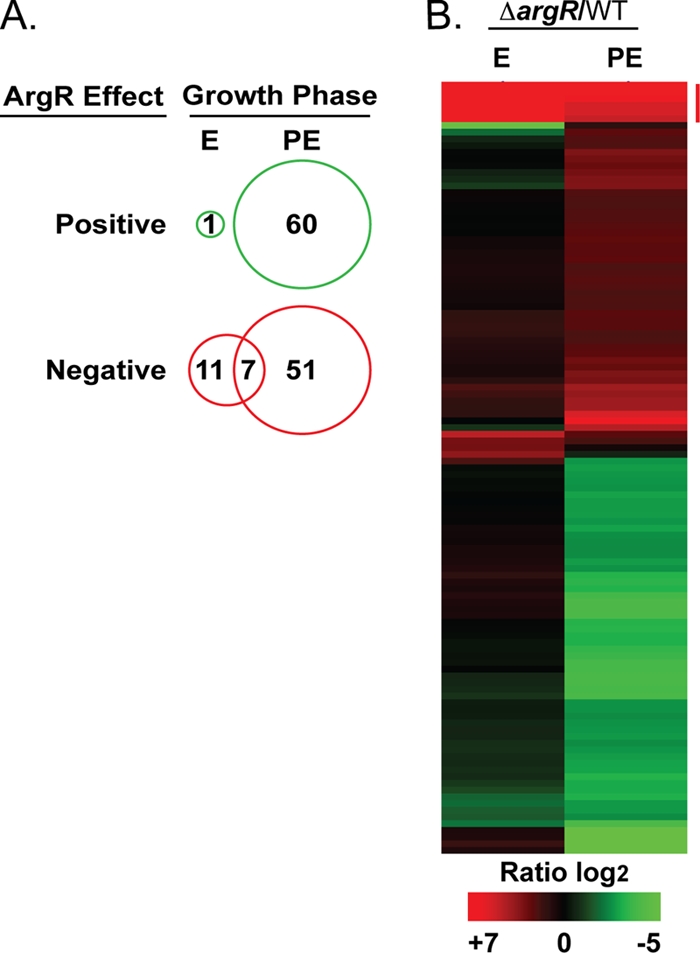
The L. pneumophila ΔargR microarray. Analysis of the transcriptional profile of a ΔargR mutant (GAH280) in comparison with its isogenic parent, wild-type (WT) strain KS79, during the exponential (E) and postexponential (PE) growth phases in AYE rich medium. ArgR-affected genes, defined by a mutant/wild-type log2 ratio ± 1.5 and P ≤ 0.005, were clustered using TIGR MeV (B). Depiction of the number of genes positively or negatively affected by ArgR in either phase of growth is shown as a Venn diagram (A). Genes negatively affected during both phases of growth are indicated by a vertical red bar (B).
Categories of ArgR-affected genes.
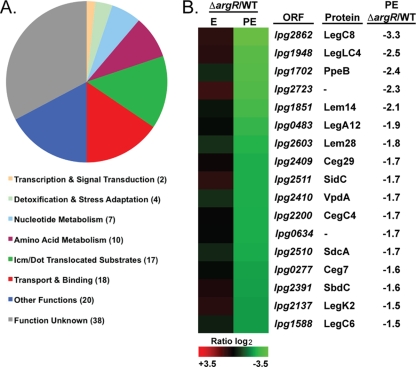
The global effects of the argR deletion were analyzed by determining the number of genes affected within specific functional categories (18) (Fig. 4A; see Table S1 in the supplemental material). In addition to amino acid metabolism, which was predicted to be affected by ArgR, the categories transport and binding, Icm/Dot-translocated substrates, nucleotide metabolism, and detoxification and stress adaptation also included a notable number of affected genes (Fig. 4A). The deletion of the argR gene affected 18 genes categorized with transport and binding (Fig. 4A). The majority of genes categorized as transport and binding genes were negatively affected by ArgR (see Table S1 in the supplemental material) including the most highly ArgR-regulated genes in the genome, lpg0491 to lpg0493 (exponential-phase, log2 ratio of +7.6 to 7.3), which encode a putative amino acid ABC transporter (18). In the postexponential phase, ArgR negatively affected components of the type IVA secretion system and TFBSS, which were also categorized with transport and binding (18). Two members of the Lvh Type IVA secretion system, lvhB8 (lpg1249, log2 ratio of 1.7) and lvHB10 (lpg1247, log2 ratio of 1.8), and three members of the Icm/Dot TFBSS, icmR (lpg0443, log2 ratio of 1.8), icmQ (lpg0444, log2 ratio of 1.7), and icmG (lpg0452, log2 ratio of 1.6) were significantly affected in the absence of ArgR. Expression of these components of the Icm/Dot TFBSS of L. pneumophila is required for efficient intracellular replication (56, 65, 66, 71). Also included the category of transport and binding were the genes most positively affected by ArgR in the genome, lpg0174 and lpg0175 (postexponential phase, log2 ratio of −5.5 and −4.0, respectively), which are predicted to encode pyoverdine biosynthetic proteins, PvcA and PvcB (18). In pseudomonads, pyoverdine is a powerful iron(III) scavenger and transporter (75), and in L. pneumophila, the pvcA/B genes are regulated by iron concentration (31). As seen previously, deletion of the pvcA and pvcB genes did not affect L. pneumophila's ability to replicate in A. castellanii (3; this study and data not shown).
FIG. 4.
Categories of ArgR-affected genes. The distribution of ArgR-affected genes in functional categories is illustrated in a pie chart, and the number of affected genes in each category is shown in parentheses (A). Heat mapping depicts ArgR-affected Icm/Dot translocated substrates along with the ORF, protein names, and the log2 ratio (B).
Deletion of the argR gene affected the transcript levels of 17 genes in the category Icm/Dot-translocated substrates (Fig. 4A and B; see Table S1 in the supplemental material). The category Icm/Dot-translocated substrates includes bacterial proteins demonstrated to be translocated into host cells during Legionella infection in an Icm/Dot TFBSS-dependent manner (9, 15, 37, 44, 53, 54, 61). To date, the translocation of 140 putative Icm/Dot substrates has been proven; however, the majority of their functions are unknown (9, 15, 37, 44, 53, 54, 61). The transcript level of 17 Icm/Dot-translocated substrates was reduced by the deletion of argR (Fig. 4B); thus, the ArgR regulator has a positive affect on the transcription of 12% of the members of this category, either directly or indirectly. Although many of the genes encoding Icm/Dot-translocated substrates are individually dispensable for intracellular multiplication, some have been shown to affect host cell trafficking. Understanding the specific contribution of Icm/Dot substrates and other ArgR-affected genes to L. pneumophila intracellular multiplication will require future investigation.
Genomic organization of the L. pneumophila ArgR regulon.
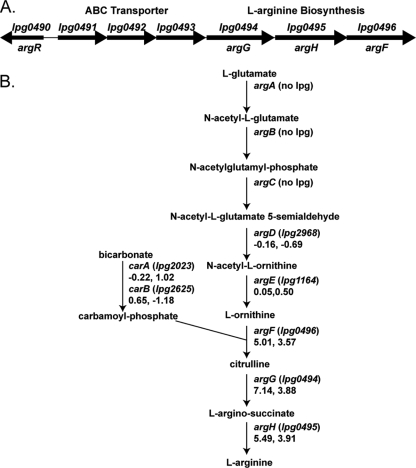
ArgR has predicted binding sites based on numerous studies in other bacteria. In E. coli, the enzymes encoded by the genes argA to argH synthesize l-arginine from l-glutamate in a process that requires the carA and carB gene products to form carbamoyl-phosphate (46) (summarized in Fig. 5 B). In E. coli, the genes required for l-arginine biosynthesis are distributed throughout the genome and transcriptionally repressed by ArgR in the presence of l-arginine, comprising the ArgR regulon (46). L. pneumophila lacks homologs of argA, argB, and argC, which explains its l-arginine auxotrophy (18, 26). This study reveals that in L. pneumophila, the mutation of argR does not significantly affect the transcript level of homologs of the predicted ArgR-regulated genes argD (lpg2968), argE (lpg1164), carA (lpg2023), and carB (lpg2625) (Fig. 5B; log2 ratio microarray data shown under gene names). Deletion of argR in L. pneumophila does, however, affect the transcript level of genes predicted to encode the terminal steps of l-arginine biosynthesis from l-ornithine; argF (lpg0496), argG (lpg0494), and argH (lpg0495), whose transcript levels are up by as much as 100-fold in the mutant (Fig. 5A; see Table S1 in the supplemental material).
FIG. 5.
Genomic organization of l-arginine biosynthesis in L. pneumophila. The ORF encoding ArgR, lpg490, is divergent from a region spanning lpg0491 to lpg0496, in which the genes exhibit strong negative regulation by ArgR (A). lpg0491 to -0493 are predicted to encode an amino acid ABC transporter, and lpg0494 to -0496 are predicted to encode the final steps of l-arginine biosynthesis (A). The biosynthetic pathway for the conversion of l-glutamate to l-arginine is shown with the L. pneumophila ORF associated with each enzymatic step, if present, in parentheses (B). The ΔargR/WT log2 ratio is shown at each step below the ORF for the exponential and postexponential phases (B).
In L. pneumophila, the genes predicted to encode the terminal steps of l-arginine biosynthesis are organized in one region of the genome along with a putative amino acid ABC transporter (lpg0491 to lpg0496). The genomic organization of this region is conserved among all sequenced L. pneumophila strains (Philadelphia-1, Corby, Lens, and Paris). The transcript levels of lpg0491 to lpg0496 are increased by the deletion of argR by between 240- and 32-fold (lpg0492 and lpg0496, respectively) in the exponential phase and between 36- and 12-fold (lpg0492 and lpg0496, respectively) in the postexponential phase (see Table S1 in the supplemental material). Thus, lpg0491 to lpg0496 constitute the most highly ArgR-repressed region of the genome and the only set of genes similarly regulated during both phases of growth (Fig. 3B, vertical red bar). In addition, lpg0491 to lpg0496 are directly downstream and divergent from the open reading frame (ORF) encoding ArgR, lpg0490 (Fig. 5A). This organization is in stark contrast to the genomic organization of l-arginine biosynthesis in bacteria such as E. coli, in which the genes are in diverse locations throughout the genome and not proximal to the gene encoding the ArgR repressor (11, 17, 39, 45, 50, 62, 82).
To determine which genes in this region may be required for growth of L. pneumophila under different conditions, we replaced selected genes with antibiotic selection markers by allelic exchange (36). Deletion of either the putative ATPase of the predicted transporter (lpg0493) or the argH homolog (lpg0495), which is predicted to encode the terminal step of arginine biosynthesis, rendered bacteria unable to grow in CDM in the absence of l-arginine, even if l-ornithine (Fig. 6A) or citrullline (data not shown) were provided in its place. The deletion of the open reading frames lpg0493 and lpg0495 did not result in a significant reduction in intracellular growth compared to the wild type (Fig. 6B). Addition of l-arginine to the infection buffer did not alter the wild-type or mutant strains' capacity for intracellular replication (data not shown). Therefore, this region is required for the utilization of l-arginine biosynthetic intermediates, as expected, but is not required for intracellular multiplication, from which we can infer that sufficient l-arginine is available in the LCV to support bacterial growth. Because the promoter of the region from lpg0491 to -0496 of the L. pneumophila genome is under strong negative control by ArgR and its transcription is predicted to respond to l-arginine, we chose it to use it as a reporter of ArgR function in subsequent studies.
FIG. 6.
Analysis of growth of the Δlpg0493 and Δlpg0495 mutants. Growth of the wild-type (Wt) strain KS79 and the Δlpg0493 (GAH318) and Δlpg0495 (GAH319) mutant strains in CDM containing l-arginine (1.0 mg/ml) (▪, ⧫, •, respectively) or in CDM without l-arginine but including l-ornithine (1.0 mg/ml) (□, ⋄, ○, respectively) was measured by absorbance at 600 nm in a TECAN Infinite M200 plate reader at 37°C for 30 h (A). The wild-type and mutant strains bearing the GFP+ plasmid pXDC31, wild-type strain GAH281 (▪), and the dotA (GAH282) (▴), ΔargR (GAH286) (▾), Δlpg0493 (GAH339) (⧫), and Δlpg0495 (GAH340) (•) mutant strains were used to infect A. castellanii, and intracellular growth was monitored by GFP fluorescence in a TECAN Infinite M200 fluorescence plate reader held at 30°C for 72 h following previously described methods (36) (B).
Transcription from the lpg0491 promoter is repressed by ArgR in the presence of l-arginine.
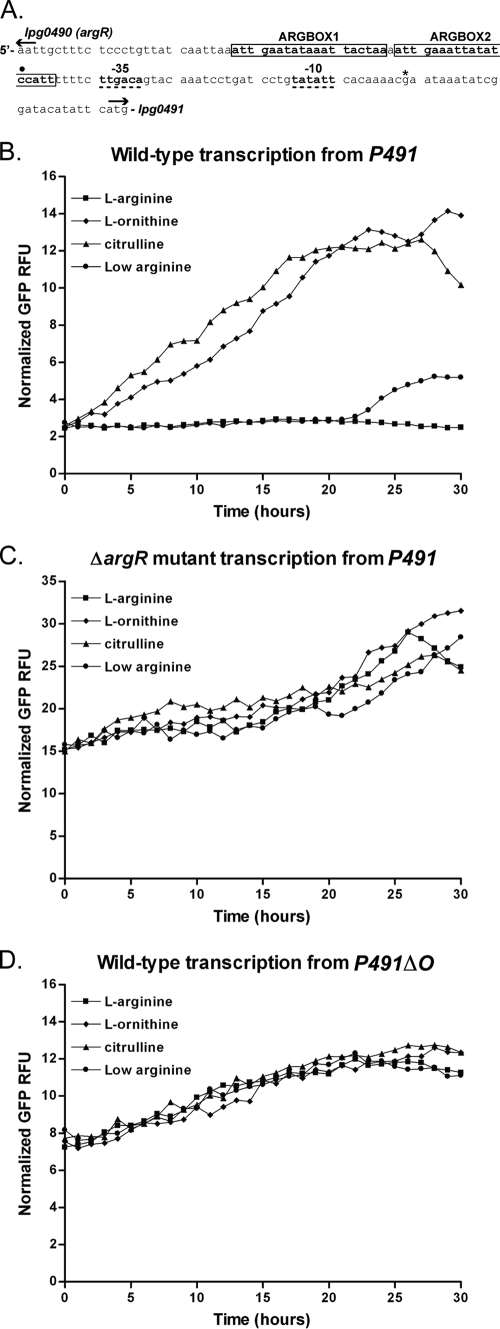
Global gene expression analysis demonstrated that transcripts of genes lpg0491 to lpg0496 are highly increased in a ΔargR mutant compared to the wild type (see Table S1 in the supplemental material). We wanted to monitor transcription from this region during growth under different environmental conditions, such as low l-arginine concentration in CDM and during intracellular growth. To accomplish this goal, the 129-bp intergenic region between argR (lpg0490) and lpg0491 was examined for possible regulatory elements (Fig. 7A). Based on its similarity to the E. coli consensus sequence (10), a putative σ70 promoter-binding site was identified that ends 34 bp upstream of the lpg0491 ORF (Fig. 7A, dashed underline). The ArgR repressor is known to bind to a conserved DNA-operator site that is composed of one or more degenerate palindromic sequences (wnGAATwwwwATTCAnw), which are called ARG boxes (13, 84). We identified two putative ARG boxes in the regulatory region preceding lpg0491 (Fig. 7A), which we predict to be an ArgR operator site.
FIG. 7.
Transcription from the lpg0491 promoter region. The intergenic region between lpg0490 and lpg0491 contains the putative −10 and −35 sites of the lpg0491 promoter (bold and dashed underline) and two putative ARG boxes (bold with rectangular outline) (A). The translational start sites for lpg0490 or lpg0491 are depicted using divergent arrows (A). The terminal 3′ base of transcriptional reporter fragments P491 and P491ΔO is depicted by an asterisk, and the terminal 5′ base of the transcriptional reporter fragment P491ΔO is depicted by a closed circle (•) (A). The 5′ end of the P491 transcriptional reporter fragment is 413 bp upstream from the translational start of lpg0491 and is beyond the scope of this figure. The transcriptional reporter fragments P491 and P491ΔO were cloned into pXDC94 (Fig. 8A) to produce pGAH140 and pGAH141, respectively. The growth in CDM (mCherry) (see Fig. S1 in the supplemental material) and GFP-reported transcription from the resulting wild-type and ΔargR strains was monitored in the context of a TECAN Infinite M200. CDM was prepared either with l-arginine (1.0 mg/ml) (▪), with a reduced concentration of l-arginine (“Low arginine”; 0.1 mg/ml) (•), without l-arginine but including l-ornithine (1.0 mg/ml) (⧫), or without l-arginine but including citrulline (1.0 mg/ml) (▴) for 30 h at 37°C (B, C, and D). Transcription data are shown for wild-type (GAH349) (B) and ΔargR mutant (GAH355) (C) strains bearing P491 reporter plasmid (pGAH140) and a wild-type strain (GAH350) bearing the P491ΔO reporter plasmid (pGAH141) (D). All conditions have been normalized to growth (mCherry) and Ptac transcription as described in Materials and Methods.
Transcriptional reporter gene fusions were designed to include the entire lpg0491 regulatory region (P491) or only the putative promoter without the predicted ArgR operator site (P491ΔO). Both fragments have the same terminal 3′ base (Fig. 7A, asterisk), 22 bp upstream the lpg0491 start codon. However, the fragments differ in their 5′ ends, which are 413 bp (outside of the data presented in Fig. 7A) and 71 bp (Fig. 7A, filled circle) upstream from the lpg0491 start codon for P491 and P491ΔO, respectively. The fragments were cloned into pXDC94, a plasmid containing a promoterless copy of the green fluorescent protein (GFP) gene to report the transcriptional activity of the cloned fragment (shown in Fig. 8A). The same plasmid also includes a copy of the gene encoding the red fluorescence protein mCherry under the control of the Ptac promoter and lacIq repressor. When IPTG is added to the growth medium, the levels of mCherry fluorescence are proportional to the number of bacteria.
Reporter plasmids derived from pXDC94, containing P491 (pGAH140) or P491ΔO (pGAH141), were introduced into the wild-type (KS79) and ΔargR (GAH280) mutant strains. The strains were grown in either CDM containing l-arginine or CDM in which l-arginine was replaced with l-ornithine, citrulline, or 10-fold less l-arginine (low arginine) in wells of a 96-well plate. The levels of absorbance (600 nm) as well as GFP and mCherry fluorescence were measured every hour for 30 h. In addition, measurements were made with isogenic strains bearing the control plasmid pXDC95, in which Ptac promotes the transcription of both GFP and mCherry fluorophores. The transcriptional profiles in Fig. 7 have been normalized to bacterial growth by mCherry (see Fig. S1 in the supplemental material), and the data presented are normalized to the levels of GFP fluorescence from an IPTG-induced Ptac promoter, as described in Materials and Methods. Thus, the levels of transcription indicated by the ratio of mCherry to GFP fluorescence shown in Fig. 7B, C, and D are independent of bacterial growth.
When wild-type L. pneumophila is grown in CDM that is replete with l-arginine (1.0 mg/ml), no increase in transcription can be measured from the P491 fragment (Fig. 7B). However, when l-arginine is absent and l-ornithine or citrulline is provided, an approximate 12-fold increase in transcription is measured from this region that begins following time zero (Fig. 7B). If the CDM contains only one-tenth of the normal concentration of l-arginine (0.1 mg/ml), no increase in transcription is observed until more than 20 h into growth, which is presumably when the CDM begins to be depleted for l-arginine (Fig. 7B). The growth of cultures, measured by mCherry, containing reduced l-arginine (“low arginine”) demonstrates a reduction in growth rate at the time corresponding to increased transcription (20 h) (see Fig. S1 in the supplemental material). Alternatively, when the same CDM growth experiment is conducted in the ΔargR mutant background, transcription from P491 is approximately 12-fold higher at time zero than in the wild type. Transcription from P491 in the ΔargR mutant background remains constitutively high regardless of the concentration of l-arginine (Fig. 7C). It is interesting to note that growth of the ΔargR mutant was more significantly reduced under the low-arginine condition than that of the wild type (see Fig. S1 in the supplemental material). The reporter fragment that does not include the putative ArgR operator site, P491ΔO, was transcriptionally active in the wild type independent of the concentration of l-arginine (Fig. 7D).
Taken together, these data show that transcription from the lpg0491 promoter region is repressed when the environment is replete with l-arginine and this repression requires a wild-type copy of the argR gene. Data from the P491ΔO fragment demonstrate that the lpg0491 promoter resides between 71 and 22 bp upstream from the start codon of the gene. These data also demonstrate that deletion of the region containing the putative ArgR operator site results in transcription that is not l-arginine repressible (Fig. 7D). Because the P491 region is sensitive to the l-arginine concentration, in an ArgR-dependent manner, it was subsequently used to determine if l-arginine availability is a transcriptional regulatory signal during intracellular growth.
Transcription of genes coregulated by argR and l-arginine during intracellular growth.
Deletion of the argR gene results in a reduced capacity for L. pneumophila intracellular growth in the unicellular protozoan A. castellanii (Fig. 6B) (36). Transcriptional repression of the promoter region preceding lpg0491 (P491) is dependent upon ArgR and l-arginine (Fig. 6). These data suggested the possibility that ArgR responds to changes in the availability of l-arginine during intracellular growth. Having characterized the response of the lpg0491 promoter region to ArgR and l-arginine (Fig. 6), we sought to determine if transcription from this region was affected during L. pneumophila infection. Using reporter plasmids derived from pXDC94 and the normalization method described in the previous section, we measured transcription from the P491 promoter region, during L. pneumophila growth in both A. castellanii and THP-1 macrophage host cells (Fig. 8B and D). Due to the low number of bacteria used in this infection method, the transcription of GFP was not detectable prior to the appearance of measurable levels of bacterial replication, which occurs at approximately 30 h postinfection for A. castellanii and 24 h postinfection for THP-1 cells. After this time, the pattern of P491 transcription can be monitored by GFP production. Following measurable intracellular growth, transcription from P491 increased over the next 5 h by between 2- and 3-fold in both host cell types (Fig. 8B and D).
To determine if derepression of ArgR-repressed genes occurs at earlier time points, we measured transcripts of lpg0491 by qRT-PCR at 3, 9, and 21 h following the infection of THP-1 macrophages compared to exponential-phase growth in rich medium. By 3 h postinfection, transcripts of lpg0491 were increased by a log2 ratio of approximately 5 (Fig. 8B), which is similar to the level of derepression observed in a ΔargR mutant compared to the wild type from the microarray (see Table S1 in the supplemental material). The lpg0491 transcript levels were maintained through the final measurement at 21 h postinfection (Fig. 8B). Therefore, an increase in transcription during intracellular growth can be observed by either the fluorescence-based (Fig. 8C and D) or qRT-PCR (Fig. 8B) method. These data, in conjunction with the CDM experiments, demonstrate that genes whose transcription is responsive to ArgR and the concentration of l-arginine are derepressed during intracellular multiplication.
DISCUSSION
Intracellular replication of L. pneumophila requires the establishment of a specialized compartment within host cells, which occurs through a series of ordered steps that interfere with normal vesicular trafficking (33, 35). L. pneumophila's ability to both survive and control this process is predicted to necessitate the sensing of and response to a myriad of signals (59). Shortly after its entry into host cells, the L. pneumophila-containing phagosome evades fusion with the lysosome to establish a prereplicative vacuole called the LCV (40). Determining the chemical and nutrient composition intracellular compartments and therefore identifying molecules that could signal this lifestyle change to a bacterium residing in the LCV is inherently challenging and may have to be achieved by proxy (5). Following our initial report that mutation of the L. pneumophila argR homolog resulted in a significant defect in intracellular growth (36), we sought to determine if the l-arginine availability is a previously unrecognized aspect of infection. In this study, we applied microarray analysis to identify the L. pneumophila ArgR regulatory targets (Fig. 3), used a promoter tightly controlled by ArgR to report its function (Fig. 7A), demonstrated repression in response to l-arginine (Fig. 7B and C), and showed derepression during intracellular growth from as early as 3 h postinfection (Fig. 8). These data strongly suggest that the concentration of l-arginine in the LCV is a transcriptional regulatory signal sensed by ArgR and thus affects L. pneumophila gene expression during intracellular growth.
Utilization of ArgR for the regulation of genes that are not traditionally associated with l-arginine could be a possible explanation for the reduced intracellular growth of an argR mutant. The Icm/Dot type IVB section system is required for the evasion of phagolysosome fusion and establishment of the LCV (56, 66). The genes encoding IcmR and IcmQ reside in their own transcriptional units, and ArgR negatively affects their transcription in the postexponential phase (see Table S1 in the supplemental material). Based on the consensus sequence of E. coli ARG boxes (84) and the ARG boxes required for repression of lpg0491 (Fig. 5 and 6), we identified putative ARG boxes in the promoter regions of both icmR and icmQ (see Table S2 in the supplemental material). The impact of l-arginine-responsive repression of icm/dot genes is unclear, but it could be involved in the proper timing and assembly of the type IVB section system. The Icm/Dot system is responsible for the translocation of protein substrates into the host cell (20). Our investigation found that ArgR positively affected transcripts of 17 of the 140 genes encoding Icm/Dot-translocated proteins during the postexponential phase (Fig. 4; see Table S1 in the supplemental material). This accounts for more than 15% of the total ArgR regulatory affect and 28% of the genes positively affected by ArgR. The genes encoding these Icm/Dot substrates are widely distributed throughout the genome, which makes their coordinate regulation in response to the deletion of the argR gene notable. Although the paradigm of ArgR regulation is transcriptional repression through steric inhibition of RNA polymerase promoter binding (19, 84), there is significant precedent for gene activation by ArgR, which also occurs through binding to promoter-proximal ARG boxes (42, 45, 48, 49). Putative ARG boxes for Icm/Dot genes and their substrates as well other ArgR-affected genes were compiled (see Table S2 in the supplemental material) and used to develop a L. pneumophila ArgR consensus binding site (see Fig. S2 in the supplemental material). For 6 of the 17 ArgR-affected genes encoding Icm/Dot-translocated substrates, putative ARG boxes were identified. The protein substrates of the Icm/Dot secretion system have been predicted to affect host cell functions, such as endocytic trafficking, in a manner that permits the establishment of the LCV and intracellular replication. Based on the results from this study, it is possible that the l-arginine concentration is a signal for the positive regulation of translocated proteins required for infection.
l-Arginine metabolism is known to be significant for many prokaryotic and eukaryotic intracellular pathogens (14, 27, 38, 57, 67, 80). This is likely due, in part, to the importance of l-arginine in host cell metabolism, especially among pathogens that replicate within macrophages. l-Arginine utilization resides at the crossroads of macrophage lineage determination (7). When activated to become MI macrophages, by lipopolysaccharide (LPS) and the appropriate cytokines, the cationic transporters (CATs) that import l-arginine and the inducible nitric oxide synthase (iNOS) are transcriptionally activated (7). The result is preferential usage of l-arginine by the iNOS pathway and conversion of l-arginine to nitric oxide (NO) and citrulline, the outcome of which can be destruction of intracellular parasites (7). Although CAT induction results in increased intracellular l-arginine, it is not a likely source of l-arginine for pathogens that reside in a vacuole because active transport of l-arginine across the vacuolar membrane is required (5). MII macrophages activate arginase to convert l-arginine into l-ornithine and urea (7). The l-ornithine produced from the arginase pathway is used to make polyamines and proline, which aid the cell in proliferation and collagen production (7). Thus, arginine utilization is a critical determinant in the formation of an MI (killing) versus MII (healing) macrophage (7). In addition, it has been proposed that macrophages use nutrient deprivation as an intracellular antimicrobial mechanism (5). Precedence for the importance of l-arginine acquisition by an intracellular pathogen was first discovered in Listeria monocytogenes. An early study found that the arginine transporter arpJ was upregulated intracellularly (43), and more recently microarray analysis showed that transcripts of L. monocytogenes arginine biosynthetic genes increase during intracellular growth (14). In addition, ArgR activates the adi genes, required for acid tolerance, in L. monocytogenes and ΔargR mutants have a 10-fold reduction in survival during murine infection (67).
Here we have highlighted the potential significance of l-arginine availability during intracellular growth and presented the first analysis of the L. pneumophila ArgR regulon. Although this is a newly discovered aspect of intracellular gene regulation for L. pneumophila, it is not without precedent in other intracellular pathogens. Understanding how l-arginine availability functions as a transcriptional signal and determining its intracellular regulatory targets will require further investigation. Additional studies are also needed to fully elucidate the biochemical aspects, associations with other regulators, and involvement in intracellular replication of the L. pneumophila ArgR transcriptional regulator.
Supplementary Material
Acknowledgments
This work was supported by award AI064481 to H.A.S. G.A.H. was supported by T32 AI007161. S.P.F. was supported by a postdoctoral fellowship from the National Sciences and Engineering Research Council of Canada.
We thank Gloria Recio for technical assistance. We also thank Courtney Plumlee for productive scientific discussions.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Khodor, S., S. Kalachikov, I. Morozova, C. T. Price, and Y. Abu Kwaik. 2009. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect. Immun. 77:374-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman, E., and G. Segal. 2008. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 190:1985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appelberg, R. 2006. Macrophage nutriprive antimicrobial mechanisms. J. Leukoc. Biol. 79:1117-1128. [DOI] [PubMed] [Google Scholar]
- 6.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72:2468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bansal, V., and J. B. Ochoa. 2003. Arginine availability, arginase, and the immune response. Curr. Opin. Clin. Nutr. Metab. Care 6:223-228. [DOI] [PubMed] [Google Scholar]
- 8.Burke, M., A. F. Merican, and D. J. Sherratt. 1994. Mutant Escherichia coli arginine repressor proteins that fail to bind L-arginine, yet retain the ability to bind their normal DNA-binding sites. Mol. Microbiol. 13:609-618. [DOI] [PubMed] [Google Scholar]
- 9.Burstein, D., T. Zusman, E. Degtyar, R. Viner, G. Segal, and T. Pupko. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5:e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 11.Caldara, M., D. Charlier, and R. Cunin. 2006. The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes and provides an integrated view of arginine regulation. Microbiology 152:3343-3354. [DOI] [PubMed] [Google Scholar]
- 12.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 13.Charlier, D., M. Roovers, F. Van Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Pierard. 1992. Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226:367-386. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 16.Chen, J., M. Reyes, M. Clarke, and H. A. Shuman. 2007. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell. Microbiol. 9:1660-1671. [DOI] [PubMed] [Google Scholar]
- 17.Cherney, L. T., M. M. Cherney, C. R. Garen, G. J. Lu, and M. N. James. 2008. Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis. J. Mol. Biol. 384:1330-1340. [DOI] [PubMed] [Google Scholar]
- 18.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 19.Czaplewski, L. G., A. K. North, M. C. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 20.de Felipe, K. S., R. T. Glover, X. Charpentier, O. R. Anderson, M. Reyes, C. D. Pericone, and H. A. Shuman. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4:e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 24.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, et al. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 25.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George, J. R., L. Pine, M. W. Reeves, and W. K. Harrell. 1980. Amino acid requirements of Legionella pneumophila. J. Clin. Microbiol. 11:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gobert, A. P., S. Daulouede, M. Lepoivre, J. L. Boucher, B. Bouteille, A. Buguet, R. Cespuglio, B. Veyret, and P. Vincendeau. 2000. l-Arginine availability modulates local nitric oxide production and parasite killing in experimental trypanosomiasis. Infect. Immun. 68:4653-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grandori, R., T. A. Lavoie, M. Pflumm, G. Tian, H. Niersbach, W. K. Maas, R. Fairman, and J. Carey. 1995. The DNA-binding domain of the hexameric arginine repressor. J. Mol. Biol. 254:150-162. [DOI] [PubMed] [Google Scholar]
- 29.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 31.Hindre, T., H. Bruggemann, C. Buchrieser, and Y. Hechard. 2008. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154:30-41. [DOI] [PubMed] [Google Scholar]
- 32.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hovel-Miner, G., S. Pampou, S. P. Faucher, M. Clarke, I. Morozova, P. Morozov, J. J. Russo, H. A. Shuman, and S. Kalachikov. 2009. σS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J. Bacteriol. 191:2461-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingmundson, A., A. Delprato, D. G. Lambright, and C. R. Roy. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365-369. [DOI] [PubMed] [Google Scholar]
- 38.Iniesta, V., L. C. Gomez-Nieto, and I. Corraliza. 2001. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J. Exp. Med. 193:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin, L., W. F. Xue, J. W. Fukayama, J. Yetter, M. Pickering, and J. Carey. 2005. Asymmetric allosteric activation of the symmetric ArgR hexamer. J. Mol. Biol. 346:43-56. [DOI] [PubMed] [Google Scholar]
- 40.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4:945-954. [DOI] [PubMed] [Google Scholar]
- 41.Karaivanova, I. M., P. Weigel, M. Takahashi, C. Fort, A. Versavaud, G. Van Duyne, D. Charlier, J. N. Hallet, N. Glansdorff, and V. Sakanyan. 1999. Mutational analysis of the thermostable arginine repressor from Bacillus stearothermophilus: dissecting residues involved in DNA binding properties. J. Mol. Biol. 291:843-855. [DOI] [PubMed] [Google Scholar]
- 42.Kiupakis, A. K., and L. Reitzer. 2002. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 184:2940-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585-597. [DOI] [PubMed] [Google Scholar]
- 44.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67:1307-1319. [DOI] [PubMed] [Google Scholar]
- 45.Larsen, R., G. Buist, O. P. Kuipers, and J. Kok. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim, D. B., J. D. Oppenheim, T. Eckhardt, and W. K. Maas. 1987. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. U. S. A. 84:6697-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 48.Lu, C. D., and A. T. Abdelal. 1999. Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J. Bacteriol. 181:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu, C. D., Z. Yang, and W. Li. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J. Bacteriol. 186:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219:241-248. [DOI] [PubMed] [Google Scholar]
- 52.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machner, M. P., and R. R. Isberg. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974-977. [DOI] [PubMed] [Google Scholar]
- 54.Machner, M. P., and R. R. Isberg. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11:47-56. [DOI] [PubMed] [Google Scholar]
- 55.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. U. S. A. 89:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McConville, M. J., D. de Souza, E. Saunders, V. A. Likic, and T. Naderer. 2007. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol. 23:368-375. [DOI] [PubMed] [Google Scholar]
- 58.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 59.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 60.Muder, R. R., V. L. Yu, and G. D. Fang. 1989. Community-acquired Legionnaires' disease. Semin. Respir. Infect. 4:32-39. [PubMed] [Google Scholar]
- 61.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 62.Ni, J., V. Sakanyan, D. Charlier, N. Glansdorff, and G. D. Van Duyne. 1999. Structure of the arginine repressor from Bacillus stearothermophilus. Nat. Struct. Biol. 6:427-432. [DOI] [PubMed] [Google Scholar]
- 63.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15:372-380. [DOI] [PubMed] [Google Scholar]
- 64.Reeves, M. W., L. Pine, S. H. Hutner, J. R. George, and W. K. Harrell. 1981. Metal requirements of Legionella pneumophila. J. Clin. Microbiol. 13:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 67.Ryan, S., M. Begley, C. G. Gahan, and C. Hill. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432-445. [DOI] [PubMed] [Google Scholar]
- 68.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 70.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U. S. A. 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 72.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6:253-255. [DOI] [PubMed] [Google Scholar]
- 73.Sinai, A. P., and K. A. Joiner. 1997. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol. 51:415-462. [DOI] [PubMed] [Google Scholar]
- 74.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 75.Stintzi, A., Z. Johnson, M. Stonehouse, U. Ochsner, J. M. Meyer, M. L. Vasil, and K. Poole. 1999. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J. Bacteriol. 181:4118-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sunnerhagen, M., M. Nilges, G. Otting, and J. Carey. 1997. Solution structure of the DNA-binding domain and model for the complex of multifunctional hexameric arginine repressor with DNA. Nat. Struct. Biol. 4:819-826. [DOI] [PubMed] [Google Scholar]
- 77.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 78.Szwajkajzer, D., L. Dai, J. W. Fukayama, B. Abramczyk, R. Fairman, and J. Carey. 2001. Quantitative analysis of DNA binding by the Escherichia coli arginine repressor. J. Mol. Biol. 312:949-962. [DOI] [PubMed] [Google Scholar]
- 79.Talaat, A. M., S. T. Howard, W. T. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talaue, M. T., V. Venketaraman, M. H. Hazbon, M. Peteroy-Kelly, A. Seth, R. Colangeli, D. Alland, and N. D. Connell. 2006. Arginine homeostasis in J774.1 macrophages in the context of Mycobacterium bovis BCG infection. J. Bacteriol. 188:4830-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tesh, M. J., S. A. Morse, and R. D. Miller. 1983. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J. Bacteriol. 154:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian, G., and W. K. Maas. 1994. Mutational analysis of the arginine repressor of Escherichia coli. Mol. Microbiol. 13:599-608. [DOI] [PubMed] [Google Scholar]
- 83.Van Duyne, G. D., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256:377-391. [DOI] [PubMed] [Google Scholar]
- 84.Wang, H., N. Glansdorff, and D. Charlier. 1998. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J. Mol. Biol. 277:805-824. [DOI] [PubMed] [Google Scholar]
- 85.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.