Abstract
Beneficial mutations in diversifying glucose-limited Escherichia coli populations are mostly unidentified. The genome of an evolved isolate with multiple differences from that of the ancestor was fully assembled. Remarkably, a single mutation in hfq was responsible for the multiple benefits under glucose limitation through changes in at least five regulation targets.
Nutrient-limited Escherichia coli populations competing for extended periods in chemostats provide a useful system for investigating evolutionary processes (6, 16, 27). Recent results suggest that glucose-limited populations exhibit a remarkable level of divergence, up to the point of individuality (15, 19). Mutations in rpoS were present in many isolates, with several different alleles coexisting within diverging populations (19). These rpoS mutations confer higher fitness under nutrient limitation by reducing transcription of genes involved in unnecessary stress responses, thereby allowing optimal expression of functions needed for transport and metabolism (14, 25). Moreover, at least three additional mutations in the mgl, malT, and mlc genes contribute to fitness in the previously analyzed rpoS isolates (26). In addition to the rpoS lineages, we previously found separate, coexisting rpoS+ isolates in the same glucose-limited populations (18, 19), with undefined mutations and fitness properties. Here we report on the mutational adaptation and fitness characteristics of one such strain, E. coli BW4001, that has a complex and previously unexplained series of adaptations under glucose limitation (18).
BW4001 is an rpoS+ isolate that was sampled after 26 days in a glucose-limited chemostat growing at a dilution rate of 0.1 h−1 (19) with evolved phenotypic properties, including (i) increased rates of glucose transport through PtsG reflected by an increased sensitivity to the glucose analog methyl α-glucoside (αMG), which is transported by PtsG (Fig. 1); (ii) higher outer membrane permeability reflected by increased sodium dodecyl sulfate (SDS) sensitivity (Fig. 1) and elevated LamB and porin levels relative to OmpA (Fig. 2 A); (iii) elevated metabolic oxidation rates (Fig. 1); (iv) decreased cell yields (Fig. 1), (v) altered cell surface properties (Fig. 1); (vi) partially reduced RpoS levels (Fig. 1); and (vii) reduced levels of negative DNA supercoiling in stationary phase (Fig. 2C). The first two phenotypic traits are in contrast to those of previously described rpoS strains that upregulated an alternative glucose uptake pathway through cumulative mutations in malT and mglD (23, 24, 26). With the expectation that multiple mutations were responsible for these phenotypes, we performed a genomic analysis to identify the molecular basis of the fitness increase in BW4001.
FIG. 1.
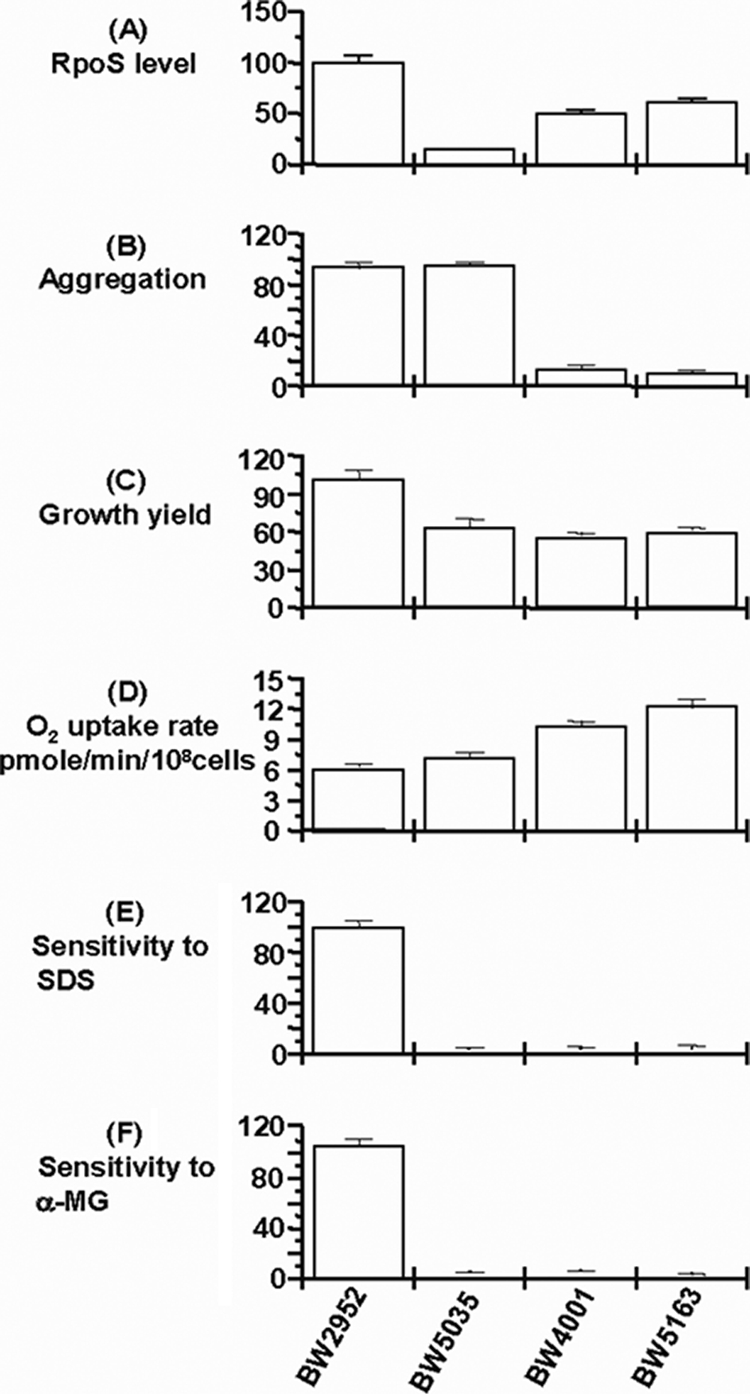
Phenotypic properties of BW4001. The evolved strain BW4001 was derived from the ancestral strain BW2952 (9, 19). BW5035 is derived from BW2952 by insertion of an ampicillin resistance-conferring cassette into the hfq gene by the protocol described previously (34). BW5163 is strain BW2952 in which the BW4001-evolved hfq allele has been transferred in two steps: a purA::tet mutation is first introduced into BW2952 (creating an adenine-auxotrophic strain BW5155) and then the purA-hfq region is introduced into BW5155 using a P1 lysate grown on BW4001 and selecting for PurA+. The transfer of the hfq4001 allele was confirmed by sequencing. (A) The RpoS level was estimated by glycogen staining. Scanning densities relative to that of the ancestor BW2952 are shown. (B) The aggregation of cells was measured based on the decrease in optical density of a standing culture as described in reference 19 and is given relative to that of the ancestor. (C) The growth yield of strains was measured based on the absorbance in L broth after overnight growth to steady state and is shown relative to that of the ancestor. (D) The oxygen uptake rate was measured using an oxygen electrode as described in reference 18. (E) The sensitivity to 2% SDS was measured in patches on L agar plates. Patches were scanned densitometrically, and the cell density relative to that of the ancestor is shown. (F) The sensitivity to 1% methyl α-glucoside was measured in patches on glycerol plates. Patches were scanned densitometrically, and the cell density relative to that of the ancestor is shown.
FIG. 2.
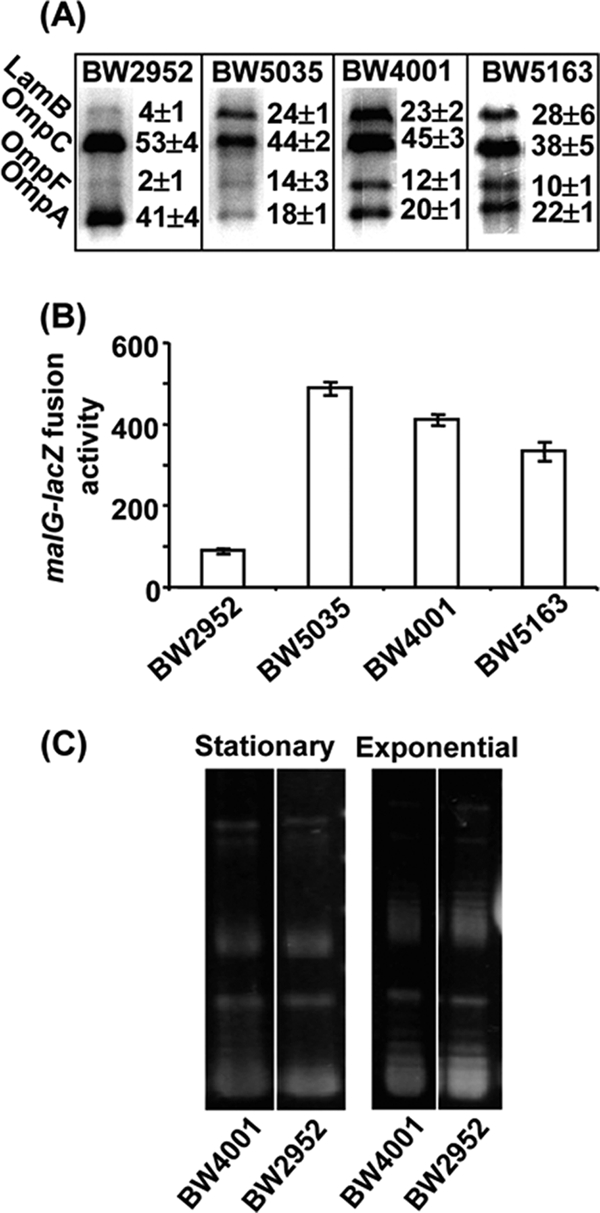
Altered regulation and DNA topology in BW4001. (A) The outer membrane proteins were extracted from bacterial cells grown under glucose limitation in a chemostat and separated by urea-SDS electrophoresis using procedures described previously (18). The major protein bands are labeled. The relative proportion of each band was estimated by densitometric analyses of stained gels and is shown next to each representative track. Means ± standard deviations are from results with four independent gels. (B) The β-galactosidase activity of a malEFG::lacZ promoter fusion was measured in bacterial cells grown exponentially in 0.2% glycerol minimal medium (MMA) at 37°C. (C) The plasmid pUC19, introduced into the strains by electroporation and extracted from bacterial cells grown in LB medium at 37°C for 2.5 h (mid-exponential phase) and 12 h (stationary phase), was resolved on 1% agarose gels containing 1.5 μg/ml chloroquine (4).
The genome of BW4001 was resequenced using 454 and Solexa methodologies (see Methods in the supplemental material) and fully assembled. In comparison to the ancestral genome (9), only one single-nucleotide polymorphism (SNP) in the hfq gene, leading to the Y25D change in the Hfq protein, was detected in BW4001; no other mutation was detected whatsoever after exhaustive analysis by Solexa, 454, and Sanger sequencing (for 78 sequence gaps and 151 suspected SNP sites). A single SNP was unexpected, given the high number of both metabolic and physiological differences between BW4001 and its ancestor (Fig. 1 and 2) (18, 19) and the presence of at least three mutations, in rpoS, mglD (or mglO), and malT (or mlc), in the previously characterized clones from the rpoS lineages sampled at the same time point as BW4001 (23, 24, 26). To confirm that the single mutation was responsible for most or all evolved phenotypes of BW4001, the hfq mutation was transferred into the ancestral genetic background by cotransduction with purA::tet as a selectable marker. The phenotypic traits of the transductant, called BW5163, matched in all respects those of BW4001 (Fig. 1 and 2), implying that the single mutation was the source of the multiple changes. The minor variations in RpoS amount, O2 uptake rate, and malG-lacZ fusion activity between BW4001 and BW5163 were not statistically significant (P of >0.05 in all cases). Moreover, the hfq transductant strain BW5163 is as fit as BW4001, in comparison to the ancestral strain (Fig. 3). Interestingly, the Y25D mutation provides a higher fitness than the hfq knockout strain BW5035, implying that a particular structural or functional change in Hfq is needed for the fitness increase under these conditions. Alternatively, the lower fitness of the hfq null mutant suggests that ancestral and Y25D Hfq proteins share some undefined properties in the cell that is missing from the null mutant.
FIG. 3.
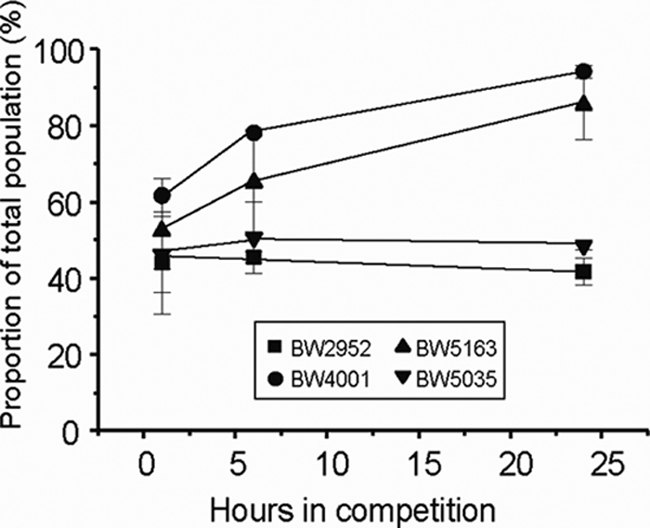
Relative fitness in strains with altered hfq genes. We compared the isolates for competitive fitness, which was measured in glucose-limited chemostats. Competitions were all performed against BW3454, an ancestral derivative marked with the metC::Tn10 selectable marker; control experiments revealed that this strain is only marginally fitter than the actual ancestor BW2952 (squares). Competing strains were mixed 50:50 after 16 h of individual acclimation in chemostats, and the changes in the proportion of each strain are shown. Mean ± standard deviations were obtained from three to five replicates.
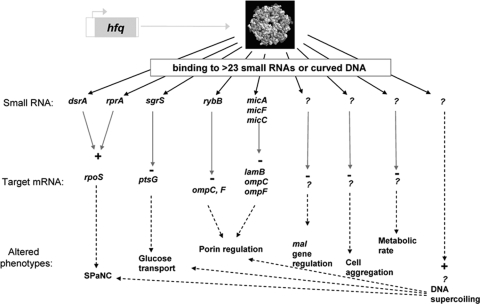
In Fig. 4, we summarize all the potential effects of the single hfq mutation in BW4001. Fitness in a glucose-limited environment can be explained through a subset of effects resulting from the extensive pleiotropic nature of hfq mutations (32). The contribution of small RNAs (sRNAs) and altered DNA topology cannot be fully disentangled. In some cases, the mutation is affecting sRNA interactions; coincidentally, the Y25D substitution was previously studied in a site-directed mutagenesis survey of hfq function (21). The Y25D mutant protein has reduced affinity for DsrA sRNA and led to partially reduced expression of RpoS; translation of rpoS mRNA is dependent upon Hfq (21). The lower level of staining by iodine and increased growth on acetate are indicators of reduced RpoS function in BW4001 (13). This partial reduction in RpoS is beneficial, as there is an inverse relationship between RpoS concentration and competitive fitness under glucose-limiting conditions (13, 29). The hfq mutation may therefore favor the expression of vegetative rather than stress response genes.
FIG. 4.
Potential relationships between fitness under glucose limitation and pleiotropic effects of an hfq mutation. We show the small RNAs potentially implicated in the regulation of mRNAs known to be involved in evolved phenotypes. Question marks represent unknown components of up- or downregulation of targets. SPaNC, self-preservation and nutritional competence (8).
The increased PtsG transport activity (17) and the associated elevated sensitivity to αMG (Fig. 1) can also be explained by the hfq mutation, because Hfq, associated with the small RNA sgrS, is involved in the destabilization of ptsG mRNA (1). The hfq mutation can result in increased PtsG expression through increased stability of ptsG mRNA, thereby increasing glucose uptake. As shown in Fig. 1, αMG sensitivity is entirely Hfq dependent.
Increased outer membrane permeability is an important factor in fitness under nutrient limitation (7) and can arise from elevated porin expression or functional changes in porins (16, 35). The porin patterns noted in both BW4001 and BW5163 were identical and resulted in elevated porin and LamB levels (Fig. 2A). Small RNAs in conjunction with Hfq control multiple targets in outer membranes, and a mutation in hfq can increase porin expression through reduced antisense translational blocks caused by, for example, MicC and MicF (33). These outer membrane changes also explain the increased sensitivity to the detergent SDS in the hfq mutant, because membrane stress sensing also involves sRNA/Hfq (30).
An elevated LamB level in the outer membrane (Fig. 2A) is beneficial under glucose limitation, because it is a glycoporin with affinity for sugars like glucose (5). Although higher LamB levels in the hfq mutant is not as easy to explain through known sRNA regulation, we, however, discovered that regulation of LamB, as part of the mal regulon (22), was also affected by Hfq. Hence, mal gene expression, measured by a malEFG-lacZ reporter fusion, was considerably higher in Hfq mutants (Fig. 2B). It was not previously recognized that regulation of mal genes, and in particular of lamB, was subject to control by small RNAs, but our results increase this possibility.
The other properties of BW4001, including elevated respiration rate, decreased growth yield, and altered aggregation properties (Fig. 1), are also Hfq dependent but through either unidentified sRNAs or sRNA-independent pathways such as curved-DNA binding (2). This change in DNA structure may confer altered expression at multiple promoters (12, 31) and, possibly, altered phenotypes. However, negative supercoiling is required for mal regulation (28), and therefore this mutant phenotype is more likely to be due to an unknown sRNA. Supercoiling sensitively alters the balance of porin expression (10), and, given that the ptsG promoter activity is affected by the histone-like protein Fis, this gene is possibly also altered by changes in supercoiling. The pleiotropy of DNA supercoiling (31) makes it difficult to exclude the possibility that all or some of the properties in Fig. 1 are either directly or indirectly affected by the Hfq influence on DNA topology (Fig. 2C). It should, however, be noted that altered DNA topology, through topA and fis mutations, was also found to confer increased fitness in glucose batch cultures over thousands of generations (4). Additionally, increased fitness may also be associated with reduced stress responses directly controlled via Hfq and other sRNAs, such as the envelope stress response (11).
Interestingly, this combination of characteristics provides an alternative fitness adaptation to that of rpoS mutants coexisting with BW4001. These evolved isolates achieved similar phenotypic changes via alternative transporter pathways and mutational paths, including mutations in rpoS, mgl, and malT. These results demonstrate that the number of mutations in a strain is clearly not an indicator of elapsed time in a chemostat and that alternative means of achieving the same adaptation (better glucose uptake) may be a source of divergence in glucose-limited populations. Syntrophic diversification can therefore arise from harnessing degeneracies in transport, metabolism, and gene regulation, allowing increased fitness through parallel adaptations.
Supplementary Material
Acknowledgments
R.M. and C.M. were supported by Australian Research Council grants to T.F. and to T.F. and P.R.R., respectively. L.W. was partly supported by the National Natural Science Foundation of China (NSFC) Key Programs grant 30530010, the Chinese National Science Fund for Distinguished Young Scholars (grant 30788001), NSFC General Program grants 30870070, 30870078, and 30771175, National 863 Program of China grants 2006AA020703, 2010AA10A203, and 2006AA06Z409, National 973 Program of China grant 2009CB522603, and National Key Programs for Infectious Diseases of China grant 2008ZX1004-009. D.S. and J.G. were supported by the French Centre National de la Recherche Scientifique, Université Joseph Fourier, and Agence Nationale de la Recherche Program “Blanc” (ANR-08-BLAN-0283-01).
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10:134-139. [DOI] [PubMed] [Google Scholar]
- 2.Azam, T. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli—sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274:33105-33113. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, D. R., S. Balasubramanian, H. P. Swerdlow, G. P. Smith, J. Milton, C. G. Brown, K. P. Hall, D. J. Evers, C. L. Barnes, H. R. Bignell, J. M. Boutell, J. Bryant, R. J. Carter, R. K. Cheetham, A. J. Cox, D. J. Ellis, M. R. Flatbush, N. A. Gormley, S. J. Humphray, L. J. Irving, M. S. Karbelashvili, S. M. Kirk, H. Li, X. H. Liu, K. S. Maisinger, L. J. Murray, B. Obradovic, T. Ost, M. L. Parkinson, M. R. Pratt, I. M. J. Rasolonjatovo, M. T. Reed, R. Rigatti, C. Rodighiero, M. T. Ross, A. Sabot, S. V. Sankar, A. Scally, G. P. Schroth, M. E. Smith, V. P. Smith, A. Spiridou, P. E. Torrance, S. S. Tzonev, E. H. Vermaas, K. Walter, X. L. Wu, L. Zhang, M. D. Alam, C. Anastasi, I. C. Aniebo, D. M. D. Bailey, I. R. Bancarz, S. Banerjee, S. G. Barbour, P. A. Baybayan, V. A. Benoit, K. F. Benson, C. Bevis, P. J. Black, A. Boodhun, J. S. Brennan, J. A. Bridgham, R. C. Brown, A. A. Brown, D. H. Buermann, A. A. Bundu, J. C. Burrows, N. P. Carter, N. Castillo, M. C. E. Catenazzi, S. Chang, R. N. Cooley, N. R. Crake, O. O. Dada, K. D. Diakoumakos, B. Dominguez-Fernandez, D. J. Earnshaw, U. C. Egbujor, D. W. Elmore, S. S. Etchin, M. R. Ewan, M. Fedurco, L. J. Fraser, K. V. F. Fajardo, W. S. Furey, D. George, K. J. Gietzen, C. P. Goddard, G. S. Golda, P. A. Granieri, D. E. Green, D. L. Gustafson, N. F. Hansen, K. Harnish, C. D. Haudenschild, N. I. Heyer, M. M. Hims, J. T. Ho, A. M. Horgan, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crozat, E., N. Philippe, R. E. Lenski, J. Geiselmann, and D. Schneider. 2005. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Death, A., L. Notley, and T. Ferenci. 1993. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 175:1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dykhuizen, D. E. 1990. Experimental studies of natural selection in bacteria. Annu. Rev. Ecol. Syst. 21:373-398. [Google Scholar]
- 7.Ferenci, T. 2008. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv. Microb. Physiol. 53:169-229. [DOI] [PubMed] [Google Scholar]
- 8.Ferenci, T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Ferenci, T., Z. M. Zhou, T. Betteridge, Y. Ren, Y. Liu, L. Feng, P. R. Reeves, and L. Wang. 2009. Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J. Bacteriol. 191:4025-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graeme-Cook, K. A., G. May, E. Bremer, and C. F. Higgins. 1989. Osmotic regulation of porin expression: a role for DNA supercoiling. Mol. Microbiol. 3:1287-1294. [DOI] [PubMed] [Google Scholar]
- 11.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the sigma(E)-mediated envelope stress response and the sigma(32)-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 189:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatfield, G. W., and C. J. Benham. 2002. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 36:175-203. [DOI] [PubMed] [Google Scholar]
- 13.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, T., S. Seeto, and T. Ferenci. 2006. Genotype-by-environment interactions influencing the emergence of rpoS mutations in Escherichia coli populations. Genetics 172:2071-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinnersley, M. A., W. E. Holben, and F. Rosenzweig. 2009. E Unibus Plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 5(11):e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurlandzka, A., R. F. Rosenzweig, and J. Adams. 1991. Identification of adaptive changes in an evolving population of Escherichia coli: the role of changes with regulatory and highly pleiotropic effects. Mol. Biol. Evol. 8:261-281. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maharjan, R., S. Seeto, and T. Ferenci. 2007. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J. Bacteriol. 189:2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maharjan, R., S. Seeto, L. Notley-McRobb, and T. Ferenci. 2006. Clonal adaptive radiation in a constant environment. Science 313:514-517. [DOI] [PubMed] [Google Scholar]
- 20.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y.-J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. I. Alenquer, T. P. Jarvie, K. B. Jirage, J.-B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikulecky, P. J., M. K. Kaw, C. C. Brescia, J. C. Takach, D. D. Sledjeski, and A. L. Feig. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 11:1206-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notley, L., and T. Ferenci. 1995. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient-stressed Escherichia coli. Mol. Microbiol. 16:121-129. [DOI] [PubMed] [Google Scholar]
- 23.Notley-McRobb, L., and T. Ferenci. 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1:33-43. [DOI] [PubMed] [Google Scholar]
- 24.Notley-McRobb, L., and T. Ferenci. 1999. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1:45-52. [DOI] [PubMed] [Google Scholar]
- 25.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notley-McRobb, L., S. Seeto, and T. Ferenci. 2003. The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc. Biol. Sci. 270:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, A., and L. Szilard. 1950. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. U. S. A. 36:708-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richet, E., and O. Raibaud. 1991. Supercoiling is essential for the formation and stability of the initiation complex at the divergent malEp and malKp promoters. J. Mol. Biol. 218:529-542. [DOI] [PubMed] [Google Scholar]
- 29.Spira, B., X. Hu, and T. Ferenci. 2008. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 154:2887-2895. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. sigma(E) regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 189:4243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travers, A., and G. Muskhelishvili. 2005. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 3:157-169. [DOI] [PubMed] [Google Scholar]
- 32.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 33.Valentin-Hansen, P., J. Johansen, and A. A. Rasmussen. 2007. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 10:152-155. [DOI] [PubMed] [Google Scholar]
- 34.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, E., and T. Ferenci. 1999. OmpF changes and the complexity of Escherichia coli adaptation to prolonged lactose limitation. FEMS Microbiol. Lett. 176:395-401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.