Abstract
Surface antigen variation in Mycoplasma agalactiae, the etiologic agent of contagious agalactia in sheep and goats, is governed by site-specific recombination within the vpma multigene locus encoding the Vpma family of variable surface lipoproteins. This high-frequency Vpma phase switching was previously shown to be mediated by a Xer1 recombinase encoded adjacent to the vpma locus. In this study, it was demonstrated in Escherichia coli that the Xer1 recombinase is responsible for catalyzing vpma gene inversions between recombination sites (RS) located in the 5′-untranslated region (UTR) in all six vpma genes, causing cleavage and strand exchange within a 21-bp conserved region that serves as a recognition sequence. It was further shown that the outcome of the site-specific recombination event depends on the orientation of the two vpma RS, as direct or inverted repeats. While recombination between inverted vpma RS led to inversions, recombination between direct repeat vpma RS led to excisions. Using a newly developed excision assay based on the lacZ reporter system, we were able to successfully demonstrate under native conditions that such Xer1-mediated excisions can indeed also occur in the M. agalactiae type strain PG2, whereas they were not observed in the control xer1-disrupted VpmaY phase-locked mutant (PLMY), which lacks Xer1 recombinase. Unless there are specific regulatory mechanisms preventing such excisions, this might be the cost that the pathogen has to render at the population level for maintaining this high-frequency phase variation machinery.
Members of the bacterial class Mollicutes, which are generally referred to as mycoplasmas, are considered among the simplest self-replicating prokaryotes carrying minimal genomes. Even having lost many biosynthetic pathways during a reductive evolution, mycoplasmas represent important pathogens of humans, animals, and plants, as they are equipped with sophisticated molecular mechanisms allowing them to spontaneously change their cell surface repertoire to persist in immunocompetent hosts (25).
The important ruminant pathogen Mycoplasma agalactiae causes contagious agalactia in sheep and goats and exhibits antigenic diversity by site-specific DNA rearrangements within a pathogenicity island-like gene locus (9, 10, 26). The so-called vpma locus constitutes a family of six distinct but related genes that encode major immunodominant membrane lipoproteins, the Vpmas (variable proteins of Mycoplasma agalactiae) (10, 11). These surface-associated proteins vary in expression at an unusually high frequency, and only one vpma gene at a time is transcribed from a single promoter present in that locus, while all other genes are silent (9, 10). An open reading frame (ORF) with homology to the λ-integrase family of site-specific recombinases was found in the vicinity of the vpma locus and was predicted to mediate DNA inversions responsible for switching the promoter from an active vpma gene to a silent one, resulting in alteration of vpma expression (9, 10). This recombinase, designated Xer1, was indeed recently demonstrated to be responsible for phase variation of Vpma proteins (4). Targeted knockouts of the xer1 gene by homologous recombination prevented Vpma switching and produced Vpma phase-locked mutants (PLMs) steadily expressing a single vpma gene without any variation. Complementation of the wild-type xer1 gene in these PLMs restored Vpma phase variation (4). Similar systems generating surface diversity by DNA inversions involving site-specific recombination have been identified in other mycoplasma species (3, 18, 26).
Site-specific recombination systems are widespread among bacteria, and the biological functions of these systems depend strongly on the participating recombination sites (RS) (16, 24, 27). Excision events between direct repeat RS usually resolve chromosome or plasmid dimers, which can arise through homologous recombination, ensuring proper segregation of newly replicated genetic material to daughter cells (1). Also, site-specific recombination mediates integration and excision of phage genomes into and out of the host chromosome (13). In contrast, site-specific inversion involving inverted repeat RS generates genetic diversity and often controls the expression of genes that are important for pathogenesis (21).
The Xer1 recombinase of M. agalactiae belongs to the λ-integrase family of site-specific recombinases (10). Members of this family share four strongly conserved amino acid residues (R-H-R-Y) within the C-terminal half of the protein. This tetrad includes the active tyrosine residue that is directly involved in the recombination reaction (8). Recombination occurs by formation and resolution of a Holliday junction intermediate involving a covalent linkage between the recombinase and the DNA through the tyrosine residue. Since energy cofactors such as ATP are not required, such recombination events can occur in the absence of replication (16, 24).
Sequence alignment of vpma genes identified a conserved 21-bp region within the 5′-untranslated region (UTR) in all vpma genes that was predicted to be involved in Xer1-mediated inversions (10). The present study clearly demonstrates that the Xer1 recombinase recognizes RS located within the 5′ UTR of vpma genes, causing cleavage and strand exchange within a conserved region of 21 bp. By placing two vpma-derived RS on a plasmid along with the xer1 gene, recombination events were demonstrated in Escherichia coli upon Xer1 induction via PCR and restriction analysis. Although the conserved 21-bp region was sufficient for inversions, additional nucleotides flanking it at the 5′ end were found to have a positive influence on the rate of recombination. An interesting outcome of these studies was that Xer1 also mediated excisions between direct repeat vpma RS in E. coli. This raised the intriguing possibility that such Xer1-mediated excisions also occur in the native M. agalactiae system. For further analysis of such excision events in the native system, we tested the feasibility of using the lacZ reporter tool in M. agalactiae, as lacZ is known to be expressed successfully in few other mycoplasma species, to study gene expression by use of promoter probe vectors (15, 19, 22, 23). We developed an excision assay based on blue-white phenotype selection to study Xer1-mediated excisions in M. agalactiae, thus displaying a novel application of the lacZ reporter gene in mycoplasmas. Successful implementation of this reporter system demonstrated Xer1-mediated excisions in the M. agalactiae type strain PG2, based on blue-white selection and PCR analysis. As expected, such excisions were not observed in the control xer1-disrupted VpmaY phase-locked mutant (PLMY), which lacks Xer1. Excisions in the native system imply that genetic material is susceptible to loss, which might be the cost for maintaining the machinery of high-frequency gene shuffling for a greater population advantage, unless there are specific regulatory mechanisms preventing such excisions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strain DH10B (Invitrogen GmbH, Lofer, Austria) was used for cloning and expression of the xer1 gene. Plasmids used for Xer1-mediated excision and inversion experiments in E. coli were derived from pBAD24 (14), and plasmid constructs for studying excisions in M. agalactiae were derived from pISM2062 (22). Plasmids p5H1.8 (11), p5H4.7 (10), and pAWC10-lac (20) have been described elsewhere. Excision studies were carried out with M. agalactiae type strain PG2 (32) and the xer1 disruptant strain PLMY (4). Cells were grown in SP4 (33) broth, and transformants were selected on SP4 agar plates containing gentamicin (50 μg/ml) and/or tetracycline (2 μg/ml), as appropriate. To monitor lacZ expression on the basis of blue-white selection, M. agalactiae cells carrying the lacZ gene were grown on SP4 agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at a concentration of 160 μg/ml.
DNA manipulations.
Preparation of plasmid and genomic DNAs and isolation of DNA fragments from agarose gels were carried out using suitable kits from Promega (Wizard SV gel and PCR clean-up system; Promega, Mannheim, Germany), Peqlab (EZNA plasmid miniprep kit I; Peqlab-Biotechnologie GmbH, Erlangen, Germany), and Qiagen (QIAamp DNA Mini kit; Qiagen GmbH, Hilden, Germany). Restriction endonucleases and nucleotides were purchased from Promega, T4 DNA ligase was purchased from Roche Diagnostics (Vienna, Austria), and Antarctic phosphatase was purchased from New England Biolabs (Frankfurt am Main, Germany). Transformation of E. coli cells was performed by electroporation with a Bio-Rad Gene-Pulser II instrument (Bio-Rad Laboratories GmbH, Vienna, Austria), using a 1.25-V voltage, 25-μF capacitance, and 200-Ω resistance. Transformation of M. agalactiae cells was carried out as described previously (5). Oligonucleotide synthesis and sequencing were carried out at VBC-Biotech Service GmbH (Vienna, Austria). Standard molecular procedures were performed as described by Sambrook et al. (28).
xer1 induction experiments in E. coli.
E. coli DH10B cells transformed with plasmids used for inversion and excision experiments were plated on LB agar plates containing ampicillin (100 μg/ml) and 5% glucose (for inhibition of basal expression from the PBAD promoter) and were grown overnight at 28°C. Individual transformants were picked, inoculated into LB broth containing ampicillin and glucose, and grown overnight at 28°C. For xer1 induction, 600 μl of the overnight culture was transferred to 60 ml LB broth containing 100 μg/ml ampicillin, and cells were grown at 37°C. Growth of bacteria was monitored by measurement of the optical density at 600 nm (OD600). At an OD600 of 0.3, xer1 expression was induced by adding 0.5% l-arabinose. Samples for plasmid preparations were removed at different time points (2 h, 4 h, 6 h, and 20 h), as appropriate. Samples for plasmid preparation of uninduced cultures serving as negative controls were taken from overnight cultures and/or at the beginning of induction. Plasmid samples were used for subsequent PCR and restriction analysis assays.
PCR amplification.
PCRs for cloning (except for amplification of the lacZ gene) were carried out in a total volume of 100 μl consisting of 50 ng template DNA, a 1 μM concentration of each primer, 25 mM MgCl2, a 0.2 μM concentration of each deoxynucleoside triphosphate (dNTP), and 5 U of Taq DNA polymerase (Promega) in 1× PCR buffer supplied by the manufacturer. Cycling parameters consisted of 1 cycle of 3 min at 94°C for initial denaturation followed by 30 cycles of 1 min of denaturation at 94°C, 1 min at various annealing temperatures (60°C for xer1, 58 to 63°C for the 200-bp vpmaY RS, 57°C for RS111Y, and 58°C for RS184U), and 1 min at 72°C for extension, with a final extension step of 5 min at 72°C.
The lacZ gene with its native Shine-Dalgarno sequence from E. coli was amplified in a 100-μl mix constituting 50 ng of plasmid pAWC10-lac, 1 μM (each) primers LacfwBN (containing BamHI and NcoI sites for subsequent cloning) and LacrvSB (containing SmaI and BglII sites), a 0.2 μM concentration of each dNTP, 1 U of Long PCR enzyme mix (Fermentas GmbH, St. Leon-Rot, Germany), and 1× PCR buffer (containing 25 mM MgCl2), supplied by the manufacturer. Cycling conditions consisted of 1 cycle of 2 min at 94°C for initial denaturation, 10 cycles of 20 s at 94°C for denaturation and 3 min at 68°C for extension, and 20 cycles of 20 s at 94°C for denaturation and 3 min 20 s at 68°C for extension, with a final extension step of 10 min at 68°C.
A three-primer hot-start PCR for detection of inversion events was accomplished in a 25-μl volume containing 25 ng of template DNA, 1 μM (each) primers P1 and P2, 0.2 μM primer P4, 2.5 mM MgCl2, a 0.2 μM concentration of each dNTP, 1 M betaine, and 5 U of Hot FIREPol DNA polymerase I (Solis BioDyne OU, Tartu, Estonia) in 1× PCR buffer, supplied by the manufacturer. Cycling parameters consisted of 1 cycle of 15 min at 95°C for initial denaturation followed by 30 cycles of 43 s of denaturation at 94°C, 43 s of annealing at 61°C, and 1 min 33 s at 72°C for extension, with a final extension step of 10 min at 72°C.
PCR for the detection of Xer1-mediated excision events in M. agalactiae was carried out in a volume of 25 μl, which consisted of 10 ng genomic DNA obtained from PG2 or PLMY transformants, 1 μM (each) primers 184Ubfw and ISR-f, 2.5 mM MgCl2, a 0.2 μM concentration of each dNTP, and 5 U of Hot FIREPol DNA polymerase I (Solis BioDyne OU, Tartu, Estonia) in 1× PCR buffer, supplied by the manufacturer. Cycling parameters consisted of 1 cycle of 15 min at 95°C for initial denaturation followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 3 min 50 s of extension at 72°C, with a final extension step of 5 min at 72°C.
Plasmid construction. (i) Construction of plasmids pBADRS21x2 and pBADXerRS21x2.
Two micrograms each of the primers Ol1new2sen and Ol1new2asen (oligonucleotides are summarized in Table 1) was mixed with 46 μl of annealing buffer (100 mM potassium acetate, 30 mM HEPES, 2 mM magnesium acetate, pH 7.4), boiled at 100°C for 5 min, and cooled down gradually to room temperature within 60 min. The resulting double-stranded 21-bp RS contained HindIII and XbaI overhangs and was ligated into HindIII and XbaI sites of pBAD24. The ligation mixture was transformed into E. coli DH10B by electroporation as described previously, resulting in plasmid pBADRS21. Another 21-bp RS was introduced into the NsiI and NarI sites of pBADRS21 to obtain a vector with two 21-bp inverted repeat RS. The second 21-bp RS, containing NsiI and NarI overhangs, was similarly obtained by annealing oligonucleotides RS21NsiI and RS21NarI and was ligated into the corresponding sites of pBADRS21 to obtain vector pBADRS21x2, containing two inverted repeats of the 21-bp RS. The xer1 gene was amplified from plasmid p5H4.7 by using primers RecATGET28 (containing an NcoI site) and XerrvSma (containing a SmaI site). The 750-bp product was digested with NcoI and SmaI and cloned at the corresponding sites of pBADRS21x2, resulting in plasmid pBADXerRS21x2.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a |
|---|---|
| RSmcsX | AATTAATCTAGAGCAATTAGTAGAAGATTGTAGCG |
| RSmcsH | AATGTAAAGCTTGGCCATTGAAGCAACTGATCCAAG |
| Rsd1cla | AATTAAATCGATGCAATTAGTAGAAGATTGTAGCG |
| Rsd1nsi | AATGTAATGCATGGCCATTGAAGCAACTGATCCAAG |
| Rsd2cla | AATGTAATCGATGCCCATTGAAGCAACTGATCCAAG |
| Rsd2nsi | AATTAAATGCATGCAATTAGTAGAAGATTGTAGCG |
| RecATGET28 | AACATTCCATGGTAGAAACCTTTATCAC |
| XerrvSma | GCTCGACCCGGGCTACTTACTATTAAGC |
| P1 | CAGACAATTGACGGCTTGACG |
| P2 | TTCTCTTACTGTCATGCCATCCG |
| P4 | TGGACTCTTGTTCCAAACTTG |
| Ol1new2sen | CTAGATTGATATTTATTAATAGATTTA |
| Ol1new2asen | AGCTTAAATCTATTAATAAATATCAAT |
| Oligo#2sense | CTAGTTGATATTTATTAATAGATTTATAAAGCATTA |
| Oligo#2asense | AACTATAAATAATTATCTAAATATTTCGTAATTCGA |
| Oligo#3sense | CTAGTTGATATTTATTAATAGATTTATAAAGCATTTTTAAGGCTATTTTATAGCCTTTAAA |
| Oligo#3asense | AACTATAAATAATTATCTSAATATTTCGTAAAAATTCCGATAAAATATCGGAAATTTTCGA |
| pBADoligo#4fw | GATCAATCTAGAGCTATTAATTTAGCACTTAATAC |
| pBADoligo#4rv | AATGTCAAGCTTTCATAAATTTATCCTTTC |
| Ol9sen | CTAGAGCTATTAATTTAGCACTTAATACATCATATAATAAATTGATATTATTAATAGATTTA |
| Ol9asen | AGCTTAAATCTATTAATAAATATCAATTTATTATATGATGTATTAAGTGCTAAATTAATAGCT |
| PrimerRSU | AATTGCTCTAGAAAGTTTAATCAAAACTTAACG |
| OligoRSXsen | CTAGATTTAGTAAGCAGTCGCTAAGCATATACTTGTACTTTTGATATTTATTAATAGATTTA |
| OligoRSXasen | AGCTTAAATCTATTAATAAATATCAAAAGTACAAGTATATGCTTAGCGACTGCTTACTAAAT |
| OligoRSUsen | CTAGAGATGTTTTTATATTCTCATAAACTCACTGTTTTTGATATTTATTAATAGATTTA |
| OligoRSUasen | AGCTTAAATCTATTAATAAATATCAAAAACAGTGAGTTTATGAGAATATAAAAACATCT |
| RS21NsiI | CGCCAAATCTATTAATAAATATCAAATGCA |
| RS21NarI | TTTGATATTTATTAATAGATTTGG |
| LacfwBN | GCACTAGGATCCCCATGGAGGAAACAGCTATGACCATGATTA |
| LacrvSB | ATCACGCCCGGGAGATCTTTATTATTTTTGACACCAGACCAACTG |
| 184Ubfw | TCATGAGGATCCAAGTTTAATCAAAACTTAAC |
| 184Unrv | ATACAACCATGGGGCCATTGAAGCAACTGATC |
| 200YBfw | TACTAGAGATCTGGCCATTGAAGCAACTG |
| 200YBrv | AAGCTTAGATCTGCAATTAGTAGAAGATTG |
| ISR-f | TATAACGCGTGATAAAGTCCGTATAATTGTG |
Restriction sites (underlined) were introduced into primers.
(ii) Construction of plasmids pBADXerIR200Y and pBADXerDR200Y.
A 200-bp fragment containing the vpmaY RS, corresponding to the 5′ UTR of the vpmaY gene, was obtained by PCR amplification as described previously, using plasmid p5H1.8 as the template and oligonucleotides RSmcsX (containing an XbaI site) and RSmcsH (containing a HindIII site) as primers. The HindIII- and XbaI-digested amplicon was then ligated into the corresponding sites of pBAD24, resulting in plasmid pBADRS. Another copy of the 200-bp fragment was introduced into plasmid pBADRS, both as a direct and as an inverted repeat, after PCR amplification from plasmid p5H1.8 by use of oligonucleotides Rsd1cla (containing a ClaI site) and Rsd1nsi (containing an NsiI site) for direct repeat RS and oligonucleotides Rsd2cla (containing a ClaI site) and Rsd2nsi (containing an NsiI site) for inverted repeat RS. The 200-bp PCR fragments were cut with ClaI and NsiI and introduced into NsiI- and NarI-cut plasmid pBADRS. NarI and ClaI produce compatible cohesive ends, and therefore NarI overhangs can be ligated to ClaI overhangs. The two plasmids were designated pBADDR (direct repeat RS) and pBADIR (inverted repeat RS). The xer1 gene was introduced into plasmids pBADDR and pBADIR as described for plasmid pBADXerRS21x2, resulting in plasmids carrying two 200-bp RS, either as inverted repeats (pBADXerIR200Y) or as direct repeats (pBADXerDR200Y), along with the xer1 gene.
(iii) Construction of plasmids pBADXerIR21Y, pBADXerIR31Y3′, pBADXerIR56Y3′, pBADXerIR57Y5′, and pBADXerIR111Y.
Oligonucleotides Ol1new2sen and Ol1new2asen were annealed to obtain RS21Y, Oligo#2sense and Oligo#2asense were annealed for RS31Y3′, Oligo#3sense and Oligo#3asense were annealed for RS56Y3′, and Ol9sen and Ol9asen were annealed for RS57Y5′. The annealed oligonucleotides carried XbaI and HindIII overhangs for cloning into the corresponding sites of pBADXerIR200Y, resulting in plasmids pBADXerIR21Y, pBADXerIR31Y3′, pBADXerIR56Y3′, and pBADXerIR57Y5′, respectively. RS111Y was obtained by PCR as described above, using plasmid p5H1.8 as the template and oligonucleotides pBADoligo#4fw (containing an XbaI site) and pBADoligo#4rv (containing a HindIII site) as primers. The HindIII- and XbaI-cut PCR product was introduced into the corresponding sites of pBADXerIR200Y, resulting in plasmid pBADXerIR111Y.
(iv) Construction of plasmids pBADXer200Y/184U, pBADXerIR200Y/56X5′, and pBADXer53U5′/57Y5′.
Plasmid pBADXer200Y/184U was obtained by replacing the XbaI- and HindIII-cloned 200-bp vpmaY RS of pBADXerIR200Y with a 184-bp vpmaU RS generated by PCR, using plasmid p5H4.7 as the template and PrimerRSU (containing an XbaI site) and RSmcsH (containing a HindIII site) as primers. The PCR fragment, restricted with HindIII and XbaI, was cloned into the corresponding sites of vector pBADXerIR200Y, resulting in plasmid pBADXerIR200Y/184U, containing a 200-bp vpmaY RS and a 184-bp vpmaU RS, aligned as inverted repeats. Similarly, plasmid pBADXerIR200Y/56X5′ was obtained by replacing the XbaI- and HindIII-cloned 200-bp vpmaY RS with a 56-bp RS originating from the vpmaX gene. Oligonucleotides OligoRSXsen and OligoRSXasen were annealed as described above. The obtained fragment, containing XbaI and HindIII overhangs, was cloned into the corresponding sites of vector pBADXerIR200Y, resulting in plasmid pBADXerIR200Y/56X5′, containing a 200-bp vpmaY RS and a 56-bp vpmaX RS, aligned as inverted repeats. Plasmid pBADXer53U5′/57Y5′ was obtained by replacing the NarI/ClaI- and NsiI-cloned 200-bp vpmaY RS of pBADXerIR57Y5′ with a 53-bp vpmaU RS generated by annealing oligonucleotides OligoRSUsen and OligoRSUasen as described above. The obtained fragment contained an MscI blunt end and a BstAPI overhang for cloning at the respective sites in plasmid pBADXerIR57Y5′, resulting in plasmid pBADXer53U5′/57Y5′, containing a 57-bp vpmaY RS and a 53-bp vpmaU RS, aligned as inverted repeats.
(v) Construction of plasmids pIL and pILDR.
The lacZ gene with its native Shine-Dalgarno sequence from E. coli was obtained by PCR as described above (see “PCR amplification”). The BamHI- and SmaI-cut 3.1-kb product was introduced into the corresponding sites in the left IS element of transposon Tn4001mod in plasmid pISM2062, resulting in plasmid pIL. The 184-bp vpmaU RS was amplified as described for the construction of pBADXer200Y/184U, except that oligonucleotides 184Ubfw (containing a BamHI site) and 184Unrv (containing an NcoI site) were used as primers. The BamHI- and NcoI-cut product was introduced into the corresponding sites of pIL, resulting in vector pILRSU, carrying the lacZ gene flanked by a 184-bp vpmaU RS at the 5′ end. The 200-bp vpmaY RS was amplified as described for construction of pBADXer200Y, except that oligonucleotides 200YBfw and 200YBrv (both containing BglII sites) were used as primers. The PCR fragment, restricted with BglII, was introduced into the BglII site of dephosphorylated plasmid pILRSU. The direct repeat orientation of this fragment was verified by sequencing. The resulting vector, pILDR, contained the lacZ gene flanked by a 184-bp vpmaU RS at the 5′ end and a 200-bp vpmaY RS at the 3′ end, aligned as direct repeats.
Restriction analysis and agarose gel electrophoresis of recombination products.
Recombination products derived from plasmid pBADXerDR200Y, containing direct repeat RS, were applied to a 1% (wt/vol) agarose gel directly or after linearization with XbaI and HindIII restriction endonucleases. Inversion events within plasmids containing two inverted repeat RS (pBADRS21x2, pBADXerRS21x2, pBADXerIR200Y pBADXerIR21Y, pBADXerIR31Y3′, pBADXerIR56Y3′, pBADXerIR57Y5′, pBADXerIR111Y, pBADXer200Y/184U, pBADXerIR200Y/56X5′, and pBADXer53U5′/57Y5′) were verified by applying the HindIII- and EcoRV-digested samples to a 1% (wt/vol) agarose gel and visualizing them under UV light after ethidium bromide staining.
RESULTS
A conserved 21-bp sequence common to all vpma genes is sufficient for Xer1-mediated inversion.
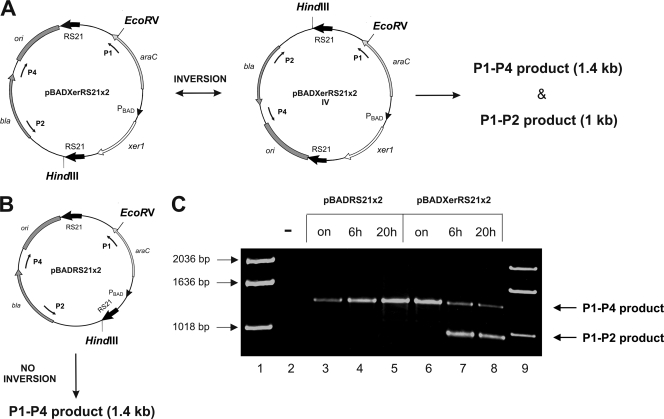
Alignment of the 5′ UTRs of all six vpma genes present in clone 55-5 of type strain PG2 identified a conserved 21-bp region (TTGATATTTATTAATAGATTT) thought to be involved in Xer1-mediated recombination (10). To verify if this 21-bp RS is indeed involved and is sufficient for recombination, two 21-bp inverted repeat RS were introduced into plasmid pBAD24, together with the xer1 gene, which was placed under the transcriptional control of the arabinose promoter. The resulting plasmid, pBADXerRS21x2, was introduced into E. coli, and recombination events were followed by testing the plasmid preparations obtained from samples removed after 6 and 20 h of xer1 induction. Uninduced samples from overnight cultures of cells carrying pBADXerRS21x2 and cells carrying pBADRS21x2 lacking the xer1 gene served as negative controls. For detection of inversion products, a restriction analysis method was used first, whereby the plasmid DNA was digested using HindIII and EcoRV restriction endonucleases and observed for the appearance of recombination products upon agarose gel electrophoresis. An inversion event between the two inverted repeat RS in pBADXerRS21x2 would place the HindIII site near the EcoRV site (Fig. 1A) and would result in the appearance of restriction fragments of 4.7 kb and 0.4 kb, in addition to the original, nonrecombined plasmid fragments of 3.4 kb and 1.7 kb, if a sufficient amount of recombined product was present after xer1 induction. However, we could not detect any inversion products by this restriction analysis method for samples taken both 6 and 20 h after xer1 induction (data not shown). To check if inversion events between two 21-bp RS occur at levels much below the detection limit of the restriction analysis assay, a more-sensitive three-primer PCR assay was developed (Fig. 1). Primer P1 (anneals to a region of the araC gene) and primer P2 (anneals to a region of the bla gene) enable the amplification of a 1-kb “recombinant” fragment only in case of an inversion between the two RS (Fig. 1A), whereas primer P1 and primer P4 (anneals to a region between the pBR322 ori and the bla gene) enable the amplification of a 1.4-kb “nonrecombinant” product from the nonrecombined vector serving as an internal positive control in the PCR assay (Fig. 1A and B). Besides the 1.4-kb P1-P4 product, samples of pBADXerRS21x2 showed a 1-kb P1-P2 amplicon after 6 and 20 h of xer1 induction (Fig. 1C, lanes 7 and 8), demonstrating that inversion occurred between the two 21-bp inverted repeat RS. This 1-kb P1-P2 recombinant amplicon was absent in the uninduced sample of pBADXerRS21x2 (Fig. 1C, lane 6) and in samples corresponding to the plasmid pBADRS21x2 (Fig. 1C, lanes 3 to 5), which carries two 21-bp inverted repeat RS but lacks the xer1 gene, clearly demonstrating the role of Xer1 recombinase in mediating inversions at the 21-bp RS. As expected, these samples showed amplification of the 1.4-kb P1-P4 nonrecombinant control product. Also, no recombination was observed in a similar PCR experiment with a control plasmid construct where only one of the two 21-bp RS was present along with the xer1 gene (data not shown). Furthermore, the 1-kb P1-P2 recombinant PCR product obtained with pBADXerRS21x2 generated a sequence, when sequenced from both sides, that correlates very well with the inversion having occurred at the 21-bp region, because the sequence corresponding to the HindIII site was evident in this 1-kb PCR product just next to the RS21 region when the product was sequenced with the P2 primer, which is plausible only if the inversion occurred in the 21-bp region. Hence, the above PCR and sequencing data are very consistent and indicate that the 21-bp region is sufficient for Xer1-mediated recombination and that this conserved region of the 5′ UTR of all vpma genes is most likely the site of cleavage and strand exchange occurring during vpma gene inversions.
FIG. 1.
Three-primer PCR inversion assay for detection of inversions within the two 21-bp RS conserved in the 5′ UTRs of all vpma genes. The schematic representation shows inversion of plasmid pBADXerRS21x2 (A), carrying two 21-bp inverted repeat RS (RS21; indicated by bold black arrows) along with the xer1 gene, in comparison with plasmid pBADRS21x2 (B), which carries the same RS but cannot undergo inversions because it lacks the xer1 gene and thus acts as a negative control. Primers P1, P2, and P4, annealing to the araC sequence (regulatory gene of l-arabinose operon), the bla sequence (ampicillin resistance gene), and the region adjacent to the ori (pBR322 origin), respectively, are indicated by thin black arrows. (C) Agarose gel electrophoresis of PCR products at different stages of arabinose induction. The presence of a 1-kb P1-P2 amplicon corresponds to inversion events in E. coli between the two RS21 sequences only upon Xer1 induction at 6 and 20 h (lanes 7 and 8) for pBADXerRS21x2, and this amplicon is absent for the uninduced overnight sample (lane 6). The latter, as well as both the induced (lanes 4 and 5) and uninduced (lane 3) samples of control plasmid pBADRS21x2, show only the 1.4-kb P1-P4 product amplified from the sequences of unrecombined plasmids. Lane 2, no-DNA template control; lanes 1 and 9, molecular size marker (1-kb ladder; Invitrogen).
vpma sequences flanking the conserved 21-bp RS enhance the amounts of recombinant products to allow detection via restriction analysis.
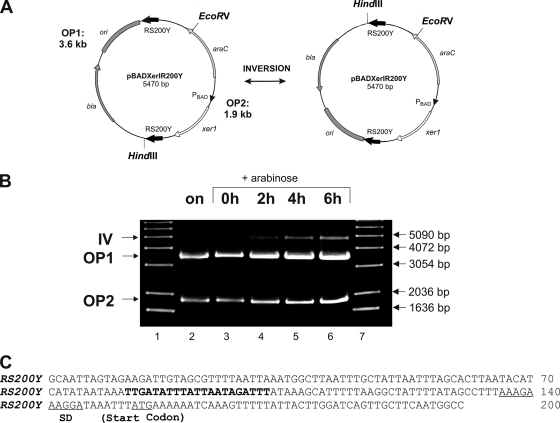
To assess if a larger RS would have an effect and would increase the amounts of inversion products such that they would be visible during the restriction analysis assay described earlier, we constructed the plasmid pBADXerIR200Y (Fig. 2A). Two 200-bp inverted repeat RS, each constituting 152 bp of the 5′ UTR of the vpmaY gene, including the conserved 21-bp region along with 48 bp of the coding sequence of vpmaY (Fig. 2C), were introduced into plasmid pBAD24 together with the xer1 recombinase gene. As described before, the inversions were studied after transformation into E. coli and upon xer1 induction. Uninduced samples of overnight cultures and samples taken before xer1 induction served as negative controls. Induction of xer1 would result in inversion of the DNA fragment between the two RS, thereby placing the HindIII site near the EcoRV site (Fig. 2A). Restriction analysis (HindIII/EcoRV) of samples removed after 2, 4, and 6 h of Xer1 induction clearly showed a 4.9-kb inversion fragment in addition to the two original, nonrecombined plasmid fragments of 3.6 kb and 1.9 kb (Fig. 2B, lanes 4 to 6). A similar inversion fragment could not be detected when the same amount of plasmid pBADXerRS21x2, containing two 21-bp RS, was used in a restriction assay alongside pBADXerIR200Y DNA (data not shown), thereby proving that additional sequences flanking the 21-bp conserved region are instrumental in increasing the amounts of Xer1-mediated recombinant products. The second HindIII/EcoRV inversion fragment of pBADXerIR200Y, which is about 0.6 kb, is not visible on the agarose gel in Fig. 2B (lanes 4 to 6) due to the cumulative effect of its small size and the small amounts of recombination products present in the total plasmid preparation. The 4.9-kb HindIII/EcoRV inversion fragment was absent in the uninduced samples (Fig. 2B, lanes 2 and 3), whereas the intensity of this fragment showed a proportional increase with Xer1 induction, starting from 2 to 6 h (Fig. 2B, lanes 4 to 6). It was further confirmed by sequence analysis that the 4.9-kb HindIII/EcoRV fragment is indeed a product of an inversion event between the two 200-bp RS. These results indicate that sequences in the vicinity of the 21-bp RS increase the amounts of Xer1-mediated inversion products and lead to their detection in the restriction analysis assay, which was not possible with 21-bp RS without flanking sequences.
FIG. 2.
Restriction analysis of inversion events occurring between two 200-bp inverted repeat RS in E. coli. (A) Schematic representation of inversion of plasmid pBADXerIR200Y, carrying two 200-bp inverted repeat RS (RS200Y; indicated by bold black arrows) along with the xer1 gene (white arrow). Induction of xer1 results in inversion of the DNA fragment flanked by the two RS200Y elements, resulting in plasmid pBADXerIR200Y IV, in which the HindIII and EcoRV sites are located close to each other. ori, pBR322 origin; bla, ampicillin resistance gene; araC, regulatory gene of l-arabinose operon. (B) Agarose gel electrophoresis of HindIII- and EcoRV-digested recombination products obtained at different time points. Two fragments, of 3.6 kb (OP1) and 1.9 kb (OP2), corresponding to the original unrecombined plasmid, were present in all samples. An inversion fragment of 4.9 kb (IV) was visible after 2 h (lane 4), 4 h (lane 5), and 6 h (lane 6) of xer1 induction, whereas it was absent in the uninduced cells grown overnight (lane 2) and in the sample taken at the start of induction (lane 3). Lanes 1 and 7, molecular size marker (1-kb ladder; Invitrogen). (C) Sequence of the RS200Y fragment obtained from the vpmaY gene of M. agalactiae. The bold letters represent the 21-bp conserved sequence found in the 5′ UTRs of all vpma genes.
Xer1 recombinase mediates excision between direct repeat RS in E. coli.
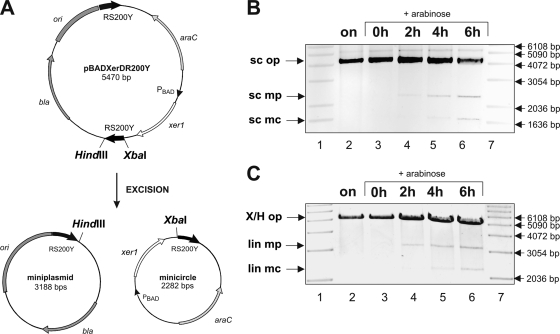
The outcome of a recombination event is determined by the alignment of the corresponding RS on a DNA segment (16). Inverted repeat RS enable inversion, whereas direct repeat RS result in excision of the interjacent DNA fragment (16). To analyze the effect of Xer1 recombinase on direct repeat RS in E. coli, plasmid pBADXerDR200Y was constructed (Fig. 3A). The xer1 gene was cloned alongside two 200-bp RS (as depicted in Fig. 2C) identical to those used for inversion studies, but this time aligned as direct repeats. Upon xer1 induction, the DNA sequence flanked by two direct repeat RS was excised and resulted in two excision products, namely, (i) a 3.2-kb replicative miniplasmid (mp) carrying the origin of replication in addition to the bla gene and (ii) a nonreplicative, 2.3-kb minicircle (mc) carrying the residual sequence (Fig. 3A). As shown in Fig. 3B (lanes 4 to 7), the supercoiled (sc) forms of the two recombination products (sc mp and sc mc) were visible after 2, 4, and 6 h of xer1 induction, along with the original unrecombined plasmid (sc op). The samples were linearized using XbaI and HindIII restriction enzymes to further confirm the sizes of the various plasmids. The 5.5-kb original unrecombined plasmid was visible in all samples, both uninduced and induced, whereas the excision products corresponding to the 3.2-kb miniplasmid and 2.3-kb minicircle were visible only upon xer1 induction (Fig. 3C, lanes 4 to 7).
FIG. 3.
Xer1 mediates excisions between two 200-bp RS. (A) Schematic representation of excision of plasmid pBADXerDR200Y, carrying two 200-bp direct repeat RS (RS200Y; indicated by bold black arrows) along with the xer1 gene (white arrow). Induction of xer1 results in excision of the DNA fragment flanked by the two direct repeat RS200Y elements, resulting in two recombination products: a miniplasmid (mp) and a minicircle (mc). ori, pBR322 origin; bla, ampicillin resistance gene; araC, regulatory gene of l-arabinose operon. (B) Inverted image of agarose gel electrophoresis of supercoiled (sc) recombination products. The supercoiled miniplasmid (sc mp) and supercoiled minicircle (sc mc) were visible 2 h (lane 4), 4 h (lane 5), and 6 h (lane 6) after xer1 induction, in addition to the band corresponding to the unrecombined original plasmid (sc op). Recombination products were not visible for the uninduced cells grown overnight (on; lane 2) and the sample taken at the start of induction (0 h; lane 3). Lanes 1 and 7, molecular size marker (1-kb ladder; Invitrogen). (C) Inverted image of agarose gel electrophoresis of HindIII- and XbaI-linearized (lin) recombination products, confirming the sizes of the original unrecombined plasmid pBADXerDR200Y (X/H op; 5.5 kb) and the minicircle and miniplasmid (lin mc [2.3 kb] and lin mp [3.2 kb]) excised out of it upon xer1 induction. Lanes 1 and 7, molecular size marker (1-kb ladder; Invitrogen).
Sequences flanking the conserved 21-bp region at the 5′ end are required for larger amounts of recombination products.
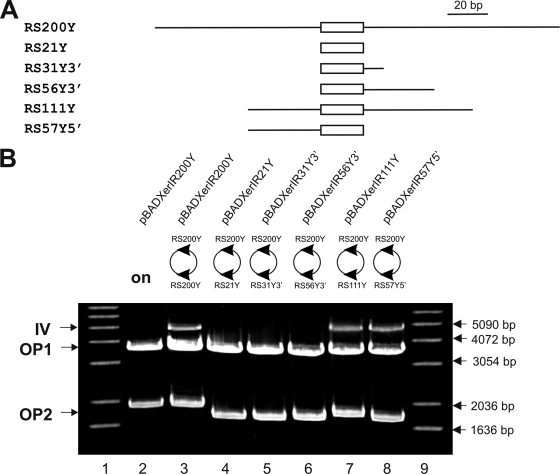
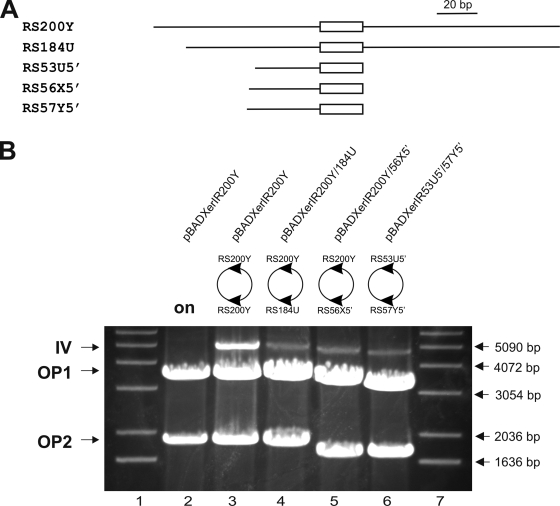
Our results indicate that the 21-bp region that is identical in the 5′ UTRs of all six vpma genes is sufficient for recombination, although a sensitive PCR-based assay is required to verify such site-specific recombinations. On the other hand, inversion between two 200-bp RS including the 21-bp region and additional flanking nucleotides allowed visualization of recombination via the restriction analysis method, indicating that a larger RS favors larger amounts of recombination products. In order to shorten the 200-bp RS and to determine if 5′- and/or 3′-flanking regions of the conserved 21-bp region are critical for the detection of sufficient amounts of recombination products, the restriction analysis method was performed using newly constructed plasmids (pBADXerIR21Y, pBADXerIR31Y3′, pBADXerIR56Y3′, pBADXerIR57Y5′, and pBADXerIR111Y) in which one of the two 200-bp RS of plasmid pBADXerIR200Y was replaced successively by shorter versions of different lengths and with different flanking regions derived from the vpmaY gene, as depicted in Fig. 4A. Inversion products between the 200-bp RS and the 21-bp RS in pBADXerIR21Y could not be detected by the restriction analysis method, even after 20 h of xer1 induction (Fig. 4B, lane 4), which confirmed our previous results for pBADXerRS21x2 (carrying two 21-bp RS) and acted as a negative control for comparison with other constructs. Also, extension of the 21-bp region at the 3′ end, by 10 nucleotides in pBADXerIR31Y3′ (Fig. 4A, RS31Y3′) and by 35 nucleotides in pBADXerIR56Y3′ (Fig. 4A, RS56Y3′), did not yield any recombination products, as shown in Fig. 4B, lanes 5 and 6, respectively. However, addition of 54 nucleotides at the 3′ end and of 36 nucleotides at the 5′ end of the 21-bp region in pBADXerIR111Y (Fig. 4A, RS111Y) allowed detection of recombination products after restriction analysis (Fig. 4B, lane 7). In comparing this result with that for pBADXerIR56Y3′ (Fig. 4A, RS56Y3′), which was negative for recombination, one of the obvious differences between the two constructs was the presence of the additional 36 nucleotides flanking the 21-bp region at the 5′ end. Based on these results, we constructed pBADXerIR57Y5′ (Fig. 4A, RS57Y5′), in which the 57-bp RS consisted of the 21-bp conserved region flanked by an additional 36 bp at the 5′ end. Indeed, when analyzed by the restriction assay, pBADXerIR57Y5′ exhibited detectable amounts of inversion products (Fig. 4B, lane 8). Taken together, these data clearly illustrate that the recombination events mediated by Xer1 recombinase are enhanced by sequences flanking the conserved 21-bp RS at the 5′ end.
FIG. 4.
Sequences flanking the 21-bp RS at the 5′ end enhance the amounts of Xer1 inversion products. One of the two 200-bp vpmaY RS in pBADXerIR200Y was replaced by shorter versions of different lengths and with different flanking regions, namely, RS21Y (in pBADXerIR21Y; the white box represents the 21-bp conserved 5′ UTR), RS31Y3′ (in pBADXerIR31Y3′), RS56Y3′ (in pBADXerIR56Y3′), RS111Y (in pBADXerIR111Y), and RS57Y5′ (in pBADXerIR57Y5′) (A), to determine the minimal RS which gives detectable amounts of inversion products (IV) during restriction analysis and agarose gel electrophoresis (B). Samples were digested with HindIII and EcoRV after 20 h of xer1 induction. Plasmid pBADXerIR200Y showed a 4.9-kb inversion band between RS200Y and RS200Y after induction (lane 3) and served as a positive control, whereas its uninduced overnight sample was negative for an inversion band (lane 2) and showed only bands that corresponded to the unrecombined parent plasmid (OP1 and OP2). Similarly, no inversion was detectable between RS200Y and RS21Y (lane 4), RS200Y and RS31Y3′ (lane 5), or RS200Y and RS56Y3′ (lane 6), whereas the appearance of 4.8-kb (lane 7) and 4.75-kb (lane 8) bands indicated inversion events between RS200Y and RS111Y and between RS200Y and RS57Y5′, respectively. Lanes 1 and 9, molecular size marker (1-kb ladder; Invitrogen).
Recombination events between RS derived from different vpma genes.
While the 21-bp RS sequence is completely conserved in the 5′ UTRs of all six vpma genes, the 5′-flanking regions do not show significant homology within different vpma genes (10). Our previous results focused on recombination events between two RS that originated from the 5′ UTR of the same gene, namely, vpmaY. However, two vpmaY RS do not reflect the native alignment of (vpma) RS in the M. agalactiae vpma locus, where Xer1-mediated recombination occurs within the RS of two different vpma genes rather than those of the same vpma gene. VpmaY was shown to be expressed in M. agalactiae clone 55-5, a clonal variant of the type strain PG2 (10, 11) in which the vpmaY gene is located downstream of the unique vpma promoter and therefore constitutes the expressed vpma gene. Our previous results indicated that a region of 36 bp flanking the 21-bp conserved sequence at the 5′ end is required for the detection of Xer1-mediated inversion products via the restriction analysis method after 20 h of xer1 induction (Fig. 4B, lane 8). This 36-bp region is specific to the 5′ UTR of the expressed vpmaY gene and has no sequence homology with the 5′ UTRs of all other unexpressed vpma genes. Since in pBADXerIR200Y both 200-bp RS originated from the 5′ UTR of the expressed vpmaY gene, this configuration does not correspond with the actual recombination scenario that might be operating in the vpma locus of M. agalactiae. Inversions within the vpma locus occur between two inverted repeat RS of different vpma genes sharing the 21-bp region but having different nucleotides flanking this region at the 5′ end. In order to verify if the two RS originating from different vpma genes would also show similar results by the restriction analysis method, plasmid constructs carrying RS from different vpma genes (Fig. 5) were constructed for further recombination experiments. In the first construct, namely, pBADXerIR200Y/184U, one of the two vpmaY 200-bp RS within pBADXerIR200Y was replaced by a 184-bp vpmaU sequence (Fig. 5A, RS184U). As expected, inversions were detected after restriction analysis and gel electrophoresis of samples removed after 20 h of induction (Fig. 5B, lane 4). Similar inversion products were observed with the construct pBADXerIR200Y/56X5′ (Fig. 5B, lane 5), carrying a 200-bp vpmaY RS and a 56-bp RS of the vpmaX gene which covers the conserved 21-bp region and 35 bp flanking it at the 5′ end (Fig. 5A, RS56X5′). So far, all of these inversions were observed in constructs where at least one of the RS was the 200-bp vpmaY RS. To verify if such inversions can be detected by restriction analysis even for those plasmids where both 200-bp vpmaY RS are replaced by shorter versions, we constructed pBADXerIR53U5′/57Y5′. In this construct, a 57-bp vpmaY RS in which the 21-bp region is extended by 36 bp at the 5′ end (Fig. 5A, RS57Y5′) and a 53-bp vpmaU RS containing the 21-bp region and 32 bp flanking it at the 5′ end (Fig. 5A, RS53U5′) were introduced alongside the xer1 gene. When the construct was subjected to induction and restriction analysis, an inversion band of 4.6 kb was visible (Fig. 5B, lane 6). Such inversions between the vpmaU RS and vpmaY RS and between the vpmaX RS and vpmaY RS not only reflect the recombination events actually operational in the vpma locus of the native M. agalactiae system but also confirm our earlier results showing that the 5′-flanking region of the 21-bp RS is indeed responsible for enhanced accumulation of recombination products.
FIG. 5.
Inversion events between different vpma RS. Plasmids carrying RS sequences derived from two different vpma genes were constructed to reflect the native recombination events operative in M. agalactiae and were transformed into E. coli to check for inversion events. (A) Schematic representation of the RS elements used in the different plasmid constructs, i.e., RS200Y and RS57Y5′ (from vpmaY), RS184U and RS53U5′ (from vpmaU), and RS56X5′ (from vpmaX). The white box depicts the 21-bp conserved region. (B) Agarose gel electrophoresis of HindIII and EcoRV digests of different samples removed after 20 h of xer1 induction. The inversion bands (IV) and the bands corresponding to the original unrecombined plasmid (OP1 and OP2) are indicated in the left margin. Plasmid pBADXerIR200Y showed a 4.9-kb inversion band between RS200Y and RS200Y after induction (lane 3) and served as a positive control, whereas its uninduced overnight sample was negative for an inversion band (lane 2) and showed only bands that corresponded to the unrecombined parent plasmid (OP1 and OP2). Inversion bands of 4.9 kb (lane 4), 4.75 kb (lane 5), and 4.6 kb (lane 6) were observed for the newly constructed pBADXerIR200Y/184U, pBADXerIR200Y/56X5′, and pBADXerIR53U5′/57Y5′ plasmids, respectively, in which the two RS originated from different vpma genes, as indicated. Lanes 1 and 7, molecular size marker (1-kb ladder; Invitrogen).
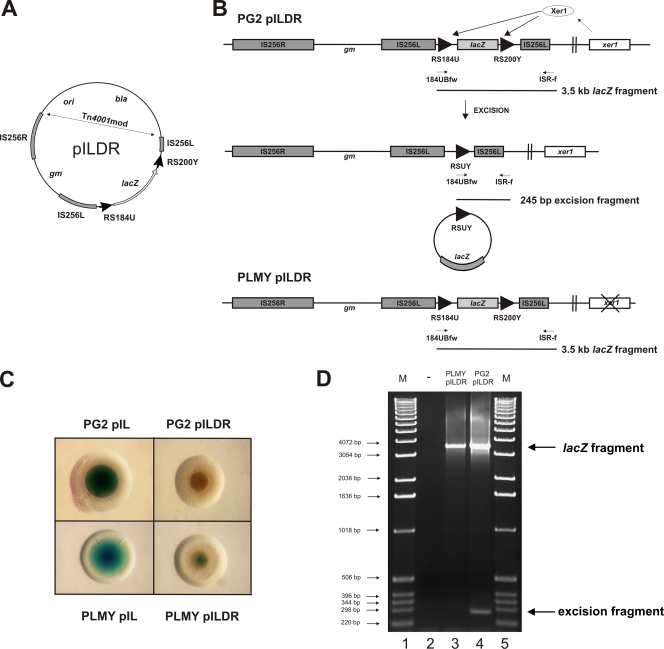
Xer1 can mediate excisions between vpma RS in M. agalactiae.
Having demonstrated excisions in E. coli by cloning two 200-bp vpmaY RS as direct repeats in plasmid pBADXerDR200Y (Fig. 3), the next step was to analyze if such excisions can occur in M. agalactiae. Since vpma genes are present in both orientations in the native vpma gene locus of M. agalactiae (4, 10), excisions between the direct repeat vpma RS are theoretically possible via the chromosomally encoded Xer1 recombinase. To analyze such excision events in the native M. agalactiae system, we developed a lacZ-based reporter system which was used to demonstrate Xer1-mediated excisions in M. agalactiae based on blue-white selection as well as PCR analysis. The lacZ gene, with its native Shine-Dalgarno sequence from E. coli, was introduced into the left IS element of transposon Tn4001mod in pISM2062 (22), resulting in recombinant plasmid pIL. Transformation of pIL into the wild-type M. agalactiae strain PG2, as well as into the xer1-disrupted PLMY strain (4), resulted in blue transformant colonies on SP4 agar plates supplemented with X-Gal (Fig. 6C). Interestingly, lacZ expression did not require the addition of a native promoter element upstream of the lacZ sequence, indicating that transcription was driven by a transposon-based promoter. After this successful demonstration of lacZ as a reporter gene in M. agalactiae, plasmid pILDR (Fig. 6A) was constructed, in which the promoterless lacZ gene was flanked by vpmaU-derived RS184U (Fig. 5A) at the 5′ end and vpmaY-derived RS200Y (Fig. 5A) at the 3′ end, with both RS elements aligned as direct repeats. Transformation of pILDR into the wild-type PG2 strain indeed resulted in white colonies (Fig. 6C), as the lacZ gene was excised from pILDR due to an excision event between the direct repeat vpmaY and vpmaU RS, mediated by the chromosomal Xer1 recombinase (Fig. 6B). Transpositional integration of pILDR in these white clones was confirmed by Southern hybridization (data not shown), using a transposon-specific probe, as described earlier (5). In contrast, pILDR transformants in the xer1-disrupted mutant strain PLMY still exhibited a faint blue colony phenotype (Fig. 6C), as they lacked Xer1 recombinase to accomplish a similar lacZ excision event (Fig. 6B). However, compared to the pIL transformants of PG2 and PLMY, these colonies showed a reduced intensity of the blue pigment (Fig. 6C). This might be due to the introduction of an extra, 184-bp vpmaU RS sequence upstream of the promoterless lacZ gene, which would lead to lower levels of lacZ transcription due to increased distance from the transposon promoter. These Xer1-mediated excisions were further confirmed by PCR analysis using primer 184Ubfw, which anneals to the 184-bp vpmaU RS, and primer ISR-f, which anneals to a region upstream of the left IS element of Tn4001mod (Fig. 6B). Since both primers anneal outside the region of recombination, the amplified product would verify the absence or presence of a lacZ gene excision event. As expected, PCRs using genomic DNA of PLMY transformed with pILDR resulted in amplification of just a single fragment, of 3.5 kb (Fig. 6D, lane 3), corresponding to the unexcised lacZ sequence flanked by the two RS, indicating that no excision occurred in this xer1 disruptant strain. In contrast, a similar PCR performed with genomic DNA obtained from PG2/pILDR transformants showed an additional band, of 245 bp (Fig. 6D, lane 4), corresponding to the shortened sequence created after Xer1-mediated recombination of the vpmaU and vpmaY RS, which leads to deletion of the interjacent lacZ gene (Fig. 6B). Furthermore, sequencing of the 245-bp product displayed an expected hybrid site (RSUY) comprising the 5′ region of the vpmaU RS, the 21-bp consensus sequence common to both RS, and the 3′ region of the vpmaY RS. These results clearly demonstrate that Xer1-mediated excisions between direct repeat vpma RS are feasible in M. agalactiae. The above data not only provide the first experimental proof of the lacZ reporter system being functional in M. agalactiae but also confirm our earlier postulates regarding the Xer1 recombination system based on the results of the recombination experiments done in E. coli.
FIG. 6.
Demonstration of Xer1-mediated excisions in M. agalactiae. (A) Illustration of plasmid pILDR, used to study Xer1-mediated excisions in M. agalactiae. The lacZ gene is flanked by a 184-bp vpmaU RS and a 200-bp vpmaY RS (RS184U and RS200Y, respectively; black arrowheads), aligned as direct repeats within the left insertion sequence of transposon Tn4001mod (IS256L). ori, pBR322 origin; bla, ampicillin resistance gene; gm, gentamicin resistance gene. (B) Schematic representation of the genomic integration of pILDR in M. agalactiae type strain PG2 (PG2 pILDR) and the xer1 disruptant PLMY (PLMY pILDR). Xer1 recombinase (white ellipse) mediates excision between RS184U and RS200Y, resulting in deletion of the interjacent lacZ sequence in PG2 but not in PLMY. Primers 184Ubfw and ISR-f (thin black arrows), used to detect excisions via PCR, and their corresponding amplicons (thin black lines) are indicated for both PG2 and PLMY. (C) Colony phenotypes of lacZ transformants. When transformed into PG2 (PG2 pIL) and PLMY (PLMY pIL), the parent plasmid pIL, which carries the lacZ gene without direct repeat RS (not shown), resulted in intense blue colonies on SP4 agar plates containing X-Gal. Introduction of plasmid pILDR in PG2 (PG2 pILDR) resulted in white colonies, indicating that the lacZ gene was lost by site-specific excision between the direct repeat RS, mediated by the M. agalactiae-encoded Xer1 recombinase (see panel B). In contrast, excision was absent in the xer1 mutant PLMY, as indicated by blue colony formation (PLMY pILDR). (D) Agarose gel electrophoresis of PCRs verifying excision events in M. agalactiae. Primers 184Ubfw and ISR-f (indicated in panel B), both annealing outside the region of recombination, were used to detect excision of the lacZ gene. Genomic DNA of PLMY/pILDR (lane 3) enabled amplification of only a 3.5-kb fragment, corresponding to the lacZ sequence, whereas PCRs using genomic DNA of PG2/pILDR transformants (lane 4) also displayed a 245-bp excision fragment corresponding to the hybrid RSUY sequence created after excision between RS184U and RS200Y, in addition to the 3.5-kb lacZ fragment. Lane 2, no-DNA template control; lanes 1 and 5, molecular size marker (1-kb ladder; Invitrogen).
DISCUSSION
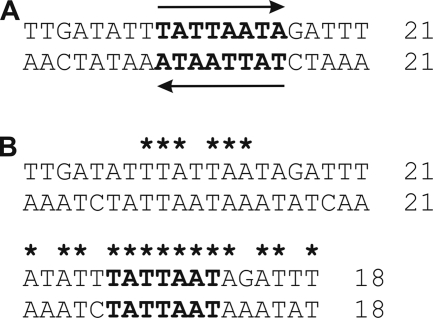
The results of this study demonstrate that a consensus sequence (TTGATATTTATTAATAGATTT) of 21 bp present in the 5′ UTRs of all vpma genes is sufficient for Xer1-mediated inversion and that recombinational strand exchange occurs within this region. In site-specific recombination systems, core RS constitute DNA sequences where recombinase binding and strand exchange take place. Typically, these core RS regions exhibit dyad symmetry, as two perfect or imperfect inverted repeats (11 to 13 bp) which act as recombinase binding elements surrounding a 6- to 8-bp overlap sequence within which strand cutting and exchange occur (7, 12, 24). Alignment of the 21-bp RS of M. agalactiae with its reverse and complemented sequence identified an 8-bp palindromic central sequence (TATTAATA) but did not reveal any other significant dyad symmetry (Fig. 7). However, if the first 3 bases of the 21-bp site are excluded, a similar alignment reveals an impressive level of dyad symmetry flanking the central 8-bp palindrome (Fig. 7). Generally, overlap sequences are nonpalindromic, and for the simplest site-specific recombination systems of the λ-integrase family, exemplified by the Cre recombinase of bacteriophage P1 and the Flp recombinase of Saccharomyces cerevisiae, this asymmetry is sufficient to provide polarity to the site (7). It has been well demonstrated that conversion of an asymmetric overlap region to a symmetric one leads to recombination events that have no polarity. A pair of such symmetric sites placed on a DNA molecule led to both excisions and inversions, independent of their orientation as direct or inverted repeats (17, 29).
FIG. 7.
Dyad symmetry in the 21-bp sequence conserved in the 5′ UTRs of vpma genes. (A) Nucleotide sequence of the conserved 21-bp region. (B) Aligned sequences of 21-bp and 18-bp (where the first three nucleotides of 21 bp are excluded) regions with the respective reverse complemented sequences. The central palindromic sequence is shown in bold, and regions with dyad symmetry are indicated by asterisks.
RS sequences involved in site-specific recombination systems of mycoplasmas show different levels of dyad symmetry. In Mycoplasma pulmonis, the site-specific HvsR recombinase catalyzes inversions at two distinct loci, causing variation in the production of restriction-modification enzymes (hsd locus) as well as phase variation of surface lipoproteins (vsa locus). Comparison of the RS of these two different loci revealed no significant sequence similarity, and the dyad symmetry was very weak for vsa RS compared to hsd RS (31). Recent studies of Mycoplasma penetrans have demonstrated that site-specific recombination mediates mpl promoter inversions, causing phase variation of individual mpl lipoprotein genes. Compared with RS of other site-specific recombinases, these inversions occur within a considerably short 12-bp inverted repeat sequence flanking the promoter region. Also, comparison of the inverted repeat sequences of individual mpl promoters revealed a consensus sequence of TAAYNNNDATTA, in which the TAA and ATTA nucleotides at both ends of the inverted repeat sequence resemble a dyad motif, whereas the central region differs and might be responsible for preventing inappropriate recombination between different mpl promoters or for differences in the inversion frequencies of individual promoters (18). In general, it appears that the RS sequences of site-specific recombination systems of mycoplasmas do not exhibit comparable dyad symmetry to that shown for other well-characterized site-specific recombination systems, so it is difficult to predict the point of strand exchange and recombinase binding. Since several RS have been characterized for mycoplasmas, identification of binding sites for recombinase enzymes and detection of the crossover region involved during strand exchange may be instrumental for further understanding of such recombination reactions.
In vivo inversion experiments with E. coli, using vpma RS from the same as well as from different vpma genes, revealed that Xer1 can act in trans and that additional nucleotides flanking the conserved 21-bp region at the 5′ end lead to enhanced accumulation of recombination products. Indeed, many site-specific recombination systems require additional elements beyond the core RS, which typically include extra binding sites for the recombinase or accessory proteins required for efficient recombination (24). However, comparison of the 5′-flanking nucleotides of the vpma RS did not show any significant sequence identity among different vpma RS, except for a box of 3 nucleotides (ATA) located 13 to 15 bases upstream of the 21-bp region. Comparing individual 5′-flanking regions with each other shows sequence identity levels of up to 82.1% (including the 21-bp RS common to all vpma genes). Future studies may concentrate on the evaluation of inversion frequencies between the RS of different vpma genes to assess if homology in this region is required to enhance the rate of recombination.
The newly developed excision assay based on the lacZ reporter gene successfully demonstrated excision events between two direct repeat vpma RS in the M. agalactiae type strain PG2. The same orientation of vpma genes in the vpma locus (10) leads to a direct repeat orientation of the RS, and any excision events between them would result in deletion of genetic material, including not only the vpma genes but also the unique vpma promoter, which is the key element for expression of these variable surface proteins. This might be critical for the survival of the pathogen in the host, and resultant clones with shorter excised versions of the vpma locus might be selected against due to reduced fitness potential. Based on their result that some PG2 clones yielded a very small proportion of smaller vpma fragments in addition to a large proportion of expected full-length amplicons during PCR amplification of the whole vpma locus, Glew et al. suggested that such excisions occur at a low frequency (10). Whether additional regulatory factors, as found in other bacterial site-specific recombination systems (2, 6, 16, 24, 30), control and favor vpma inversions over excisions needs to be elucidated. Considering the minimal genome of M. agalactiae, it is possible that such stochastic excisions in a proportion of the population could be the cost that the pathogen pays to maintain this antigenic variation machinery for an overall population advantage.
Acknowledgments
This work was supported by grant P18668-B05 (to W.J. and R.R.) from the Austrian Science Fund (FWF).
We thank Martina Zimmermann for her excellent technical assistance and Michael Szostak (IBMH), Angela Witte (Department of Microbiology, Immunobiology and Genetics, University of Vienna), and Christine Citti (INRA-ENVT, Toulouse, France) for helpful discussions.
Footnotes
Published ahead of print on 18 June 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Barre, F. X., and D. J. Sherratt. 2002. Xer site-specific recombination: promoting chromosome segregation, p. 149-161. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 2.Bayliss, C. D. 2009. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 33:504-520. [DOI] [PubMed] [Google Scholar]
- 3.Bhugra, B., L. L. Voelker, N. Zou, H. Yu, and K. Dybvig. 1995. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol. Microbiol. 18:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Chopra-Dewasthaly, R., C. Citti, M. D. Glew, M. Zimmermann, R. Rosengarten, and W. Jechlinger. 2008. Phase-locked mutants of Mycoplasma agalactiae: defining the molecular switch of high-frequency Vpma antigenic variation. Mol. Microbiol. 67:1196-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra-Dewasthaly, R., M. Zimmermann, R. Rosengarten, and C. Citti. 2005. First steps towards the genetic manipulation of Mycoplasma agalactiae and Mycoplasma bovis using the transposon Tn4001mod. Int. J. Med. Microbiol. 294:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colloms, S. D., J. Bath, and D. J. Sherratt. 1997. Topological selectivity in Xer site-specific recombination. Cell 88:855-864. [DOI] [PubMed] [Google Scholar]
- 7.Duyne, G. D. 2002. A structural view of tyrosine recombinase site-specific recombination, p. 93-113. In N. L. Craig (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 8.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flitman-Tene, R., S. Mudahi-Orenstein, S. Levisohn, and D. Yogev. 2003. Variable lipoprotein genes of Mycoplasma agalactiae are activated in vivo by promoter addition via site-specific DNA inversions. Infect. Immun. 71:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glew, M. D., M. Marenda, R. Rosengarten, and C. Citti. 2002. Surface diversity in Mycoplasma agalactiae is driven by site-specific DNA inversions within the vpma multigene locus. J. Bacteriol. 184:5987-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glew, M. D., L. Papazisi, F. Poumarat, D. Bergonier, R. Rosengarten, and C. Citti. 2000. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect. Immun. 68:4539-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainge, I., and M. Jayaram. 1999. The integrase family of recombinase: organization and function of the active site. Mol. Microbiol. 33:449-456. [DOI] [PubMed] [Google Scholar]
- 13.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667-678. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halbedel, S., and J. Stülke. 2006. Probing in vivo promoter activities in Mycoplasma pneumoniae: a system for generation of single-copy reporter constructs. Appl. Environ. Microbiol. 72:1696-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallet, B., and D. J. Sherratt. 1997. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 21:157-178. [DOI] [PubMed] [Google Scholar]
- 17.Hoess, R. H., A. Wierzbicki, and K. Abremski. 1986. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 14:2287-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horino, A., T. Kenri, Y. Sasaki, N. Okamura, and T. Sasaki. 2009. Identification of a site-specific tyrosine recombinase that mediates promoter inversions of phase-variable mpl lipoprotein genes in Mycoplasma penetrans. Microbiology 155:1241-1249. [DOI] [PubMed] [Google Scholar]
- 19.Janis, C., C. Lartigue, J. Frey, H. Wroblewski, F. Thiaucourt, A. Blanchard, and P. Sirand-Pugnet. 2005. Versatile use of oriC plasmids for functional genomics of Mycoplasma capricolum subsp. capricolum. Appl. Environ. Microbiol. 71:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jechlinger, W., J. Glocker, W. Haidinger, A. Matis, M. P. Szostak, and W. Lubitz. 2005. Modulation of gene expression by promoter mutants of the lambdacI857/pRM/pR system. J. Biotechnol. 116:11-20. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, C. J. 2002. Bacterial site-specific DNA-inversion systems, p. 230-271. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 22.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L., K. Dybvig, V. S. Panangala, V. L. van Santen, and C. T. French. 2000. GAA trinucleotide repeat region regulates M9/pMGA gene expression in Mycoplasma gallisepticum. Infect. Immun. 68:871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments, p. 2363-2376. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 25.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ron, Y., R. Flitman-Tene, K. Dybvig, and D. Yogev. 2002. Identification and characterization of a site-specific tyrosine recombinase within the variable loci of Mycoplasma bovis, Mycoplasma pulmonis and Mycoplasma agalactiae. Gene 292:205-211. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski, P. 1986. Site-specific recombinases: changing partners and doing the twist. J. Bacteriol. 165:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Senecoff, J. F., and M. M. Cox. 1986. Directionality in FLP protein-promoted site-specific recombination is mediated by DNA-DNA pairing. J. Biol. Chem. 261:7380-7386. [PubMed] [Google Scholar]
- 30.Sherratt, D. J., L. K. Arciszewska, G. Blakely, S. Colloms, K. Grant, N. Leslie, and R. McCulloch. 1995. Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 347:37-42. [DOI] [PubMed] [Google Scholar]
- 31.Sitaraman, R., A. M. Denison, and K. Dybvig. 2002. A unique, bifunctional site-specific DNA recombinase from Mycoplasma pulmonis. Mol. Microbiol. 46:1033-1040. [DOI] [PubMed] [Google Scholar]
- 32.Solsona, M., M. Lambert, and F. Poumarat. 1996. Genomic, protein homogeneity and antigenic variability of Mycoplasma agalactiae. Vet. Microbiol. 50:45-58. [DOI] [PubMed] [Google Scholar]
- 33.Tully, J. G. 1995. Culture medium formulation for primary isolation and maintenance of mollicutes, p. 33-39. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. 1. Academic Press, San Diego, CA. [Google Scholar]