Abstract
The molecular basis for the recognition of glucose as a germinant molecule by spores of Bacillus megaterium QM B1551 has been examined. A chromosome-located locus (BMQ_1820, renamed gerWB) is shown to encode a receptor B-protein subunit that interacts with the GerUA and GerUC proteins to form a receptor that is cognate for both glucose and leucine. GerWB represents the third receptor B protein that binds to glucose in this strain. Site-directed mutagenesis (SDM) experiments conducted on charged proline and aromatic residues predicted to reside in the transmembrane domains of a previously characterized receptor B protein, GerVB, reveal the importance to receptor function of a cluster of residues predicted to reside in the middle of the transmembrane 6 (TM6) domain. Reductions in the region of 70- to 165-fold in the apparent affinity of receptors for glucose in which Glu196, Tyr191, and Phe192 are individually replaced by SDM indicate that some or all of these residues may be directly involved in the binding of glucose and perhaps other germinants to the germinant receptor.
Spores of Bacillus species are formed via the process of sporulation in response to nutrient starvation and are characterized by metabolic dormancy and extreme properties of resistance to a range of stress factors. Reinitiation of metabolism and the generation of a new vegetative cell necessitate the dormant spore to undergo the process of germination, which can be stimulated by a number of biochemical and/or physical means (19, 33).
Two pathways to germination that are probably the most relevant under physiological conditions have been identified. The first of these has been identified in Bacillus subtilis only recently (35). In this pathway an inner-membrane-located serine/threonine kinase (PrkC) triggers germination upon binding of meso-diaminopimelate-containing muropeptides, with the inference being that these peptidoglycan fragments may signal the presence of growing bacteria in the surrounding milieu.
In terms of research activities, the more established physiological route to germination is mediated by germinant receptors that, as with the serine/threonine kinase mentioned above, are located within the inner membrane that separates the protoplast and protective outer layers of the spore (13, 20, 22, 23, 47). Orthologues of the GerA family of receptors have evolved to recognize a diverse range of amino acids, nucleotides, and sugars, the presence of which signals to the spore that environmental conditions may be conducive to growth. Structural genes encoding the germinant receptors are typically arranged in tricistronic operons, and molecular-genetic and biochemical evidence indicates that the receptor is a complex of all three protein subunits (3, 14, 41).
Although we can state with reasonable confidence that the germinant receptors provide cognate binding sites for defined germinant molecules, interaction with which leads to initiation of the germination cascade, the precise function of the germinant receptors is not yet clear. Whether they have a direct role, for example, in transporting small molecules released from the spore core very early in the germination process, has yet to be determined (32, 38). Similarly, the molecular mechanism and/or transduction process that permits germinant-activated receptor proteins, which are present only in extremely low abundance, to stimulate the opening of significantly more plentiful calcium dipicolinate (Ca2+-DPA) channels (42, 43) and possibly ion channel proteins in some species (24, 39) is entirely unknown.
Molecular-genetic and bioinformatic evidence has been presented, however, to suggest that the B-protein subunit of the receptor presents the site for the receptor-ligand interaction (6, 16, 29). In particular, experiments conducted in our own laboratory using cross-homologue chimeric constructs and site-directed mutagenesis (SDM) experiments with two closely related receptor B proteins have begun to yield insights into the structure-function relationships in the Bacillus megaterium GerU receptor (4, 5). GerU structural genes, which are plasmid-borne on the wild-type QM B1551 strain, are required to initiate the B. megaterium QM B1551 germinative response to glucose, proline, leucine, and inorganic salts (6, 36). Subsequent analysis revealed that whereas spores bearing the GerUB protein can initiate germination in response to glucose and leucine, those bearing the GerVB protein can additionally respond to proline and high concentrations of inorganic salts (6).
The molecular basis for this difference in germinant recognition was examined and revealed that spores bearing a B-protein chimera, comprising the first one to eight transmembrane (TM) domains of GerUB plus TM9 to TM10 of GerVB, gain a germinative response to proline (4). However, while a single residue in TM10 of GerVB (leucine 345) was shown to be crucial to the proline-mediated germinative response, amino acid substitution and kinetic analyses indicated that this position is not directly involved in binding to the germinant (5). Indeed, while a number of residues have been identified as being important to receptor function, particularly in response to proline- and leucine-stimulated germination, the residues involved directly in binding to germinants—and, in particular, those involved in binding to glucose—have yet to be identified. This communication reports on an investigation performed to elucidate further the proteins involved in the recognition of glucose as a germinant by B. megaterium spores and to identify the receptor residues that may be directly involved in the receptor-ligand interaction.
MATERIALS AND METHODS
Bacterial strains and media.
Bacillus megaterium strains were routinely cultured at 30°C on LB agar or broth containing antibiotics where appropriate (1 μg/ml erythromycin and 25 μg/ml lincomycin for macrolide-lincosamide-streptogramin B resistance [MLSr], 100 μg/ml spectinomycin, 12.5 μg/ml tetracycline). The Escherichia coli strains used for site-directed mutagenesis (XL1-Blue [Stratagene]) or preparation of plasmids for transformation of B. megaterium (Top10 [Invitrogen]) were cultured at 37°C in LB medium supplemented with 50 μg/ml carbenicillin.
Construction of Bacillus megaterium mutant strains.
All the Bacillus megaterium strains employed in this study (Table 1) are derivatives of strain PV361, a plasmidless variant of the wild-type QM B1551 strain that lacks the operon for GerU and the gerVB structural gene required to initiate germination in response to single trigger compounds (6, 36). Strain GC500, the gerWB (BMQ_1820) insertion-deletion strain, was constructed by ligating a 970-bp internal fragment of the gerWB gene, digested at engineered EcoRI and HindIII sites at the 5′ and 3′ ends, with pGEM-3Z, which was digested with the same enzymes. The resultant plasmid was used as a template to perform an inverse PCR that introduced a deletion of 37 bp between positions 488 and 525 of the cloned fragment. The purified PCR product was subsequently blunt-end ligated with a spectinomycin resistance (Spr) cassette excised from plasmid pDG1726 and was used to transform E. coli, from which plasmid pGEM-ΔgerWB::Spr was isolated. The ΔgerWB::Spr cassette was amplified by PCR, using primers designed to incorporate EcoRI sites at the 5′ and 3′ ends of the cassette, and ligated with MfeI-digested pUCTV2. Plasmid pUCTV-ΔgerWB::Sp was introduced into B. megaterium PV361 by polyethylene glycol-mediated protoplast transformation (5). A Tets Spr transformant, which had undergone double homologous recombination to introduce an insertion-deletion at the gerWB locus, was isolated. The correct construction of the strain (GC500) was confirmed by PCR and sequencing.
TABLE 1.
Strains and plasmids used in this study
| B. megaterium strain or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| QM B1551 | Wild-type strain | P. S. Vary |
| PV361 | Plasmidless derivative of QM B1551 that lacks GerU receptor genes | P. S. Vary |
| GC500 | PV361 ΔgerWB::Spr | This study |
| GC500 transformantsb | ||
| GC501 | GerU* (containing gerUA, gerUC, and gerVB) | This study |
| GC502 | GerU (containing gerUA, gerUC, and gerUB) | This study |
| GC503 | gerUA and gerUC | This study |
| GC504 | gerUA, gerUC, and gerWB arranged as a tricistronic operon with GerU regulatory sequences | This study |
| GC505 | GerU* with GerVB E19Q | This study |
| GC506 | GerU* with GerVB D93N | This study |
| GC507 | GerU* with GerVB E112Q | This study |
| GC508 | GerU* with GerVB H119N | This study |
| GC509 | GerU* with GerVB E196Q | This study |
| GC510 | GerU* with GerVB E196D | This study |
| GC511 | GerU* with GerVB K282A | This study |
| GC512 | GerU* with GerVB H341N | This study |
| GC513 | GerU* with GerVB P27A | This study |
| GC514 | GerU* with GerVB P114A | This study |
| GC515 | GerU* with GerVB P193A | This study |
| GC516 | GerU* with GerVB P309A | This study |
| GC517 | GerU* with GerVB P343A | This study |
| GC518 | GerU* with GerVB P349A | This study |
| GC519 | GerU* with GerVB F84A | This study |
| GC520 | GerU* with GerVB Y86A | This study |
| GC521 | GerU* with GerVB Y89A | This study |
| GC522 | GerU* with GerVB F90A | This study |
| GC523 | GerU* with GerVB Y92A | This study |
| GC524 | GerU* with GerVB F186A | This study |
| GC525 | GerU* with GerVB Y191A | This study |
| GC526 | GerU* with GerVB F192A | This study |
| GC527 | GerU* with GerVB F194A | This study |
| GC528 | GerU* with GerVB F200A | This study |
| GC529 | GerU* with GerVB F280A | This study |
| GC530 | GerU* with GerVB Y288A | This study |
| GC531 | GerU with GerUB E196Q | This study |
| GC532 | GerU with GerUB Y191A | This study |
| GC533 | GerU with GerUB F192A | This study |
| Plasmids | ||
| pGEM-3Z | E. coli cloning vector; Ampr | Promega |
| pDG1726 | Spr cassette | 12 |
| pUCTV2 | E. coli/Bacillus shuttle plasmid with orits Tetr | 45 |
| pHT315 | B. megaterium host plasmid; MLSr | 1 |
Spr, spectinomycin resistance; Ampr, ampicillin (β-lactam) resistance; Tetr, tetracycline resistance; MLSr, macrolide-lincosamide-streptogramin B resistance; orits, temperature-sensitive origin of replication.
Strains were transformed to MLSr with plasmid pHT315 carrying the described GerU receptor variant.
The chimeric receptor operon comprising the gerUA-gerUC-gerWB structural genes was created using an overlap PCR technique. Two PCR amplicons, the first encompassing coding and putative upstream regulatory sequences for gerUA and gerUC and the second encompassing the gerWB gene and predicted ribosome binding site and a downstream rho-independent terminator sequence, were purified and mixed to provide the template for a subsequent round of PCR. The resulting DNA fragment comprised the gerUA, gerUC, and gerWB genes arranged as a GerA-type receptor operon with appropriate regulatory sequences. The 5′ and 3′ ends of the purified amplicon were digested with BamHI, ligated with pHT315 (a low-copy-number B. thuringiensis-derived vector that is highly stable in B. megaterium in the absence of selective pressure [1, 18]) restricted with the same enzyme, and used to transform E. coli. The resultant plasmid was used to transform B. megaterium strain GC500 to MLSr, giving strain GC504.
SDM procedures were conducted using a Stratagene Lightning site-directed mutagenesis kit, as directed by the manufacturer. Primers for SDM were designed using the QuikChange primer design program (Stratagene). All primer sequences are available upon request. Plasmid pHT315 with appropriate receptor operons cloned at the BamHI site served as the template for SDM procedures. Transformant E. coli isolates carrying receptor genes with the correct mutations were identified by purifying plasmid DNA and then sequencing the target gene. Receptor operons carrying the correct mutation were sequenced in their entirety prior to transformation of B. megaterium GC500 to MLSr. Protoplasting and transformation of B. megaterium were performed as described previously (5).
Spore preparation and storage.
Bacillus megaterium spores were prepared by inoculating 200 ml supplemented nutrient broth (5), containing erythromycin and lincomycin where appropriate, with 1 ml of a mid-log-phase culture of the designated strain. The cultures were incubated (30°C, 225 rpm) for 72 h before spores were harvested by repeated rounds of centrifugation (4,300 × g for 7 min, 4°C), removal of the upper vegetative debris layer, and resuspension in ice-cold water. This wash procedure was repeated until the suspension was observed to be free (99%) of vegetative cells, debris, and germinated spores, as judged by phase-contrast microscopy. Spore suspensions (optical density at 600 nm [OD600], ∼100) were stored on ice and protected from light.
Germination assays.
Aliquots of spores suspended in water (OD600, ∼100) were heat activated at 60°C for 10 min and then briefly cooled on ice. Germination assays were conducted in 96-well plates using a Perkin-Elmer EnVision-Xcite multilabel plate reader fitted with a 600-nm photometric filter. Heat-activated spores were added to buffer (5 mM Tris-HCl, pH 7.8) that had been preheated to 30°C and that contained the appropriate concentration of germinant, and germination was monitored by measuring the decrease in OD600 every minute for at least 60 min. The plates were sealed with transparent adhesive film to reduce evaporative losses, shaken orbitally for 10 s every minute to keep the spores in suspension, and maintained at 30°C for the duration of the assay. The optical density at the start of the germination assays (OD600, ∼0.4) corresponds to approximately 108 spores ml−1, and this was observed to fall by approximately 65% upon complete germination of the spore population. The progress of germination was also routinely monitored by phase-contrast microscopy. Experiments were conducted in triplicate with at least two different spore preparations.
The germination rates (v [OD units min−1]) for kinetic analyses (see Table 7) were determined from the slope of the linear segment of optical density changes over time that follows the initial lag phase upon addition of germinant. Apparent Km and Vmax values were subsequently determined using the ligand-binding macro of the SigmaPlot (version 11) program (Systat Software Inc.). Hyperbolic curves yielded by plotting the germinant concentration versus percent spore germination were analyzed using the same function to determine the concentration of germinant required to stimulate the germination of 50% of the spore population [K0.5(germ)].
TABLE 7.
Kinetic analysis of spore germination in response to glucose where GerVB is substituted at defined residuesa
| Strain | Substitution | Km (mM) | Vmax(OD/min) |
|---|---|---|---|
| GC501 | None | 3.1 | 0.077 |
| GC509 | E196Q | 513 | 0.068 |
| GC510 | E196D | 305 | 0.058 |
| GC525 | Y191A | 215 | 0.052 |
| GC526 | F192A | 229 | 0.079 |
| GC515 | P193A | 123 | 0.055 |
| GC517 | P343A | 728 | 0.074 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 90 min with various glucose concentrations (10, 25, 50, 100, 175, 250, 400, 600, 800, and 1,000 mM), and germination was monitored as described in Materials and Methods. Strain GC517 was additionally germinated in 1,200 and 1,400 mM glucose. Strain GC501 was germinated in glucose at concentrations ranging from 0.1 to 10 mM. Kinetic parameters were determined as described in Materials and Methods. The values presented for each strain are derived from three independent experiments conducted with the same spore preparation; similar values were obtained with other spore preparations. The calculated standard error associated with all measurements is less than 10%.
DNA sequencing and bioinformatics analyses.
DNA sequencing was performed by the Department of Biochemistry sequencing facility (University of Cambridge). DNA sequence analysis was performed using CLC Combined Workbench 5 software (CLC bio). Protein topology and transmembrane helix predictions were made using the HMMTOP program (40), which is available on the ExPASy server (Swiss Institute of Bioinformatics; http://expasy.org/). The B. megaterium QM B1551 genomic sequence (GenBank accession no. CP001983) can be accessed at http://www.bios.niu.edu/b_megaterium/index.html.
RESULTS
Receptor proteins involved in the spore germinative response to glucose.
Structural genes encoding Bacillus spore germinant-receptor proteins are typically organized as tricistronic operons. Mutational analyses of these operons indicate that all three proteins are required to form a functional germinant receptor. The Bacillus megaterium plasmid-borne GerU receptor operon differs slightly in that two close receptor B-protein homologues, encoded by gerUB and gerVB, respectively, can interact with proteins encoded by gerUA and gerUC to form a receptor that is cognate for glucose. Despite the assertion made above that germinant receptors typically require all three components for their function, Bacillus megaterium PV361 spores (which lack all GerU structural genes) complemented with only gerUA and gerUC retain a moderate germinative response to glucose (stimulating approximately 20% germination when glucose is added as a single trigger germinant and with the response rising to 60% when it is added as a cogerminant with leucine) (5).
These data indicate that the GerUA and GerUC proteins might be interacting with a previously unidentified B-protein subunit to form a functional receptor. A BLAST search of the B. megaterium QM B1551 genome revealed the presence of a locus (BMQ_1820) predicted to encode a 366-residue, 40.9-kDa protein with 10 TM domains that shares 82% identity at the amino acid level with GerVB. The locus is located on the chromosome and is preceded by an open reading frame (BMQ_1819) predicted to encode a germinant-receptor protein-C subunit. Sequence alignments with other receptor C proteins and analyses of the DNA in this region indicate that the putative product of BMQ_1819 may be truncated at the N terminus by a deletion (data not shown). Additionally, there is no A-protein homologue encoded either up- or downstream of this apparent bicistronic operon, which indicates further that the genetic organization of the operon may have been disrupted at some point.
To investigate the possibility that the product of locus BMQ_1820 might be involved in the spore germinative response to glucose, an insertion-deletion mutant strain (GC500) was constructed and complemented with various plasmid-borne receptor genes. The germinative response of the various mutants (Table 2)—and, in particular, that of strain GC504—indicate that the product of BMQ_1820 can interact with GerUA and GerUC proteins to form a germinant receptor that is cognate for both glucose and leucine. We henceforth refer to this locus as gerWB, in view of the sequence and functional similarity of the protein that it encodes (GerWB) to the GerUB and GerVB germinant-receptor B proteins. Additionally, the gerWB null mutant complemented with gerUA and gerUC (strain GC503) shows no germinative response to glucose, indicating that there are no further B-protein subunits encoded on the chromosome that can interact with the GerUA and GerUC proteins (at least when they are expressed from a plasmid) to form a glucose receptor. These data are also consistent with the hypothesis that functional germinant receptors in Bacillus species comprise A-, B-, and C-protein subunits.
TABLE 2.
Rates of germination of B. megaterium GC500 (ΔgerWB) spores complemented with various germinant receptor genesa
| Strain | Genotype | Rate of spore germination with indicated germinantb |
|||||
|---|---|---|---|---|---|---|---|
| Buffer | GPLK | Glucose | Proline | Leucine | KBr | ||
| GC500 | ΔgerWB | <1 | <1 | <1 | <1 | <1 | <1 |
| GC503 | ΔgerWB pHT-gerUA-gerUC | <1 | 4 | <1 | <1 | <1 | 4 |
| GC504 | ΔgerWB pHT-gerUA-gerUC-gerWB | 1 | 100 | 91 | <1 | 85 | 4 |
| GC501 | ΔgerWB pHT-gerUA-gerUC-gerVB | <1 | 100 | 100 | 100 | 43 | 100 |
| GC502 | ΔgerWB pHT-gerUA-gerUC-gerUB | <1 | 100 | 76 | 7 | 60 | 8 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 60 min with 10 mM germinant (50 mM KBr). GPLK is a mixture of glucose, proline, and leucine (each at 10 mM) and KBr (50 mM). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the OD600 loss (65%) for spores of strain QM B1551 in 10 mM glucose, which was set equal to 100. This value is equivalent to 100% germination after 60 min incubation. Values are the means of at least duplicate experiments; the standard deviation was ≤5% of the mean.
Finally, analyses of gerWB null mutant spores complemented with the various receptor constructs described previously reveal that deletion of gerWB has little influence on the published results and conclusions (4, 5) (data not shown), with the exception of those for the aforementioned strain GC503 (pHT-gerUA gerUC), which no longer shows a germinative response to glucose or leucine.
Identification of putative glucose-binding residues.
Identification and functional characterization of the gerWB locus complete, as far as we know, the assignment of the receptor components involved in the B. megaterium germinative response to glucose. The construction of strain GC500, which lacks all known glucose germinant-receptor genes, therefore provides a genetic background improved over that used in previous studies (4, 5) with which complementation-based analyses aimed at revealing the consequences of amino acid substitutions directed to the various receptor B proteins may be conducted.
We have previously used SDM to examine the role of residues positioned at nonconserved regions of GerUB and GerVB in an attempt to gain insight into the molecular basis for differences in the germinant recognition profile between the two receptor proteins (4, 5). The same approach was deemed unlikely to be successful when an attempt is made to identify the residues involved in binding to glucose, since the high degree of similarity and the common response to glucose indicate that at least some residues involved in this interaction may be conserved in GerUB, GerVB, and GerWB.
Instead, a survey of the literature pertaining to essential ligand-binding residues in a variety of integral membrane proteins involved in the transport of sugars, amino acids, and ions was conducted and revealed the recurring importance of charged and aromatic residues located on α helices that span the membrane (TM helices) (8, 9, 21, 25, 27, 34, 37). Transmembrane-located proline residues, which serve to introduce distortions and/or regions of local flexibility in the alpha helix, are also often of functional importance, particularly where conformational changes at proline-containing molecular hinges are employed to transduce extracellular signals across the membrane (7, 30, 31).
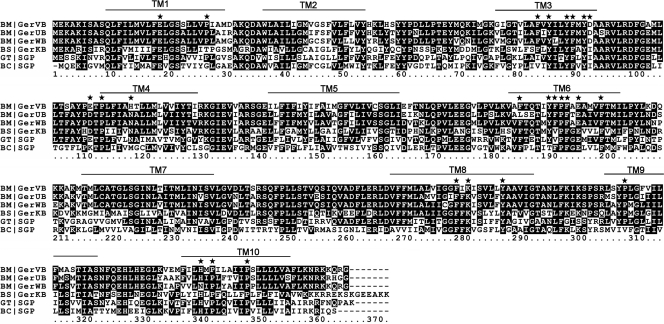
With this in mind, the predicted topology of GerVB was examined and revealed that a number of charged, proline, and aromatic residues are predicted to reside in TM segments of the protein. Several of these residues are conserved in germinant-receptor B proteins from different species of Bacillus, which indicates that they may be of functional significance (Fig. 1). Due to the relatively large proportion of TM aromatic residues, we decided to substitute first the TM-located charged and proline residues in an attempt to identify functionally important regions of the receptor, before focusing our attention on those aromatic residues which, on the basis of secondary structure predictions, reside in the same TM domains. In order to minimize the introduction of size-related structural perturbation, the four acidic residues were replaced by the corresponding amide residues (E19Q, D93N, E112Q, and E196Q), whereas both histidines were replaced by asparagine. The single basic residue, Lys282, was replaced by alanine, as were all six TM-located proline residues.
FIG. 1.
ClustalW alignment of various Bacillus species germinant-receptor B proteins. B. megaterium GerVB, GerUB, and GerWB are cognate for glucose; B. subtilis GerKB is also thought to be involved in mediating glucose-associated germinative responses. Predicted TM helices, as judged by HMMTOP (40) analysis of GerVB, are highlighted. Residues denoted by stars were subject to amino acid substitution in this work. BM, B. megaterium QM B1551; BS, B. subtilis 168; GT, Geobacillus thermodenitrificans NG80-2; BC, B. cereus E33L.
Analysis of the germinative response of mutant strains to germinant concentrations that are saturating for wild-type spores reveals that the E196Q substitution (GC509) is associated with the almost complete loss of the germinative response to glucose and proline (Table 3). Replacement of Glu by Asp at this position permits only a weak germinative response to glucose (21%) but abolishes the germinative response to all other germinants. Considered together, these data indicate that both the size and the charge of the residue at this position are important to receptor function. Unfortunately, antisera raised against a 14-amino-acid stretch of GerVB proved to be nonspecific (data not shown); therefore, we cannot ascertain the effects that these substitutions have on receptor abundance and/or ability to insert into the membrane correctly. However, the retention of a strong germinative response to inorganic salts and essentially complete germination in response to a mixture of germinants indicates that at least some receptor protein assumes the correct fold and is inserted into the membrane.
TABLE 3.
Rates of germination of spores with GerVB with substitutions at predicted transmembrane-located charged and proline residuesa
| Strain | Domain | Substitution | Rate of spore germination with indicated germinantb |
|||||
|---|---|---|---|---|---|---|---|---|
| Buffer | GPLK | Glucose | Proline | Leucine | KBr | |||
| GC501 | None | 1 | 100 | 99 | 100 | 57 | 100 | |
| GC505 | TM1 | E19Q | <1 | 100 | 94 | 96 | 51 | 92 |
| GC506 | TM3 | D93N | <1 | 90 | 68 | 53 | 4 | 62 |
| GC507 | TM4 | E112Q | 3 | 97 | 82 | 87 | 8 | 67 |
| GC508 | TM4 | H119N | <1 | 99 | 78 | 80 | 1 | 7 |
| GC509 | TM6 | E196Q | <1 | 96 | 6 | 1 | 21 | 74 |
| GC510 | TM6 | E196D | <1 | 97 | 21 | <1 | <1 | 5 |
| GC511 | TM8 | K282A | <1 | 94 | 81 | 87 | 25 | 79 |
| GC512 | TM10 | H341N | <1 | 98 | 82 | 82 | 40 | 69 |
| Strains with proline substitutions | ||||||||
| GC513 | TM1 | P27A | <1 | 99 | 69 | 77 | <1 | 3 |
| GC514 | TM4 | P114A | 1 | 59 | 29 | 34 | 5 | <1 |
| GC515 | TM6 | P193A | 4 | 88 | 15 | 30 | 45 | 80 |
| GC516 | TM9 | P309A | <1 | 100 | 86 | 93 | 13 | 86 |
| GC517 | TM10 | P343A | <1 | 88 | 3 | <1 | <1 | 6 |
| GC518 | TM10 | P349A | <1 | 79 | 58 | 64 | 34 | 60 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 60 min with 10 mM germinant (50 mM KBr). GPLK is a mixture of glucose, proline, and leucine (each at 10 mM) and KBr (50 mM). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the OD600 loss (65%) for spores of strain QM B1551 in 10 mM glucose, which was set equal to 100. This value is equivalent to 100% germination after 60 min incubation. Values are the means of three independent experiments conducted with the same spore preparations; the standard deviation was ≤5% of the mean. Similar values were obtained with other spore preparations.
Analysis of mutant strains bearing GerVB with proline residue replacements reveals that Pro343 is of particular importance, since the germinative response of strain GC517 to all four main single-trigger compounds is essentially eliminated (Table 3). This residue is predicted to reside on TM10 and is in close proximity to Leu345, a position previously identified as being sensitive to structural perturbation when it is replaced by bulky amino acids (5). A strong germinative response to a mixture of all four germinants again provides evidence that at least some functional receptor protein is present.
More detailed analyses of spores bearing the P343A mutation reveal that when each of the individual nutrient germinants was combined with KBr, a strong germinative response was induced (Table 4). Combinations of nutrient germinants in the absence of KBr also induce germinative responses to various degrees, except where glucose and proline are combined (<1% germination). Cooperative or synergistic effects in the presence of multiple germinants have been observed previously (4), and it may be that some germinants, particularly leucine and KBr, can facilitate or stabilize the binding of other germinants. This could entail direct chemical interactions between different germinant molecules in a shared binding site, or alternatively, binding of the first germinant may induce a conformational change in the receptor that facilitates binding to a different site for another germinant. Either model would explain why combinations of different germinants can effectively trigger germination in mutant strains where some or all single-trigger compounds are no longer effective, particularly where ligand-binding sites (or access to binding sites) are disrupted by unfavorable substitutions.
TABLE 4.
Effects of germinant mixtures on the germination properties of GC517 (GerVB P343A) sporesa
| Germinant | Rate of spore germinationb |
|---|---|
| Buffer | <1 |
| Glu, Pro, Leu, and KBr | 96 |
| Pro, Leu, and KBr | 97 |
| Glucose and proline | <1 |
| Glucose and leucine | 67 |
| Glucose and KBr | 92 |
| Proline and leucine | 23 |
| Proline and KBr | 79 |
| Leucine and KBr | 81 |
Spores were incubated in 5 mM Tris-HCl, pH 7.8, for 60 min with the respective germinants (10 mM each; 50 mM KBr). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the OD600 loss (65%) for spores of strain QM B1551 in 10 mM glucose, which was set equal to 100. This value is equivalent to 100% germination after 60 min incubation. Values are the means of three independent experiments conducted with the same spore preparations; the standard deviation was ≤5% of the mean.
The observation that Pro343 and, to a lesser extent, Pro349 are phenotypically important is also consistent with the idea, observed in other membrane proteins, that these proline residues might serve as molecular brackets, positioning and maintaining the structural integrity of this region of the helix for intra- or intermolecular interactions and/or conformational changes (17).
Spores bearing the P114A substitution are also notable in that they demonstrate the weakest response to a mixture of glucose, proline, and leucine (each 10 mM) and KBr (50 mM) of all strains tested in this work (59%). Proline 114 is predicted to reside at the end of TM4 (Fig. 1), positioned toward the outside of the membrane, where it may be structurally important in defining the end of the helix. Thus, it seems more likely that substitution of this putative helical cap may exert a deleterious effect on the overall receptor structure, as opposed to having a role directly in binding to germinants.
Substitution of Pro193 (GC515) is also observed to exert a deleterious effect on receptor function, in particular, the germinative response to glucose. The defective germination phenotypes of spores bearing the Glu196 and Pro193 substitutions—both located toward the middle of TM6 and highly conserved in receptor B proteins of many Bacillus species (Fig. 1)—indicate that this domain, like TM10, is of functional and/or structural importance. In view of this observation, we decided to replace all five aromatic residues predicted to reside in TM6 with alanine, in an attempt to identify potential ligand-binding residues. Of the other TM domains rich in aromatic residues, perhaps the most highly conserved in receptor B-protein subunits from different species of Bacillus are those in TM3, which were also replaced by alanine. The two aromatic residues predicted to flank Lys282 in the middle of TM8 (Phe280 and Tyr288), which are also highly conserved, were also selected for substitution.
Analysis of the germinative responses to glucose of these various mutant strains is striking (Table 5), with replacement of Tyr191 (GC525) and Phe192 (GC526), both of which reside in TM6, by alanine essentially abrogating the germinative response to glucose. The F192A substitution, like the E196Q substitution described above, also eliminates the germinative response to proline, which, when these findings are considered together, might indicate that binding sites for these germinants overlap. Alternatively, substitutions at this position may introduce structural modifications that impair access to the respective germinant-binding sites. Regardless, the efficient germinative responses observed with a mixture of germinants once again indicate that at least some correctly folded and membrane-inserted receptor is present in the spore.
TABLE 5.
Rates of germination of spores with GerVB with substitutions at predicted transmembrane-located aromatic residuesa
| Strain | Domain | Substitution | Rate of spore germination with indicated germinantb |
|||||
|---|---|---|---|---|---|---|---|---|
| Buffer | GPLK | Glucose | Proline | Leucine | KBr | |||
| GC501 | None | 1 | 100 | 99 | 100 | 57 | 100 | |
| GC519 | TM3 | F84A | 4 | 81 | 63 | 69 | 5 | 50 |
| GC520 | TM3 | Y86A | <1 | 98 | 86 | 93 | 31 | 92 |
| GC521 | TM3 | Y89A | 5 | 95 | 62 | 35 | 6 | 56 |
| GC522 | TM3 | F90A | 3 | 99 | 84 | 90 | 36 | 97 |
| GC523 | TM3 | Y92A | 3 | 95 | 56 | 25 | 6 | 37 |
| GC524 | TM6 | F186A | <1 | 96 | 92 | 94 | 65 | 90 |
| GC525 | TM6 | Y191A | <1 | 82 | 3 | 31 | <1 | 3 |
| GC526 | TM6 | F192A | <1 | 98 | 2 | <1 | 35 | 46 |
| GC527 | TM6 | F194A | 14 | 97 | 91 | 95 | 82 | 100 |
| GC528 | TM6 | F200A | 5 | 99 | 92 | 98 | 81 | 99 |
| GC529 | TM8 | F280A | 45 | 93 | 79 | 85 | 78 | 90 |
| GC530 | TM8 | Y288A | <1 | 99 | 85 | 91 | 17 | 88 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 60 min with 10 mM germinant (50 mM KBr). GPLK is a mixture of glucose, proline, and leucine (each at 10 mM) and KBr (50 mM). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the OD600 loss (65%) for spores of strain QM B1551 in 10 mM glucose, which was set equal to 100. This value is equivalent to 100% germination after 60 min incubation. Values are the means of three independent experiments conducted with the same spore preparations; the standard deviation was ≤5% of the mean.
All other mutant strains with aromatic substitutions are observed to retain a strong germinative response to glucose, although the responses to other germinants are variable. The F280A substitution (GC529) appears to be significant, however; although they were stable upon storage in ice-cold water, heat-activated spores are observed to germinate reasonably efficiently (45%) in 5 mM Tris-HCl buffer alone. The tendency for these spores to germinate in the absence of appreciable levels of germinant might indicate that Phe280 is in the vicinity of the active site of the receptor or may have an important structural role in receptor stability.
Kinetic analysis of glucose-defective germination mutants.
Strains with substitutions to GerVB that were identified by preliminary screening as showing defective germinative responses to glucose were selected for more detailed kinetic analyses in an attempt to gain some insight into the structural basis for the defect. Despite showing extremely weak germinative responses when they were exposed to glucose concentrations that are saturating for wild-type spores, all six mutant strains tested were observed to retain a strong germinative response (>90% germination) when they were exposed to sufficiently high concentrations of glucose, as ascertained by determination of K0.5(germ) values (Table 6). Control strains that lack all known GerU structural genes (GC500) and those complemented with only the A and C receptor components (GC503) do not show any response to the maximum concentration of glucose (or other nutrient germinants) tested, indicating that receptors containing mutated GerVB proteins are responsible for the observed germinative responses.
TABLE 6.
Concentrations of nutrient germinants required to stimulate 50% spore germination in strains with substituted GerVBa
| Strain | Relevant genotypec |
K0.5(germ) (mM)b |
||
|---|---|---|---|---|
| Glucose | Proline | Leucine | ||
| GC500 | gerU negative | NAd | NA | NA |
| GC501 | gerU positive (gerUA-gerUC-gerVB) | 0.33 | 0.34 | 49 |
| GC503 | gerUA-gerUC | NA | NA | NA |
| GC509 | GerVB E196Q | 72 | NA | 100 |
| GC510 | GerVB E196D | 55 | NA | NA |
| GC525 | GerVB Y191A | 33 | 18 | NA |
| GC526 | GerVB F192A | 25 | 127 | ND (21)e |
| GC515 | GerVB P193A | 16 | 30 | 34 |
| GC517 | GerVB P343A | 72 | ND (23) | NA |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 90 min with various concentrations of glucose (10, 25, 50, 100, 175, 250, 400, 600, 800, and 1,000 mM), proline (25, 50, 75, 100, 150, 200, 250, and 300 mM), and leucine (1, 5, 10, 20, 50, 80, 100, and 120 mM); and germination was monitored as described in Materials and Methods. Strain GC517 was additionally germinated in 1,200 and 1,400 mM glucose. Strain GC501 was germinated in more dilute solutions (0.1 to 10 mM) of glucose and proline.
The concentrations of germinant required to stimulate 50% spore germination [K0.5(germ)] were calculated after incubation for 60 min for glucose and proline, after which no further germination was observed, and 90 min incubation for leucine. The values presented for each strain are derived from three independent experiments conducted with the same spore preparation; similar values were obtained with other spore preparations. The calculated standard error associated with all measurements is less than 10%.
All strains are ΔgerWB::Spr.
NA, not applicable (spore germination <5% after 90 min incubation).
ND, not determined. Values in parentheses indicate the apparent rate of spore germination (in percent) after 90 min incubation at the highest concentration of germinant tested.
Michaelis-Menten parameters were derived for select strains from hyperbolic curves of glucose-mediated germinative rates (Table 7). In all cases examined, Km values were subject to large (40- to 200-fold) increases, whereas maximal germinative rates (Vmax) were observed to deviate only slightly from the wild-type values, from which we can infer that the affinity of the receptor for glucose is probably being adversely affected by the replacement of residues that participate in glucose binding or perhaps substitutions that affect the conformation of the binding site.
Of the residues examined in functionally important TM6, the replacement of Glu196 by glutamine (E196Q) is observed to decrease the affinity of the receptor for glucose by approximately 165-fold (Km values, 513 mM versus 3.1 mM for the wild type). This is also reflected in the glucose concentration required to stimulate 50% spore germination within 60 min of exposure (72 mM versus 0.33 mM for the wild type), which reflects a 218-fold increase in germinant concentration. The introduction of a negatively charged residue with a smaller side chain at this position (E196D) is associated with an approximately 100-fold reduction in the affinity of the receptor for glucose, highlighting the sensitivity of this position to even subtle substitutions.
The apparent Km values for receptors with alanine replacements at the key TM6 aromatic residues (Tyr191 and Phe192) are similar (215 mM and 219 mM, respectively), representing an approximate 70-fold decrease in affinity compared to that for the wild-type response. The replacement of Pro193 by alanine results in an apparent 40-fold decrease in receptor affinity for glucose (Km, 123 mM), indicating that a proline-induced kink or proline-mediated flexibility toward the center of TM6 is important for efficient receptor function. The only other residue examined in detail, Pro343, causes the largest shift in the apparent affinity of the receptor (an approximately 200-fold reduction), although the maximal germinative rate is similar to that for wild-type spores. Thus, the loss of the proposed proline-flanking molecular bracket in TM10 appears to disrupt the germinant-binding site, as opposed to interruption of subsequent signal transduction.
Germinative responses of glucose-defective strains to other germinants.
Mutant strains carrying substitutions that strongly affect the glucose-mediated germinative response were observed in most cases to show diminished responses to other nutrient germinants (Table 6). Indeed, whereas the glucose-mediated response is retained when Glu196 is replaced by either glutamine or aspartate, albeit with a considerable increase in the K0.5(germ) value, the proline response is completely abolished even at the highest concentration tested (300 mM). On the other hand, spores bearing substitutions at other TM6 residues (Y191A, F192A, P193A) retain strong germinative responses when they are exposed to relatively high concentrations of proline (>85% germination; data not shown), although significant increases in K0.5(germ) values are evident. Spores bearing the P343A substitution, however, which is predicted to reside in TM10, show only a weak germinative response (23%) at the highest concentration tested (300 mM).
Similarly, while leucine is only a weak germinant for PV361-derived spores complemented with GerVB [K0.5(germ), ∼50 mM], it is evident that substitutions that severely affect the glucose-mediated response are also largely deleterious to leucine-triggered germination. With the exception of the P193A mutant, all glucose-deficient strains demonstrate an increased requirement for leucine to stimulate a 50% germinative response (E196Q) or fail to respond at all to the highest concentration of leucine tested (120 mM). The germinative response to inorganic salts also appears to be affected in these mutant strains, at least in response to 50 mM KBr (Tables 3 and 5), although this has not been examined in detail due to the potential for the introduction of nonspecific effects at high concentrations of inorganic salts.
SDM of TM6 residues in GerUB.
The GerUB protein shares 80% sequence identity with GerVB at the amino acid level and, like GerVB, is capable of initiating a glucose-mediated germinative response. It seems reasonable, therefore, to assume that residues identified on TM6 of GerVB as perhaps having an important role in receptor-ligand binding would fulfill the same role in GerUB. This hypothesis was tested by preparing spores complemented with GerUB bearing individually the Y191A, F192A, and E196Q amino acid substitutions.
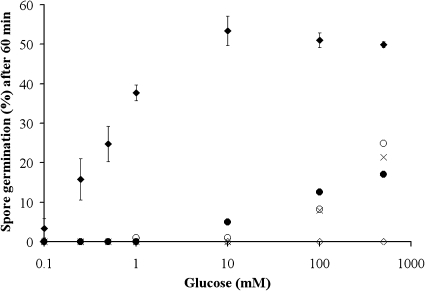
Analysis of the germinative properties of these strains (Fig. 2) reveals fractional germinative responses, even when spores complemented with native GerUB (GC502) are exposed to relatively high concentrations of germinant. However, whereas the germinative response of spores complemented with the native GerUB protein is saturated by 10 mM glucose, which stimulates 53% germination, spores complemented with any of the mutated GerUB receptors barely initiate a germinative response at all (<5%) at the same concentration of germinant. Indeed, relatively weak germinative responses are attained even with 500 mM glucose (GerUB E196Q, 24%; Y191A, 17%; F192A, 21%). The observation that spores lacking GerUB (GC503) do not germinate under any condition tested indicates that the mutated GerUB variants are responsible for the germinative responses at relatively high concentrations of glucose.
FIG. 2.
Germination rates of B. megaterium (ΔgerWB) spores complemented with GerU receptor genes in which the GerUB protein has been subjected to site-directed mutagenesis. Spores were germinated in buffer (5 mM Tris-HCl, pH 7.8) plus glucose at various concentrations (0.1, 0.25, 0.5, 1, 10, 100, and 500 mM glucose). ⧫, GerUB; ⋄, GerUA GerUC; ○, GerUB E196Q; •, GerUB Y191A; ×, GerUB F192A. The results are the averages of three independent experiments conducted with single batches of spores; similar values were obtained with other spore preparations. Error bars represent the standard deviation from the mean (omitted for clarity on the mutant strains [standard deviation < 5%]).
DISCUSSION
This communication describes a number of new observations concerning receptor proteins involved in initiating the B. megaterium QM B1551 spore germinative response. A chromosome-located structural gene, gerWB, has been characterized via mutational and genetic complementation analyses as encoding a spore germinant-receptor protein-B subunit that can interact with the GerUA and GerUC proteins to confer a receptor complex that is cognate for both glucose and leucine. Observations that PV361 spores complemented with only gerUA and gerUC structural genes retain a glucose response whereas gerWB mutant spores do not indicate that the GerWB protein is expressed and present in the spore. Whether the resultant protein contributes to the wild-type QM B1551 response to glucose and/or leucine, where it would presumably face competition from the GerUB and GerVB proteins for putative interaction sites with the GerUA and GerUC proteins, has yet to be established.
We also report for the first time on the identification of residues that significantly affect the GerU-mediated germinative response to glucose. SDM was used to introduce amino acid substitutions to various charged, proline, and aromatic residues predicted to reside in TM regions of GerVB. Whereas the probable structural importance of Pro343 in TM10, where it might promote helical interactions that facilitate access to crucial ligand-binding residues, is consistent with previous observations as to the importance of this domain to efficient receptor function (5), the importance of TM6 has additionally been identified.
A number of observations lead us to postulate that residues predicted to reside in TM6 of GerVB may be directly involved in binding of germinants. The decreases in the apparent affinity of the receptor when Glu196, Tyr191, and Phe192 are individually replaced are of magnitudes similar to those reported for various prokaryotic amino acid/sugar transporters where ligand-binding residues are replaced (11, 26). Similarly, the chemical nature of the candidate ligand-binding residues, i.e., charged and/or aromatic amino acids, is consistent with the nature of residues observed to be directly involved in binding to ligands in various single-component transporters (10, 21, 34), although we acknowledge that any comparisons between spore germinant-receptor proteins and bacterial transporters have to be made with caution. Finally, amino acids at the equivalent positions in other germinant-receptor proteins are highly conserved, including the B. subtilis GerKB protein, which appears to be cognate for glucose (2, 15) (Fig. 1), and, indeed, of functional importance, as demonstrated for the homologous GerUB protein.
It is evident also that substitutions to residues that are crucial to glucose-mediated germinative responses exert deleterious effects on the responses to other nutrient germinants. These observations and the results of kinetic analyses previously conducted with combinations of germinants (4, 5) are consistent with a receptor model in which the respective germinant-binding sites are close to the receptor and may in some cases overlap. Alternatively, it is entirely feasible that residues identified as being important to receptor function are not involved in ligand binding and that, instead, substitution introduces structural perturbations that adversely affect the conformation of germinant-binding sites. Similarly, the possibility that some or all of these substitutions result in reduced receptor abundance, perhaps by interfering with the ability to insert correctly into the membrane, which might be expected to lead to an increased requirement for germinants in order to trigger germination, should also be recognized.
Definitive identification of the residues that are directly involved in binding to germinants and, indeed, mechanistic insights into spore germinant-receptor function will require appropriate structural information at the level of atomic resolution. This seems to be some way off since, in our experience, receptor B proteins are recalcitrant to overexpression in E. coli, presenting a major bottleneck to crystallization trials.
It may be possible, however, to derive structural information from other sources. The spore germinant-receptor B proteins are members of the amino acid, polyamine, and organocation (APC) superfamily of secondary transporters (spore germination protein [SGP] family; transporter classification no. 2.A.3.9) (16), the first high-resolution structures of which have recently been solved (10, 11, 34). Despite limited sequence identity at the amino acid level, the overall fold of these APC proteins (in which TM6 to TM10 are structurally related and whose folds are inverted with respect to those of TM1 to TM5) is very similar to the core structure observed in four other families of unrelated transport proteins (9, 28, 44, 46). This raises the question as to whether members of the SGP family might also adopt this common structural fold, particularly since the predicted 10 α-helical domains that define the topology of the SGP family are structurally analogous (based on protein alignments) to TM1 to TM10 of other APC proteins.
It is intriguing that helices that comprise TM6 and TM1 in the known APC crystal structures are involved in substrate binding via characteristic short nonhelical sections that expose main-chain carbonyl and amide groups that promote hydrogen bonding with a substrate(s) and facilitate subsequent conformational changes (10, 11, 34). TM10 is also shown to participate in the substrate-binding cavity of the arginine/agmatine antiporter AdiC, where it is thought to undergo a conformational shift to assist with the occlusion of bound substrate to the periplasm (10). Thus, the functional importance of TM6 and TM10 to protein-B subunits of the B. megaterium GerU receptor is at least consistent with the notion that some members of the SGP family may adopt a fold similar to that of APC transporters.
Additionally, residues in TM3 and TM8 are also involved in substrate binding in various APC transporters. An aromatic residue (Trp293), for example, located toward the center of TM8 of the arginine/agmatine antiporter AdiC, forms part of the floor of the substrate-binding cavity (10). It is not inconceivable that Phe280, which is located at a similar position in TM8 of GerVB and which is highly conserved in other receptors, performs a similar role, and this is why the receptor structure appears to adopt an activated state upon its replacement by alanine, as evidenced by the high degree of spore germination initiated in buffer alone. The determination of whether these observations are entirely coincidental or the first evidence that an APC transporter-like fold is shared by members of the SGP family is a current focus of our research.
Acknowledgments
This work was supported by a grant awarded to G.C. and C.R.L. by the United Kingdom Home Office.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 2.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabrera-Martinez, R. M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie, G., M. Lazarevska, and C. R. Lowe. 2008. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J. Bacteriol. 190:2014-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie, G., and C. R. Lowe. 2008. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J. Bacteriol. 190:8009-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie, G., and C. R. Lowe. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 189:4375-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordes, F. S., J. N. Bright, and M. S. Sansom. 2002. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323:951-960. [DOI] [PubMed] [Google Scholar]
- 8.Cosgriff, A. J., G. Brasier, J. Pi, C. Dogovski, J. P. Sarsero, and A. J. Pittard. 2000. A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport. J. Bacteriol. 182:2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faham, S., A. Watanabe, G. M. Besserer, D. Cascio, A. Specht, B. A. Hirayama, E. M. Wright, and J. Abramson. 2008. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, Y., H. Jayaram, T. Shane, L. Kolmakova-Partensky, F. Wu, C. Williams, Y. Xiong, and C. Miller. 2009. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460:1040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, X., F. Lu, L. Zhou, S. Dang, L. Sun, X. Li, J. Wang, and Y. Shi. 2009. Structure and mechanism of an amino acid antiporter. Science 324:1565-1568. [DOI] [PubMed] [Google Scholar]
- 12.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 13.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igarashi, T., and P. Setlow. 2005. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J. Bacteriol. 187:2513-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irie, R., Y. Fujita, and M. Kobayashi. 1996. Nucleotide sequence and gene organisation of the gerK spore germination locus of Bacillus subtilis 168. J. Gen. Appl. Microbiol. 42:141-153. [Google Scholar]
- 16.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146(Pt 8):1797-1814. [DOI] [PubMed] [Google Scholar]
- 17.Kini, R. M., and H. J. Evans. 1995. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem. Biophys. Res. Commun. 212:1115-1124. [DOI] [PubMed] [Google Scholar]
- 18.Kunnimalaiyaan, M., D. M. Stevenson, Y. Zhou, and P. S. Vary. 2001. Analysis of the replicon region and identification of an rRNA operon on pBM400 of Bacillus megaterium QM B1551. Mol. Microbiol. 39:1010-1021. [DOI] [PubMed] [Google Scholar]
- 19.Moir, A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526-530. [DOI] [PubMed] [Google Scholar]
- 20.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. 1994. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 77:9S-16S. [PubMed] [Google Scholar]
- 21.Nassif, H., H. Al-Ali, S. Khuri, and W. Keirouz. 2009. Prediction of protein-glucose binding sites using support vector machines. Proteins 77:121-132. [DOI] [PubMed] [Google Scholar]
- 22.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. GerO, a putative Na+/H+-K+ antiporter, is essential for normal germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:3822-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pi, J., P. J. Wookey, and A. J. Pittard. 1993. Site-directed mutagenesis reveals the importance of conserved charged residues for the transport activity of the PheP permease of Escherichia coli. J. Bacteriol. 175:7500-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirch, T., M. Quick, M. Nietschke, M. Langkamp, and H. Jung. 2002. Sites important for Na+ and substrate binding in the Na+/proline transporter of Escherichia coli, a member of the Na+/solute symporter family. J. Biol. Chem. 277:8790-8796. [DOI] [PubMed] [Google Scholar]
- 27.Quick, M., and H. Jung. 1998. A conserved aspartate residue, Asp187, is important for Na+-dependent proline binding and transport by the Na+/proline transporter of Escherichia coli. Biochemistry 37:13800-13806. [DOI] [PubMed] [Google Scholar]
- 28.Ressl, S., A. C. Terwisscha van Scheltinga, C. Vonrhein, V. Ott, and C. Ziegler. 2009. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature 458:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Sammons, R. L., A. Moir, and D. A. Smith. 1981. Isolation and properties of spore germination mutants of Bacillus subtilis-168 deficient in the initiation of germination. J. Gen. Microbiol. 124:229-241. [Google Scholar]
- 30.Sansom, M. S. 1992. Proline residues in transmembrane helices of channel and transport proteins: a molecular modelling study. Protein Eng. 5:53-60. [DOI] [PubMed] [Google Scholar]
- 31.Sansom, M. S., and H. Weinstein. 2000. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol. Sci. 21:445-451. [DOI] [PubMed] [Google Scholar]
- 32.Setlow, B., P. G. Wahome, and P. Setlow. 2008. Release of small molecules during germination of spores of Bacillus species. J. Bacteriol. 190:4759-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 34.Shaffer, P. L., A. Goehring, A. Shankaranarayanan, and E. Gouaux. 2009. Structure and mechanism of a Na+-independent amino acid transporter. Science 325:1010-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah, I. M., M. H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson, D. M., D. Lach, and P. S. Vary. 1993. A gene required for germination in Bacillus megaterium is plasmid-borne, p. 197-207. In E. Balla and G. Berencsie (ed.), DNA transfer and gene expression in microorganisms. Intercept, Budapest, Hungary.
- 37.Sujatha, M. S., Y. U. Sasidhar, and P. V. Balaji. 2005. Insights into the role of the aromatic residue in galactose-binding sites: MP2/6-311G++** study on galactose- and glucose-aromatic residue analogue complexes. Biochemistry 44:8554-8562. [DOI] [PubMed] [Google Scholar]
- 38.Swerdlow, B. M., B. Setlow, and P. Setlow. 1981. Levels of H+ and other monovalent cations in dormant and germinating spores of Bacillus megaterium. J. Bacteriol. 148:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thackray, P. D., J. Behravan, T. W. Southworth, and A. Moir. 2001. GerN, an antiporter homologue important in germination of Bacillus cereus endospores. J. Bacteriol. 183:476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 41.Vepachedu, V. R., and P. Setlow. 2007. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol. Lett. 274:42-47. [DOI] [PubMed] [Google Scholar]
- 42.Vepachedu, V. R., and P. Setlow. 2004. Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon. FEMS Microbiol. Lett. 239:71-77. [DOI] [PubMed] [Google Scholar]
- 43.Vepachedu, V. R., and P. Setlow. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187:5677-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weyand, S., T. Shimamura, S. Yajima, S. Suzuki, O. Mirza, K. Krusong, E. P. Carpenter, N. G. Rutherford, J. M. Hadden, J. O'Reilly, P. Ma, M. Saidijam, S. G. Patching, R. J. Hope, H. T. Norbertczak, P. C. Roach, S. Iwata, P. J. Henderson, and A. D. Cameron. 2008. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittchen, K. D., and F. Meinhardt. 1995. Inactivation of the major extracellular protease from Bacillus megaterium DSM319 by gene replacement. Appl. Microbiol. Biotechnol. 42:871-877. [DOI] [PubMed] [Google Scholar]
- 46.Yamashita, A., S. K. Singh, T. Kawate, Y. Jin, and E. Gouaux. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437:215-223. [DOI] [PubMed] [Google Scholar]
- 47.Zuberi, A. R., A. Moir, and I. M. Feavers. 1987. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene 51:1-11. [DOI] [PubMed] [Google Scholar]