Abstract
The genetic relatedness of Vibrio cholerae O1/O139 isolates obtained from 100 patients and 146 of their household contacts in Dhaka, Bangladesh, between 2002 and 2005 was assessed by multilocus variable-number tandem-repeat analysis. Isolate genotypes were analyzed at five loci containing tandem repeats. Across the population, as well as within households, isolates with identical genotypes were clustered in time. Isolates from individuals within the same household were more likely to have similar or identical genotypes than were isolates from different households, but even within a household, isolates from different individuals often had different genotypes. When household contacts were sampled regularly for 3 weeks after the illness of the household index patient, isolates with genotypes related to the index patient appeared in contacts, on average, ∼3 days after the index patient, while isolates with unrelated genotypes appeared in contacts ∼6 days after. Limited data revealed that multiple isolates from the same individual collected within days of each other or even from a single stool sample may have identical, similar, or unrelated genotypes as well. Our results demonstrate that genetically related V. cholerae strains cluster in local outbreaks but also suggest that multiple distinct strains of V. cholerae O1 may circulate simultaneously within a household.
Vibrio cholerae is the etiologic agent of cholera, a secretory diarrheal disease with a high mortality rate in humans if untreated (25). Serogroups of V. cholerae, a motile, Gram-negative, curved rod, can be defined serologically by the O side chain of the lipopolysaccharide (LPS) component of the outer membrane (9). V. cholerae is found in a variety of forms in aquatic ecosystems (41, 42), and more than 200 different serogroups have been isolated, mostly from environmental sources (45). However, the vast majority of V. cholerae strains that cause the clinical disease cholera belong to serogroup O1 or O139 (37, 42). V. cholerae O1, the historical agent of epidemic and pandemic cholera and the current leading cause of cholera both globally and in Bangladesh (42), is classified into two major biotypes, classical and El Tor (44), and two major serotypes, Ogawa and Inaba (48). The current global pandemic is caused by V. cholerae O1 El Tor. A second pathogenic serogroup, O139, emerged in the Bengal region in 1992 by horizontal transfer of new LPS biosynthesis-encoding genes into the El Tor biotype (1, 4). This new serogroup continues to cocirculate with El Tor V. cholerae O1 serotypes Ogawa and Inaba as a cause of disease in humans, although it accounts for a smaller proportion of all cholera now than in its first years of circulation (16, 20). Recently, comparative genomics has revealed an extensive amount of lateral gene transfer between strains, suggesting that genomic classification may be an alternative to serogrouping for classifying pathogenic V. cholerae strains (11).
Toxigenic V. cholerae may be present in environmental sources in regions of endemicity and emerge, often seasonally, to cause cholera in humans (12, 18). Once an outbreak has begun, organisms from one infected individual are more infectious for the next individual, a property termed hyperinfectivity, and these forms may be able to pass directly from human to human through fecal-oral contamination (35). However, because vibrio organisms are difficult to isolate from implicated environmental or domestic water sources (28, 29), little is known about the diversity of V. cholerae in inocula that cause human infection.
Established laboratory methods for differentiating V. cholerae strains, apart from serogrouping and serotyping, include rRNA restriction fragment length polymorphism (ribotyping), pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST). These methods, however, have a limited capacity to differentiate between pathogenic V. cholerae strains, as clinical isolates are relatively genetically monomorphic. For instance, V. cholerae O1 comprises approximately 30 ribotypes (39); however, only a few ribotypes are common in clinical isolates, ribotypes evolve slowly, and all isolates of a given pathogenic V. cholerae serotype in a local area over a period of multiple years often belong to a single ribotype (8, 14, 17). In a broad sampling of 154 V. cholerae isolates from Bangladesh and worldwide over several decades, only 15 ribotypes were identified, and of these, many were found in nonpathogenic environmental isolates only; only five ribotypes were associated with the V. cholerae O1 El Tor biotype that currently predominates as the cause of clinical disease, while pathogenic isolates of serogroup O139 were indistinguishable from each other by ribotype (19).
PFGE, in which restriction endonuclease digestion of genomic DNA generates mutation-sensitive banding patterns, is often more sensitive than ribotyping in detecting strain variation (7, 34, 51) and detects extensive genetic variation within nonpathogenic V. cholerae serogroups (3, 46). However, PFGE types change slowly and are useful primarily for distinguishing between strains in different pandemics or between different continental branches of those pandemics. In an analysis of 180 mostly western-hemisphere isolates (7), PFGE differences had developed from a prior pandemic strain over the 30 years since its arrival in Latin America, but a new strain that had been causing disease for 2 years still had only a single PFGE type across the 64 isolates analyzed. Similarly, in a Japanese study (2), although 19 PFGE types were identified among O1 isolates, the majority of the domestic isolates, along with several imported isolates, belonged to a single PFGE type.
Further differentiation between V. cholerae isolates is achievable by MLST, which characterizes isolates by internal DNA sequences in selected housekeeping genes (32). Nevertheless, epidemic strains also cluster tightly in this typing scheme (5, 32) and the method has been useful primarily for determining relationships between nontoxigenic strains (36) or for linking regional outbreaks (which typically appear monoclonal by these methods) with the pandemic strain responsible (5, 33).
Although these methods have distinguished major pandemic clones from other nonpathogenic human and environmental isolates of V. cholerae, the near clonality of pathogenic O1 and O139 strains means that established methods may not provide sufficiently robust differentiation of these genetically similar pathogenic strains to answer important epidemiological questions. Therefore, there is a need for other methods that can distinguish among clinical O1 and O139 isolates and track the epidemiology of outbreaks in a restricted geographic area on a shorter time scale.
Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) is one method that may be useful for differentiating between pathogenic V. cholerae O1 and O139 strains that would be indistinguishable by other techniques (15). This method examines short repeating DNA segments at various locations in the genome that can vary in number at each location and uses the number of repeats at each varying locus as a fingerprint to distinguish between isolates.
Escherichia coli is the paradigm organism for demonstrating the value of the MLVA method. Noller et al. (38) showed that E. coli O157 isolates that were indistinguishable by MLST could be distinguished to some extent by PFGE but that MLVA distinguished between isolates that had the same PFGE type and did so in a manner consistent with the known epidemiology of the isolates (38a). In addition, machine-scored VNTR assays have been demonstrated to be robust and portable and to discriminate clearly between isolates by using relatively few loci, therefore limiting the effect of compounding genotyping errors (6).
For V. cholerae, five VNTR loci have been identified (15), and the initial application of MLVA at those loci has demonstrated distinct populations of clinical isolates of V. cholerae in different geographic regions within Bangladesh and India (23, 47). Predominant isolates in each of two rural Bangladeshi regions varied gradually over a time scale of months to years (47), and isolates collected from India over a 15-year period varied widely, with individual MLVA types clustering in time and place—some with widespread dissemination and others with limited local occurrence only (23). MLVA has also been used to classify hybrid and altered V. cholerae variants and to demonstrate their genetic distance from the pandemic El Tor strain (10). Use of the MLVA method for epidemiologic study of cholera requires that V. cholerae VNTR alleles remain reasonably stable during bacterial replication in patients or in laboratory culture after isolation. Some degree of stability of two of the five loci used in V. cholerae MLVA has been demonstrated previously by serial passage in vitro through four overnight cultures (15). In this study, we used MLVA to examine V. cholerae O1 and O139 isolates obtained from infected patients and their household contacts—including multiple isolates from the same individual and isolates from multiple individuals within the same household—in a large city where cholera is endemic.
MATERIALS AND METHODS
Clinical sample collection.
Between March 2002 and June 2005, patients >6 months of age presenting to the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B) with acute watery diarrhea, most of whom were residents of Dhaka city, were screened for V. cholerae infection by stool sample culture. If cultures were positive for V. cholerae O1 or O139, written informed consent was sought from patients and all available household contacts (defined as individuals sharing a cooking pot with the patient for 3 or more antecedent days), and consenting individuals were enrolled in a study approved by ethical review boards of the ICDDR,B and Massachusetts General Hospital (27). Household contacts were monitored for 3 weeks, and once-daily rectal swabs were collected from them for culture 1, 2, 3, 4, 5, 6, 13, and 20 days after the index patient's presentation at the hospital. For the epidemiological analysis reported here, all culture-confirmed index patients who had V. cholerae O1 or O139 infection in at least one of their household contacts during the 3 weeks of follow-up were retrospectively selected. All available O1 and O139 V. cholerae isolates from these patients and their household contacts (sometimes including serial isolates obtained over multiple days from a single contact, as well as multiple contacts per index patient) were included in the analysis. Stool samples from an additional nine patients with cholera were collected in August 2009 for separate analysis.
Sample processing.
Rectal swab specimens from household contacts were collected in Cary-Blair transport medium. These samples, as well as initial stool samples from suspected index patients, were cultured on taurocholate-tellurite-gelatin agar. After overnight incubation, suspected V. cholerae colonies were confirmed by slide agglutination with specific monoclonal antibodies to identify the serogroup (O1 or O139) and the O1 serotype (Ogawa or Inaba). A single colony was picked for each stool sample or rectal swab specimen. Nine additional stool samples collected in 2009 were mixed with glycerol and frozen at −80°C (24) and then shipped to the United States, where 17 to 20 colonies per specimen were selected after growth on thiosulfate-citrate-bile salt-sucrose agar.
Isolates from both parts of the study were stored in glycerol at −80°C and then recultured on Luria-Bertani (LB) agar. Individual colonies were selected and placed in 200 μl of LB for overnight culture. In addition, for each of three arbitrarily chosen clinical isolates, a 96-well plate with 200 μl of LB broth per well was inoculated with 95 individual colonies per isolate, and daily for 30 days, 2 μl of each culture was transferred into 200 μl of fresh broth on a new plate. After culturing (on day 2 or 31), DNA was isolated from 5 μl of culture using Prepman (ABI) by following the manufacturer's instructions.
V. cholerae O1 and O139 isolates were then genotyped at each of five previously identified VNTR loci (15, 47). Each locus was amplified by PCR using the previously described forward primer (47) and a new end-labeled reverse primer (see Table 1). The labeled fragments were separated using a 3730xl ABI Automatic Sequencer. The size was determined using internal lane standards (LIZ600; ABI, Foster City, CA) with the Gene Mapper v4.0 program (ABI) and the formulae in Table 1. This new method was tested in 54 instances, in all of which it and the previously used sequencing method produced identical results. Alleles were identified by the number of repeats at a locus (rather than by the arbitrary numeric allele labels used in prior publications [23, 47]). Numbers of repeats were listed sequentially for the five VNTR loci (VC0147, pVC0437, VC1650, VCA0171, and VCA0283) to generate an isolate genotype (e.g., the genotype 9 4 6 21 14 indicates nine repeats at locus VC0147, four at promoter of VC0437, etc.).
TABLE 1.
PCR primers and formulae used to determine V. cholerae VNTR repeat numbers
| Locus | Dye-primer | Expected rangea (bp) | Formulab |
|---|---|---|---|
| VC0147 | Tetc-ACGTGCAGGTTCAACCGTG | 186-224 | (x − 150)/6 |
| TTGTCATGGCTTGGATTTGG | |||
| VC0437 | Tet-GTTGCCGCCATCACCAGCTTG | 265-301 | (x − 245)/6 |
| CGTTAGCATCGAAACTGCTG | |||
| VC1650 | Tet-CCGCTAACTGAGTGACCGC | 370-440 | (x − 307)/9 |
| CTACCAAGCGGCGGTTAAGCTG | |||
| VCA0171 | Famd-AGGCGCCTGATGACGAATCC | 316-442 | (x − 270)/6 |
| GCTGAAGCCTTTCGCGATCC | |||
| VCA0283 | Fam-GGAGGTAGCTACGAATTCTAC | 118-244 | (x − 95)/6 |
| GTACATTCACAATTTGCTCACC |
The expected range of the sizes of the fragments produced by amplification using the primers is shown.
In each formula, x is the size of the fragment for each individual isolate and locus pair. Genemapper v4.0 produces sizes in hundredths of a base pair. When the formulas are applied, the value is rounded to the nearest whole number to determine the number of repeats.
Tet, 6-tetamidite.
Fam, 6-carboxyfluorescein.
For selected isolates, MLST and PFGE were also performed. MLST was performed by following the published protocol for nine loci: dnaE, lap, recA, pgm, gyrB, cat, chi, rstR, and gmd (22). PFGE was performed by following the Centers for Disease Control and Prevention PulseNet protocol for V. cholerae using the enzyme NotI (13).
Statistical and analytical methods.
The relatedness of V. cholerae isolates was assessed using several different measures with a range of specificities. Alleles were compared at each of the five individual VNTR loci, and the relatedness of MLVA genotypes was assessed by using eBURST (http://eburst.mlst.net) (21) to divide isolates into clonal complexes within which all genotypes could be connected through a chain of single-locus variants. In addition, isolates were compared on the simple basis of the serogroup and, for O1 isolates, serotype. For the isolates selected for PFGE and MLST analyses, PFGE data were evaluated using criteria proposed by Tenover et al. (49), with isolates whose banding patterns differed by three or fewer bands called closely related, and MLST sequences of these selected isolates were also compared pairwise.
Using each of the relatedness measures, comparisons were made between isolates from different households, between an isolate from an index patient and the first isolate from each contact within the corresponding household, and between initial and subsequent isolates from the same individual. When the isolate from the index patient in a household was missing or could not be recultured (N = 3), the first contact isolate was redefined as that household's index isolate. Average numbers of days between collections for isolates within a household or from an individual were compared between groups with different degrees of relatedness, using unpaired two-sample t tests without assuming uniform variance with P < 0.05 as the threshold of statistical significance. Computations and statistical analyses were performed using Excel, R, and Stata 9.0.
RESULTS
Isolates from 100 index patients were successfully analyzed. In addition, initial isolates from 146 distinct household contacts (range, 0 to 5 contacts per index patient) were analyzed, along with 68 additional isolates obtained on days after the day of initial positive culture from 50 contacts (1 of whom had been redefined as an index patient because of a missing isolate from the original index patient). Of the 214 nonindex V. cholerae isolates that were collected on follow-up surveillance days, there were 56 isolates identified on day 1 after presentation of the household index patient, 51 on day 2, 33 on day 3, 24 on day 4, 16 on day 5, 17 on day 6, 13 on day 13, and 4 on day 20.
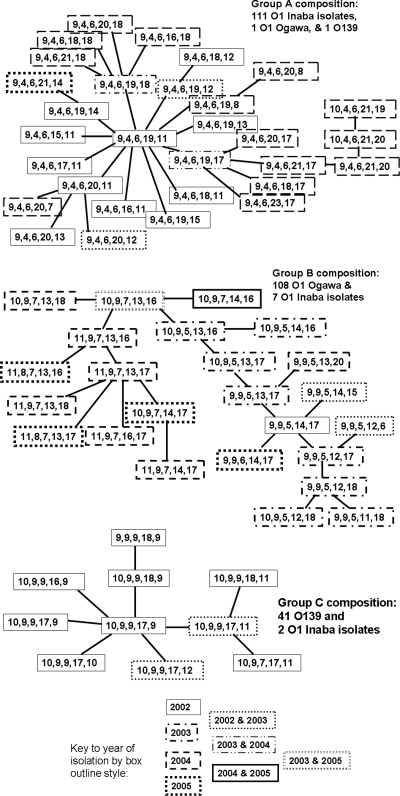
There was extensive serological and VNTR genotypic variation among the 314 V. cholerae isolates studied (see Table S1 in the supplemental material). There were 130 isolates of the O1 Ogawa serotype and 129 of O1 Inaba, with the remaining 55 isolates belonging to serogroup O139 (41, 42, and 17, respectively, among the index isolates). When all five VNTR loci were considered, there were 50 distinct genotypes among the 100 index isolates and 83 distinct genotypes among all 314 isolates. The numbers of distinct alleles among the isolates at loci VC0147, VC0437, VC1650, VCA0171, and VCA0283 were 5, 5, 6, 14, and 20, respectively. When eBURST was used to analyze the genotypes, six clonal complexes were identified. The three largest eBURST complexes and their close correspondence to serological grouping are shown in Fig. 1. Although data about the year of isolation were not given to the eBURST program, the trees it produced have 2002 genotypes centrally located and all of the other genotypes are connected to those central 2002 genotypes through a sequence of genotypes isolated in the same or subsequent years, suggesting the evolution of isolates over time. For example, all of the 2002 genotypes in group A are connected to each other through other 2002 genotypes only, while a path connecting all 2003 genotypes must pass through 2002 genotypes and a path connecting the 2004 genotypes must pass through both the 2002 and 2003 genotypes. In addition to the six complexes, eBURST identified 10 singleton genotypes, each occurring in one or two isolates.
FIG. 1.
The three largest clonal complexes from eBURST analysis. Each line between genotypes represents a change in a single locus.
Differences in allelic variability between VNTR loci.
As suggested by the dissimilar numbers of distinct alleles identified for different VNTR loci, less variability was seen in the three large-chromosome loci (VC0147, VC0437, VC1650) than in the two small-chromosome loci (VCA0171, VCA0283). In instances where relatedness of isolates would be anticipated—namely, between isolates from index patients and contacts within the same household, particularly when both isolates belonged to the same serogroup and serotype—the majority of pairs at any locus had matching alleles, but alleles were more likely to differ at the small-chromosome loci than at any of the three large-chromosome loci; this is shown in Table 2. Using the chi-square test, fewer contact isolates differed from the index isolate in their household at VC0147, VC0437, or VC1650 than at VCA0171 or VCA0283, both overall (P = 0.00002) and when the isolates were matched by serogroup and serotype (P = 0.000001). In contrast, in instances where allelic similarity would not be expected because the serotype or serogroup differed between the index and contact isolates, alleles also differed in most (71 of 95) single-locus comparisons and there was no significant difference in the proportions of matching alleles at small- versus large-chromosome loci (P = 0.09). An alternative approach to evaluating differences in locus variability is to consider those contact isolates that differed from the index isolate in the household. Of the 80 contact isolate genotypes that did not fully match the index isolate genotype, only two differed at a large-chromosome locus while matching at both of the small-chromosome loci; in contrast, 31 of 80 differed at at least one small-chromosome locus and not at any of the large-chromosome loci.
TABLE 2.
Number and percentage of initial V. cholerae isolates in household contacts differing from the index genotype in that household at each VNTR locus
| Condition (no. of isolates) | No. (%) of isolates differing at: |
||||
|---|---|---|---|---|---|
| Large-chromosome loci |
Small-chromosome loci |
||||
| VC0147 | VC0437 | VC1650 | VCA0171 | VCA0283 | |
| Overall (146) | 35 (24) | 30 (21) | 36 (25) | 61 (42) | 58 (40) |
| Contact isolate matches index serogroup and serotype (127) | 25 (20) | 16 (13) | 19 (15) | 45 (35) | 44 (35) |
| Contact isolate does not match index serogroup or serotype (19) | 10 (53) | 14 (74) | 17 (89) | 16 (84) | 14 (74) |
Stability of loci over time in vitro.
In order to determine if the numbers of repeats in the VNTR loci were stable over time, and therefore useful for epidemiological analyses in the same time frame, the stability of the loci after serial passage in LB broth was examined according to a protocol used to measure the stability of tandem repeats in V. parahaemolyticus (6). As shown in Table 3, serial passage of 95 lineages from each of three distinct clinical isolates for 30 days produced only 18 lineages that had an allele distinct from the original allele among the 1,425 tests (95 lineages by three isolates by five loci). No novel alleles were observed at the first or second locus (VC0147 or VC0437), and the third large chromosomal locus (VC1650) had a single lineage with a novel allele. In contrast, the fourth locus (VCA0171) had a total of 13 lineages with novel alleles (4.6%) and the fifth locus (VCA0283) had 4 (1.4%). Fourteen of those 18 novel alleles had one repeat more or less, and the remaining 4 had two repeats more or less, than the original number of repeats. There were 7 novel alleles with an increased number of repeats and 11 with a decreased number of repeats (not statistically different; binomial, P = 0.24). However, when the proportion of novel alleles at the large-chromosome loci was compared with that at the small-chromosome loci, the small-chromosome loci were significantly (chi square, P = 1.12 × 10−6) more likely to have novel alleles.
TABLE 3.
Number of novel alleles in VNTR loci detected among 95 independent lineages after 30 days of serial passage of three clinical isolates of V. cholerae in LB broth
| Strain | No. of novel alleles in VNTR locus: |
||||
|---|---|---|---|---|---|
| VC0147 | VC0436 | VC1650 | VCA0171 | VCA0283 | |
| 297.1 | 0 | 0 | 0 | 6 | 0 |
| 328.0 | 0 | 0 | 0 | 3 | 3 |
| 137.3 | 0 | 0 | 1 | 4 | 1 |
Because of these suggestions of greater variability at the two small-chromosome loci, subsequent MLVA of genotype relatedness focused on comparisons of three-locus genotypes using only the three large-chromosome loci (the first three alleles listed when five-locus genotypes are presented); however, five-locus genotype comparisons were also performed as a secondary analysis. When only these three loci were considered, the number of distinct genotypes among our 314 isolates was reduced to 19.
Interhousehold variation over time.
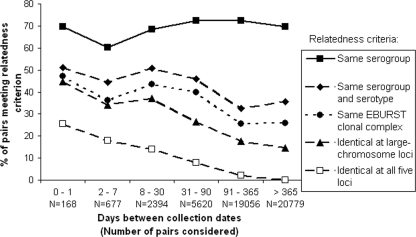
The relatedness of isolates from different households over the course of the study was analyzed in relation to the time between isolations. By any of a variety of measures of relatedness, ranging from loose (same serogroup) to strict (identical five-locus genotype) criteria, pairs of isolates from different households were more likely to correspond if they were collected closer together in time, with the probability of correspondence peaking around 30 days (Fig. 2). When serogroups and serotypes of O1 isolates were compared, 51.2% of the isolate pairs isolated within ≤1 day of each other across our entire data set matched, while only 35.5% of the pairs isolated more than a year apart matched. When measured by the strict criterion of allelic correspondence at all five VNTR loci, 23.2% of the pairs separated by ≤1 day matched, whereas only 0.12% of the pairs separated by more than a year matched. Similar analysis was performed on only the 100 index isolates (not shown); exclusion of contact isolates yielded trends similar to those seen for all of the isolates.
FIG. 2.
Variations over time in the relatedness, by various criteria, of paired V. cholerae isolates from distinct Dhaka households.
Intrahousehold variation.
Variation of genotypes within households was also considered, with the expectation that household contacts who developed V. cholerae infection within days of the index patient in their household would be infected by V. cholerae strains that were genetically identical or very similar to the index isolate. In fact, this was not always the case: 13% of the household contact isolates differed from the index isolate in the same household by serotype or serogroup, 24% belonged to different eBURST clusters, nearly one-third differed at one of the large-chromosome VNTR loci, and more than half differed from the index isolate of their household at at least one of the five VNTR loci (Table 4). Contact isolates that were genotypically or serologically unrelated to the index isolate tended to occur later in time, relative to the index isolate, than did contact isolates that were related, no matter which criterion was used to judge relatedness; differences were statistically significant for comparisons by serogroup (P = 0.04), eBURST cluster (P = 0.0003), three-locus (large-chromosome-locus) genotype (P = 0.0006), and five-locus genotype (P = 0.02) but not serotype (P = 0.08).
TABLE 4.
Relatedness of initial contact isolates to the index isolate in the household
| Condition | No. (%) isolatesa | Mean no. of days after index case (95% CI)b |
|---|---|---|
| Same serogroup and serotype | 127 (87) | 3.2 (2.6-3.9) |
| O1 serogroup, different serotype | 10 (7) | 3.2 (1.6-5.8) |
| Different serogroup | 9 (6) | 9.2 (3.5-14.9) |
| Identical at large-chromosome loci | 100 (68) | 2.6 (2.1-3.2) |
| Different at large-chromosome locus | 46 (32) | 5.8 (4.1-7.4) |
| Same 5-locus genotype | 67 (46) | 2.7 (2.1-3.4) |
| Nonidentical, same eBURST clonal complex | 44 (30) | 2.5 (1.5-3.5) |
| Unrelated by eBURST | 35 (24) | 6.6 (4.7-8.6) |
Total n = 146.
CI, confidence interval.
Intraindividual variation.
Because unexpected variation within households was seen in V. cholerae isolates, variation within individuals was also evaluated. Serial isolates from a single infected individual were obtained from only a small number of individuals: a total of 68 follow-up isolates from 50 individuals were obtained between 1 and 11 (median 2) days after the first isolates had been obtained from the same individuals. Similarly to isolates from different individuals within a household, serial isolates obtained on different days from the same individual differed by serogroup 3% of the time and at a large-chromosome VNTR locus one-third of the time (31%), with the majority of the isolates (62%) differing in some way in the five-locus genotype. Serial isolates obtained closer together in time were marginally more likely to be related than serial isolates separated by several days (Table 5), although the differences in average time intervals were not statistically significant. Of note, 6 of the 42 nonidentical follow-up isolates were third or fourth isolates from an individual and matched some prior isolate but not the first isolate from that individual. These were treated as unrelated in Table 5 but suggest that the passage of identical or related isolates over multiple days by an individual occurs somewhat more frequently than our aggregate data identify, and their failure to appear on the first day of culture may be due to our collection of only one isolate per day in the presence of what often appeared to be multistrain infections.
TABLE 5.
Relatedness of subsequent isolates to the first isolate from the same individual
| Condition | No. (%) of isolatesa | Mean no. of days after index case (95% CI)b |
|---|---|---|
| Same serogroup and serotype | 66 (97) | 1.9 (1.5-2.2) |
| O1 serogroup, different serotype | 0 (0) | |
| Different serogroup | 2 (3) | 9.5 (−10-29) |
| Identical at large-chromosome loci | 47 (69) | 1.9 (1.6-2.1) |
| Different at large-chromosome locus | 21 (31) | 2.6 (1.3-4.0) |
| Same 5-locus genotype | 26 (38) | 2.0 (1.6-2.3) |
| Nonidentical, same eBURST clonal complex | 28 (41) | 1.7 (1.3-2.1) |
| Unrelated by eBURST | 14 (21) | 3.2 (1.1-5.3) |
Total n = 68.
CI, confidence interval.
Among the 42 follow-up isolates that did not completely match the initial isolate from the same individual, there were 29 distinct genotypes, many of which were also seen elsewhere among our population-wide 314 isolates. Of these genotypes, eight (28%) were also isolated from some other individual within the same household during the 3-week follow-up period, another nine (31%) were isolated in a different household within 1 month (mean 10.4 days), another three (10%) were isolated in another household with more than 1 month of separation, and the remaining nine (31%) occurred in no other isolate in our study. Often, however, only one household was enrolled per month, so these data are likely to underrepresent the frequency of overlap of infecting genotypes between households.
MLVA, MLST, and PFGE data.
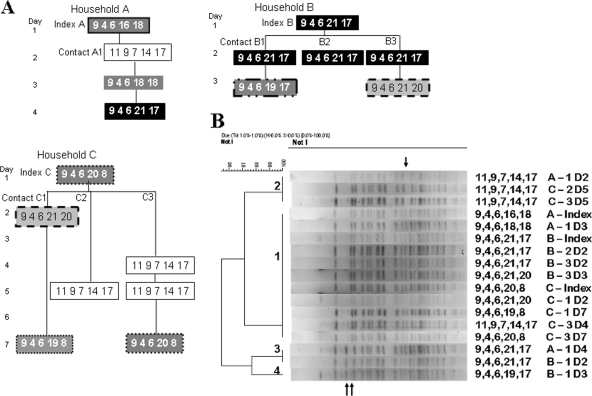
For three large households that had been enrolled in our study within a short time, each of the index and contact isolates was genotyped by MLST and PFGE, as well as by MLVA. As Fig. 3 A illustrates, the MLVA genotype of the isolates could be the same or different in individuals within the same household. Focusing on the three more-stable, large-chromosome loci, contact A1 in household A, for example, had a very different genotype than the index patient on day 2 but a genotype that matched the index genotype on days 3 and 4. Similarly, in household C, the genotype of the index patient did not match the first isolate of either contact C2 or C3 but it did match the two isolates of C1 and the third isolate of C3. The two distinct three-locus genotypes present within the household were the same for household A as for household C. In contrast, when analyzed by MLST, every isolate among these three households had exactly the same sequence type; i.e., all 4,364 sequenced base pairs were identical between any two isolates. Similarly, when the isolates were genotyped by PFGE with NotI digestion (Fig. 3B), only three variable bands were seen, and thus, by the criteria of Tenover et al. (49), these variants are considered closely related, consistent with the MLST data.
FIG. 3.
(A) MLVA genotypes of V. cholerae isolates collected from three households in February and March 2004. The genotype of each isolate is displayed as a five-digit number; identical genotypes have the same background and surrounding line. All of the isolates illustrated belong to serogroup O1 serotype Inaba. (B) PFGE gel of NotI-digested DNA from the same isolates displayed in panel A. The variable bands are indicated by arrows, and each lane is identified by the MLVA genotype and the source household, individual, and day.
Relatedness between multiple isolates from the same stool sample.
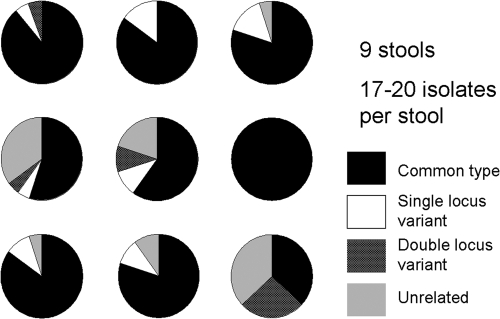
The data in Fig. 3A suggested the possibility that multiple strains circulated in the same household at the same time and that even a single household member could shed more than one strain when sampled on different days. This raised the question of whether individual patients might be infected simultaneously with more than one strain. Therefore, further analysis of intraindividual variation was performed by selecting multiple colonies from the same stool specimens collected in 2009 from each of nine individual patients. Analysis of 17 to 20 isolates from each individual stool sample showed that most of the individual stool samples had multiple MLVA genotypes present at the same time (see Table S2 in the supplemental material). As shown in Fig. 4, only one stool sample had the same genotype by MLVA in all of the colonies picked from the same stool sample; the other eight had at least two genetically distinct genotypes, and many had several distinct genotypes. Single-locus variants (compared to the type that made up the majority or in one case the plurality of isolates) were seen in seven of the stool specimens, and double-locus variants were seen in four of the specimens; it is worth noting that none of the double-locus variants had a variant allele in common with the single-locus variants in the same stool sample. Unrelated isolates (with a different three-locus genotype and in a different clonal complex) were observed in six of the stool samples. Although the number of single stool specimens is small (nine), and although each single stool specimen was sampled more thoroughly (17 to 20 isolates per stool sample) than the individual colonies sampled over multiple days in contacts (2 to 4 days of isolates per contact) in the earlier part of the study, the fraction of stool samples with multiple genotypes at the same time is consistent with the data from isolates taken individually from sequential stool samples: 66% of the single stool specimens had isolates with unrelated genotypes, while 21% of the individuals who had single isolates from multiple stool samples over time had isolates with unrelated genotypes, both consistent with an approximately 5 to 7% independent chance per isolate of being unrelated to the predominant isolate. Similarly, using a stricter delineation, 89% of the single stool specimens had isolates with distinct genotypes of some kind, while 62% of the individuals who had isolates from multiple stool samples over time had isolates with distinct genotypes.
FIG. 4.
Proportions of genetic variants among colonies selected at the same time from a single stool sample. Each pie diagram indicates a separate stool sample; each sector is in proportion to the frequency with which the type of genetic variant was found.
DISCUSSION
Our results demonstrate substantial variability of MLVA genotypes among pathogenic V. cholerae O1/O139 isolates that are difficult to differentiate by other commonly used typing methods such as PFGE or MLST. This variability is present not only within a large city where cholera is endemic, as anticipated based on previous work, but also, more surprisingly, within single households and even within individuals. Still, the majority of isolates from the same household or same individual appear closely related by MLVA, whether judged by eBURST complex or by three-locus genotype, and when they are isolated within a short time span of weeks to months, a large minority of isolates from separate households appear closely related as well.
Clustering in time of MLVA relatedness was seen population wide among our isolates. This trend is suggestive of successive and sometimes overlapping local outbreaks of distinct cholera strains that each cause multiple clustered infections within a population and then recede. Similar patterns have previously been observed by MLVA analysis elsewhere in Bangladesh (47) and are also consistent with observed fluctuations between Ogawa and Inaba serotype predominance (26) (perhaps due in part to the development of serotype-specific immunity [31] and in part to serotype conversion [8]) and with the dramatic emergence and subsequent fading to low levels of the novel serogroup O139 (40). In our data, not only identical MLVA genotypes but also different but apparently related genotypes were more likely to appear within a few days to months of each other than less-similar genotypes, consistent with a combination of processes contributing to genotypic diversity: emergent environmental strains replacing others in clinical predominance, together with gradual evolution within the clinically prevalent strains. In some instances, a fluctuation from O139 predominance to O1 Ogawa or Inaba predominance clearly reflects unrelated outbreaks. However, the gradual divergence of both three-locus and five-locus MLVA genotypes within the population over time, the time-divergent relationships within the eBURST clusters observed in Fig. 1, and the minor changes in MLVA typing observed during 30 days of serial culture are all consistent with previous observations (23) that suggest that small, persistent genotypic changes may be occurring as strains circulate through a human population.
Stability of MLVA types.
Within a household, we had anticipated that we would attribute infections to only a single strain of V. cholerae; we expected that an infected individual would shed only a single strain and that infections in other household members over subsequent days would match the first individual's infection. By MLVA, we found this to be true less than half of the time, although PFGE and MLST detected fewer differences. The possibility that some of this variation within households and individuals reflects instability or inaccuracy of MLVA genotypes, rather than simultaneous infection by multiple strains of V. cholerae, must be considered. Also, in evaluating the significance of the high degree of variation observed by MLVA, it is important to clarify what constitutes a “strain” of V. cholerae. We propose that a strain could be reasonably defined as an isolate or set of isolates that differ from related isolates by a stably inherited marker that can be used for epidemiological studies. With this definition, the pertinent question in interpreting MLVA results is whether MLVA types are stably inherited or intrinsically variable.
Many aspects of our data suggest that the observed variations represent the concurrent circulation of multiple, stably inherited MLVA genotypes. The correspondence of the three major eBURST complexes to the O1 Inaba, O1 Ogawa, and O139 groups lends credence to the significance of VNTR variations. Our experiment involving 30 days of serial culture in vitro demonstrates that changes in MLVA genotype in culture tend to be confined to specific loci and are too infrequent to account for the variation seen in clinical isolates from one day to the next in households or individuals. Also, the substantial fraction of intrahousehold and intraindividual isolate pairs that differed not only by MLVA but even by serotype and/or serogroup demonstrates at least some concurrent presence of multiple strains within single households and infected individuals. Although shifts between O1 Ogawa and O1 Inaba have been described (48), the serogroup is certainly sufficiently stable to be a strain marker, and 9 of the 19 observed serological mismatches between index and contact isolates were between the O1 and O139 serogroups, not merely between serotypes of O1. The MLVA results for multiple isolates from the same stool sample, in which multiple genotypes were usually present at the same time, also support the hypothesis that multiple strains may infect a single individual.
Examining the day-to-day variation in the genotypes of isolates within households also provides evidence that dissimilarities between index and contact isolates represent real strain differences. In the overall study population, it was observed that more-closely related contact isolate variants tended to occur fewer days after the index case than less-related variants—a difference that is also present as a nonsignificant trend in the data for multiple variants from a single individual. This observation would be most consistent with multiple exposures occurring simultaneously or close in time for household members or individuals. Further evidence of the cocirculation of multiple distinct and stable strains (rather than clonal evolution within an individual) is the sequence of isolations made in the example households illustrated in Fig. 3. If MLVA were intrinsically hypervariable, it is unlikely that clonally evolving isolates would match other isolates from the same individual or isolates from the subsequent household, as was seen; the more logical conclusion is that two different strains were circulating simultaneously.
Despite these patterns that support some amount of true strain differences within households, MLVA as an approach to differentiating strains of V. cholerae is new and relatively untested, and our data from in vitro culture do reveal some degree of instability in some of the VNTR loci analyzed. Until we understand more definitively the behavior of VNTR loci relative to other accepted strain markers, and until our findings regarding strain diversity are further validated in laboratory studies and epidemiological samples, it remains possible that some of the genotype differences we detected are a reflection of unstable fluctuations in repeat numbers, either during transmission and passage in human subjects or during the culture and genotyping process. The three large-chromosome loci appear to be a more reliable means than the other two loci of identifying stably inherited strain differences.
Potential origins and significance of MLVA variation.
The observed MLVA diversity within individuals and within households has multiple possible explanations. One possibility is that typical infecting inocula of V. cholerae, at least in the setting of this study where cholera is endemic in Bangladesh, contain multiple genotypic variants and that individuals are infected simultaneously by multiple strains of V. cholerae. This has not been previously described, but it would not be entirely unsurprising given the large inocula of organisms necessary to produce cholera, i.e., at least 108 organisms in most volunteer studies (30), 104 organisms under ideal conditions with decreased stomach acidity (43), and 102 or 103 as a typical exposure in settings where cholera is endemic, which produces illness in <10% of the individuals exposed (30). Another possible explanation for strain variation is that mutation and selection of V. cholerae strains are occurring during passage through the gut or during human-human transmission within households. A third possibility is that V. cholerae exposure is so common in the setting of this study that multiple, unrelated infecting events occur within the 3-week surveillance window; however, the fact that household contacts are far more likely to become infected on day 1, 2, or 3 after the index infection than on later days, even when the contact isolates differed from the index strain, supports a single infection event in each household. These possibilities are not mutually exclusive, since inocula containing multiple strains could initiate the infection, after which each of the strains could vary during the infection.
Another important question regarding MLVA variation is its relationship to other aspects of the growth or virulence of the organism. VNTR changes may be associated with other changes also accruing in the genome, based on which different genotypic variants detected by MLVA could differ also in features related to pathogenicity or other clinical features. Further work is required to epidemiologically characterize key MLVA strains and study their relationship to clinical illness.
Conclusions.
MLVA of multiple V. cholerae isolates from within the same household suggests that a single household and even a single individual may shed genetically distinct vibrios within a short period of time. If this is true, it has important implications for understanding the epidemiology of cholera, since it may be the result of either more-diverse or more-frequent exposure to pathogenic V. cholerae organisms than has previously been recognized. Determination of the extent and importance of the genetic variation associated with the multiple cocirculating MLVA genotypes of V. cholerae, including any associated changes in pathogenesis or transmission, will require additional laboratory and epidemiological efforts.
Supplementary Material
Acknowledgments
This research was supported in part by NIAID grants UO1 AI058935 (S.B.C.), U01 AI077883 (E.T.R.), and RO3 AI063079 (F.Q.) and by the University of Maryland Clinical Research Unit of the Food and Waterborne Diseases Integrated Research Network, which is funded by the National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health, under contract N01-AI-40014. E.A.K. is a recipient of a Fogarty International Clinical Research Scholars award (D43 TW005572 and R24 TW007988).
The work reported here was performed at the International Centre for Diarrhoeal Disease Research, Mohakhali, Dhaka, Bangladesh, and the University of Maryland School of Medicine, Baltimore.
Footnotes
Published ahead of print on 28 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albert, M. J., M. Ansaruzzaman, P. K. Bardhan, A. S. G. Faruque, S. M. Faruque, M. S. Islam, D. Mahalanabis, R. B. Sack, M. A. Salam, A. K. Siddique, M. D. Yunus, and K. Zaman. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. [PubMed] [Google Scholar]
- 2.Arakawa, E., T. Murase, S. Matsushita, T. Shimada, S. Yamai, T. Ito, and H. Watanabe. 2000. Pulsed-field gel electrophoresis-based molecular comparison of Vibrio cholerae O1 isolates from domestic and imported cases of cholera in Japan. J. Clin. Microbiol. 38:424-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakhshi, B., H. M. Barzelighi, M. Adabi, A. R. Lari, and M. R. Pourshafie. 2009. A molecular survey on virulence associated genotypes of non-O1 non-O139 Vibrio cholerae in aquatic environment of Tehran, Iran. Water Res. 43:1441-1447. [DOI] [PubMed] [Google Scholar]
- 4.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Call, D. R., L. Orfe, M. A. Davis, S. Lafrentz, and M. S. Kang. 2008. Impact of compounding error on strategies for subtyping pathogenic bacteria. Foodborne Pathog. Dis. 5:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, D. N., F. M. Khambaty, I. K. Wachsmuth, R. V. Tauxe, and T. J. Barrett. 1994. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:1685-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, S., K. Ghosh, A. Raychoudhuri, A. Pan, M. K. Bhattacharya, A. K. Mukhopadhyay, T. Ramamurthy, S. K. Bhattacharya, and R. K. Nandy. 2007. Phenotypic and genotypic traits and epidemiological implication of Vibrio cholerae O1 and O139 strains in India during 2003. J. Med. Microbiol. 56:824-832. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, S. N., and K. Chaudhuri. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65-79. [DOI] [PubMed] [Google Scholar]
- 10.Choi, S. Y., J. H. Lee, Y. S. Jeon, H. R. Lee, E. J. Kim, M. Anssaruzzaman, N. Bhuiyan, H. Endtz, S. Niyogi, B. Sarkar, G. Nair, B. Nguyen, N. Hien, C. Czerkinsky, J. Clemens, J. Chun, and D. W. Kim. 2010. Multilocus variable-number tandem repeat analysis of Vibrio cholerae O1 El Tor strains harbouring classical toxin B. J. Med. Microbiol. 59(Pt. 7):763-769. [DOI] [PubMed] [Google Scholar]
- 11.Chun, J., C. J. Grim, N. A. Hasan, J. H. Lee, S. Y. Choi, B. J. Haley, E. Taviani, Y. S. Jeon, D. W. Kim, J. H. Lee, T. S. Brettin, D. C. Bruce, J. F. Challacombe, J. C. Detter, C. S. Han, A. C. Munk, O. Chertkov, L. Meincke, E. Saunders, R. A. Walters, A. Huq, G. B. Nair, and R. R. Colwell. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442-15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constantin de Magny, G., R. Murtugudde, M. R. Sapiano, A. Nizam, C. W. Brown, A. J. Busalacchi, M. Yunus, G. B. Nair, A. I. Gil, C. F. Lanata, J. Calkins, B. Manna, K. Rajendran, M. K. Bhattacharya, A. Huq, R. B. Sack, and R. R. Colwell. 2008. Environmental signatures associated with cholera epidemics. Proc. Natl. Acad. Sci. U. S. A. 105:17676-17681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper, K. L., C. K. Luey, M. Bird, J. Terajima, G. B. Nair, K. M. Kam, E. Arakawa, A. Safa, D. T. Cheung, C. P. Law, H. Watanabe, K. Kubota, B. Swaminathan, and E. M. Ribot. 2006. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3:51-58. [DOI] [PubMed] [Google Scholar]
- 14.Dalsgaard, A., M. N. Skov, O. Serichantalergs, P. Echeverria, R. Meza, and D. N. Taylor. 1997. Molecular evolution of Vibrio cholerae O1 strains isolated in Lima, Peru, from 1991 to 1995. J. Clin. Microbiol. 35:1151-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danin-Poleg, Y., L. A. Cohen, H. Gancz, Y. Y. Broza, H. Goldshmidt, E. Malul, L. Valinsky, L. Lerner, M. Broza, and Y. Kashi. 2007. Vibrio cholerae strain typing and phylogeny study based on simple sequence repeats. J. Clin. Microbiol. 45:736-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, S., and S. Gupta. 2005. Diversity of Vibrio cholerae strains isolated in Delhi, India, during 1992-2000. J. Health Popul. Nutr. 23:44-51. [PubMed] [Google Scholar]
- 17.Dutta, B., R. Ghosh, N. C. Sharma, G. P. Pazhani, N. Taneja, A. Raychowdhuri, B. L. Sarkar, S. K. Mondal, A. K. Mukhopadhyay, R. K. Nandy, M. K. Bhattacharya, S. K. Bhattacharya, and T. Ramamurthy. 2006. Spread of cholera with newer clones of Vibrio cholerae O1 El Tor, serotype Inaba, in India. J. Clin. Microbiol. 44:3391-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque, S. M., S. K. Roy, A. R. Alim, A. K. Siddique, and M. J. Albert. 1995. Molecular epidemiology of toxigenic Vibrio cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J. Clin. Microbiol. 33:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. U. S. A. 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garg, P., A. Aydanian, D. Smith, J. G., Morris, Jr., G. B. Nair, and O. C. Stine. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh, R., G. B. Nair, L. Tang, J. G. Morris, N. C. Sharma, M. Ballal, P. Garg, T. Ramamurthy, and O. C. Stine. 2008. Epidemiological study of Vibrio cholerae using variable number of tandem repeats. FEMS Microbiol. Lett. 288:196-201. [DOI] [PubMed] [Google Scholar]
- 24.Green, H. P., J. A. Johnson, J. P. Furuno, S. M. Strauss, E. N. Perencevich, E. Lautenbach, D. Lee, and A. D. Harris. 2007. Impact of freezing on the future utility of archived surveillance culture specimens. Infect. Control Hosp. Epidemiol. 28:886-888. [DOI] [PubMed] [Google Scholar]
- 25.Guerrant, R. L., B. A. Carneiro-Filho, and R. A. Dillingham. 2003. Cholera, diarrhea, and oral rehydration therapy: triumph and indictment. Clin. Infect. Dis. 37:398-405. [DOI] [PubMed] [Google Scholar]
- 26.Harris, A. M., F. Chowdhury, Y. A. Begum, A. I. Khan, A. S. Faruque, A. M. Svennerholm, J. B. Harris, E. T. Ryan, A. Cravioto, S. B. Calderwood, and F. Qadri. 2008. Shifting prevalence of major diarrheal pathogens in patients seeking hospital care during floods in 1998, 2004, and 2007 in Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 79:708-714. [PMC free article] [PubMed] [Google Scholar]
- 27.Harris, J. B., R. C. Larocque, F. Chowdhury, A. I. Khan, T. Logvinenko, A. S. Faruque, E. T. Ryan, F. Qadri, and S. B. Calderwood. 2008. Susceptibility to Vibrio cholerae infection in a cohort of household contacts of patients with cholera in Bangladesh. PLoS Negl. Trop. Dis. 2:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huq, A., R. R. Colwell, R. Rahman, A. Ali, M. A. Chowdhury, S. Parveen, D. A. Sack, and E. Russek-Cohen. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huq, A., R. B. Sack, A. Nizam, I. M. Longini, G. B. Nair, A. Ali, J. G. Morris, Jr., M. N. Khan, A. K. Siddique, M. Yunus, M. J. Albert, D. A. Sack, and R. R. Colwell. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71:4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelle, K., M. Pascual, and M. Yunus. 2006. Serotype cycles in cholera dynamics. Proc. Biol. Sci. 273:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotetishvili, M., O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. G. Morris, Jr. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. H., K. H. Han, S. Y. Choi, M. E. Lucas, C. Mondlane, M. Ansaruzzaman, G. B. Nair, D. A. Sack, L. von Seidlein, J. D. Clemens, M. Song, J. Chun, and D. W. Kim. 2006. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 55:165-170. [DOI] [PubMed] [Google Scholar]
- 34.Lizárraga-Partida, M. L., and M. L. Quilici. 2009. Molecular analyses of Vibrio cholerae O1 clinical strains, including new nontoxigenic variants isolated in Mexico during the cholera epidemic years between 1991 and 2000. J. Clin. Microbiol. 47:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohapatra, S. S., D. Ramachandran, C. K. Mantri, R. R. Colwell, and D. V. Singh. 2009. Determination of relationships among non-toxigenic Vibrio cholerae O1 biotype El Tor strains from housekeeping gene sequences and ribotype patterns. Res. Microbiol. 160:57-62. [DOI] [PubMed] [Google Scholar]
- 37.Morris, J. G., Jr. 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179-191. [DOI] [PubMed] [Google Scholar]
- 38.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem-repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popovic, T., C. Bopp, O. Olsvik, and K. Wachsmuth. 1993. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J. Clin. Microbiol. 31:2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramamurthy, T., S. Yamasaki, Y. Takeda, and G. B. Nair. 2003. Vibrio cholerae O139 Bengal: odyssey of a fortuitous variant. Microbes Infect. 5:329-344. [DOI] [PubMed] [Google Scholar]
- 41.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 42.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 43.Sack, D. A., C. O. Tacket, M. B. Cohen, R. B. Sack, G. A. Losonsky, J. Shimko, J. P. Nataro, R. Edelman, M. M. Levine, R. A. Giannella, G. Schiff, and D. Lang. 1998. Validation of a volunteer model of cholera with frozen bacteria as the challenge. Infect. Immun. 66:1968-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safa, A., G. B. Nair, and R. Y. Kong. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46-54. [DOI] [PubMed] [Google Scholar]
- 45.Shimada, T., E. Arakawa, K. Itoh, T. Okitso, A. Matsushima, Y. Asai, S. Yamai, T. Nakazato, G. Nair, M. Albert, and Y. Takeda. 1994. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 28:175-178. [Google Scholar]
- 46.Singh, D. V., M. H. Matte, G. R. Matte, S. Jiang, F. Sabeena, B. N. Shukla, S. C. Sanyal, A. Huq, and R. R. Colwell. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stine, O. C., M. Alam, L. Tang, G. B. Nair, A. K. Siddique, S. M. Faruque, A. Huq, R. Colwell, R. B. Sack, and J. G. Morris, Jr. 2008. Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerg. Infect. Dis. 14:831-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stroeher, U. H., L. E. Karageorgos, R. Morona, and P. A. Manning. 1992. Serotype conversion in Vibrio cholerae O1. Proc. Natl. Acad. Sci. U. S. A. 89:2566-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Zhou, H. J., B. W. Diao, Z. G. Cui, B. Pang, L. J. Zhang, and B. Kan. 2009. Comparison of automated ribotyping and pulsed-field gel electrophoresis for subtyping of Vibrio cholerae. Lett. Appl. Microbiol. 48:726-731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.