Abstract
In Pseudomonas syringae, the type III secretion system (T3SS) is essential for disease in compatible hosts and for eliciting the hypersensitive response in incompatible hosts. P. syringae pathovars secrete a variable number of type III effectors that form their secretomes. The secretome of Pseudomonas syringae pv. phaseolicola 1448a (Pph1448a) currently includes 22 experimentally validated effectors, one HrpL-regulated candidate for which translocation results have been inconsistent, two translocated candidates for which in planta expression has not been established, one bioinformatically identified candidate, and six candidates that have been experimentally discarded. We analyzed the translocation and/or expression of these and other candidates to complete the Pph1448a effector inventory, bringing this inventory to 27 bona fide effectors, including a new one that does not belong to any of the previously described effector families. We developed a simple process for rapidly making single and double knockout mutants and apply it to the generation of an effector mutant collection that includes single knockouts for the majority of the Pph1448a effector inventory. We also generated two double mutant strains containing effectors with potentially redundant functions and analyzed the virulence of the single and double mutant strains as well as strains expressing each of the effectors from a plasmid. We demonstrate that AvrB4-1 and AvrB4-2, as well as HopW1-1 and HopW1-2, are fully redundant and contribute to virulence in bean plants, thus validating this approach for dissecting the contribution of the Pph1448a type III effector inventory to virulence. We also analyzed the effect that the expression of these four effectors from Pseudomonas syringae pv. tomato DC3000 (PtoDC3000) has during its interaction with Arabidopsis thaliana, establishing that AvrB4-1, but not the others, determines a restriction of bacterial growth that takes place mostly independently of the salicylic acid (SA)-signaling pathway.
Type III secretion systems (T3SS) are complex and specialized machineries that inject effector proteins directly into the host cell cytosol (2). In Pseudomonas syringae, T3SS-mediated secretion is essential for disease in compatible hosts and for eliciting the hypersensitive response (HR) in incompatible hosts (1). P. syringae pathovars secrete a variable number of type III effectors that form their so-called secretomes and are expressed within the plant under the control of the alternative sigma factor HrpL (47). Understanding how the T3SS determines pathogenicity requires the functional characterization of the complete type III effector inventory. However, this characterization has been partially hindered by the fact that mutation of individual effectors, usually the most straightforward approach, rarely causes virulence attenuation (14). Thus, reports showing the contribution of the type III effector to virulence in P. syringae pathovars have resorted to ectopic expression in homolog-lacking related strains (40), plasmid-cured derivatives (21), double mutants (6, 28), or polymutants (3, 26). In relation to this, we have previously established the use of the competitive index (CI) in mixed infections (13, 42) as a more sensitive virulence assay for P. syringae pathovars than traditional assays (31). Using CIs, we demonstrated for the first time the individual contribution of AvrPto, an otherwise thoroughly characterized type III effector from Pseudomonas syringae pv. tomato (9, 17, 18, 27, 36, 39, 40, 46), to pathogen growth within its natural host (31). Therefore, analysis of effector mutants by use of the CI may provide the means to establish the quantitative contribution of the members of P. syringae T3SS secretomes to virulence. In addition, genetic analysis of the effects of combinations of effector mutations on virulence has already proven a useful approach to establishing the contribution of the members of the P. syringae pv. tomato DC3000 secretome to virulence by revealing a functional overlap (6, 26, 28). Thus, generation of knockouts in all individual effector genes of a given secretome, achieved in such a manner as to allow for easy combination of these strains into double or multiple mutant strains, is a desirable task, albeit a cumbersome one, considering the size of most secretomes.
The secretome of the fully sequenced wild-type (wt) representative of the Pseudomonas syringae pv. phaseolicola 1448a strain (Pph1448a) has previously been analyzed, using a differential fluorescence induction screen (7) and bioinformatics (44), to identify effector genes. Our laboratory contributed to establishing this secretome through the development and application of a very sensitive assay for T3SS-mediated translocation based on CI assays (30). This assay represents an improvement over the sensitivity of the commonly used AvrRpt2 reporter assay. When fused to a T3SS-secreted protein, AvrRpt281-255 is translocated inside the host cell, eliciting a hypersensitive response (HR), dependent on the resistance protein RPS2 (32). By using CIs to measure the bacterial growth reduction associated with the AvrRpt2-RPS-mediated defense response, we detected translocation for two out of four Pph1448a effector candidates previously discarded by other assays, HopAJ1 and HopAK1 (30), and demonstrated translocation for two out of five previously untested candidates, HopAH2 and A0129. However, although in planta expression has been shown to take place in an HrpL-dependent manner for HopAJ1 and HopAK1 (7), it has not been established for HopAH2 and A0129. Effector nomenclature guidelines recommend that the abbreviation for the pathovars as well as the name of the strain should be included within the effector name (29). For simplicity, we include this indication only when effectors from other pathovars are mentioned. In summary, to date, 22 effectors in Pph1448a have been experimentally validated (7, 30, 44), one HrpL-regulated candidate has given inconsistent translocation results (AvrE1) (7), two translocated candidates have not been analyzed for expression in planta (HopAH2 and A0129) (30), one bioinformatically identified candidate has not been experimentally tested (AvrB4-2) (23), and six additional candidates have been proposed but experimentally ruled out (PSPPH3757, HopAN1, HopAJ2, HopW1-2, HopV1, and HopJ1) (7, 30).
In this work, we analyzed the translocation and/or expression of these and other candidate effectors to close the type III effector inventory of Pph1448a. Our results indicate that the Pph1448a complete type III secretome is formed by 27 validated effectors, including a new one, HopAY1, which does not belong to any of the previously described effector families. The work includes the development of a simplified process for quick generation of single and double knockout mutants and its application to constructing a collection of single mutants for almost all members of the Pph1448a type III secretome. Additionally, we generated two double mutant strains containing effectors with potentially redundant functions and analyzed the virulence of the four single and two double mutant strains as well as the double mutants expressing each of the effectors from a plasmid. We demonstrate that AvrB4-1 and AvrB4-2, as well as HopW1-1 and HopW1-2, are fully redundant and contribute to the virulence of Pph1448a. The tools and approach used in this work set the groundwork for dissecting the contribution of the entire Pph1448a type III secretome to virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this work are listed in Table 1. Escherichia coli and Pph1448a derivative strains were grown at 37°C and 28°C, respectively, with aeration in Luria-Bertani (LB) medium. Antibiotics were used at the following concentrations: for ampicillin, 100 μg/ml for E. coli DH5α and 100 μg/ml for liquid cultures and 300 μg/ml for plates for Pph1448a derivative strains; for kanamycin (Km), 50 μg/ml for E. coli DH5α and 15 μg/ml for Pph1448a derivative strains; and for cycloheximide, 2 μg/ml.
TABLE 1.
Bacterial strains used and effector mutant collection generated in this work
| Bacterial strain | Description | Antibiotic resistance | Source or reference |
|---|---|---|---|
| E. coli DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 ΔlacU189 φ80 ΔlacZDM15 | None | 16 |
| P. syringae pv. tomato DC3000 | Wild-type strain | Rifr | 10 |
| P. syringae pv. phaseolicola | |||
| 1448a | Race 6; wild-type strain | Nalr | 43 |
| IOM7 | ΔhrpL | Nalr Kmr | 33 |
| AZJ11 | ΔhopW1-1::nptII | Nalr Kmr | This work |
| AZJ12 | ΔhopW1-1 | Nalr | This work |
| AZJ13 | ΔhopW1-2::nptII | Nalr Kmr | This work |
| AZJ14 | ΔhopW1-1 ΔhopW1-2::nptII | Nalr Kmr | This work |
| AZJ15 | ΔavrB4-1::nptII | Nalr Kmr | This work |
| AZJ16 | ΔavrB4-2::nptII | Nalr Kmr | This work |
| AZJ17 | ΔavrB4-2 | Nalr | This work |
| AZJ18 | ΔavrB4-1::nptII ΔavrB4-2 | Nalr Kmr | This work |
| AZJ19 | ΔhopAB1::nptII | Nalr Kmr | This work |
| AZJ20 | ΔhopX1::nptII | Nalr Kmr | This work |
| AZJ21 | ΔavrB2::nptII | Nalr Kmr | This work |
| AZJ22 | ΔhopR1::nptII | Nalr Kmr | This work |
| AZJ23 | ΔhopAY1::nptII | Nalr Kmr | This work |
| AZJ24 | ΔhopAW1::nptII | Nalr Kmr | This work |
| AZJ25 | ΔhopAS1::nptII | Nalr Kmr | This work |
| AZJ26 | ΔhopAJ1::nptII | Nalr Kmr | This work |
| AZJ27 | ΔhopAU1::nptII | Nalr Kmr | This work |
| AZJ28 | ΔhopAK1::nptII | Nalr Kmr | This work |
| AZJ29 | ΔhopAE1::nptII | Nalr Kmr | This work |
| AZJ30 | ΔhopD1::nptII | Nalr Kmr | This work |
| AZJ31 | ΔhopQ1::nptII | Nalr Kmr | This work |
| AZJ32 | ΔhopG1::nptII | Nalr Kmr | This work |
| AZJ33 | ΔhopI1::nptII | Nalr Kmr | This work |
| AZJ34 | ΔavrD1::nptII | Nalr Kmr | This work |
| JRP1 | ΔavrB4-1 ΔavrB4-2 | Nalr | This work |
Plasmids and cloning procedures.
All plasmids used in this work are listed in Table 2. To obtain pAME9.18, pAME9.19, pAME9.20, and pAME9.21, the open reading frames (ORFs) of avrB4-2, hopW1-2, PSPPH1154, and avrE1 without their Stop codons and with their putative ribosome-binding sites were PCR amplified and cloned into pAME9 (30), using Pph1448a as a template and the corresponding primers (primers and cloning sites are listed in Table 3 ). To clone avrE1, only the first 978 bp of the ORF were used. Primers were designed to generate translational fusions of the proteins encoded by these genes to the sequence encoding amino acids 81 to 255 of AvrRpt2, to be expressed under the control of the PnptII promoter. Attention was paid during primer designing to avoid cross amplification of highly homologous genes.
TABLE 2.
Plasmids used and effector knockout vector collection generated in this work
| Plasmid | Description | Antibiotic resistance | Source or reference |
|---|---|---|---|
| pGEM-T | Cloning vector | Ampr | Promega |
| pKD4 | pANTSγ derivative containing an FRT-flanked kanamycin resistance gene | Ampr Kmr | 11 |
| pFLP2 | Contains a flipase gene | Ampr | 19 |
| pGEM-T-KmFRT EcoRI | pGEM-T derivative containing a Km resistance gene flanked by FRT and EcoRI sites | Ampr Kmr | This work |
| pGEM-T-KmFRT BamHI | pGEM-T derivative containing a Km resistance gene flanked by FRT and BamHI sites | Ampr Kmr | This work |
| pGEM-T-KmFRT XhoI | pGEM-T derivative containing a Km resistance gene flanked by FRT and XhoI sites | Ampr Kmr | This work |
| pAZJ6 | pGEM-T derivative carrying the ΔavrB2::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ7 | pGEM-T derivative carrying the ΔhopR1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ8 | pGEM-T derivative carrying the ΔavrB4-1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ9 | pGEM-T derivative carrying the ΔhopW1-1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ10 | pGEM-T derivative carrying the ΔhopX1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ11 | pGEM-T derivative carrying the ΔhopAE1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ12 | pGEM-T derivative carrying the ΔhopAB1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ13 | pGEM-T derivative carrying the ΔhopAW1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ14 | pGEM-T derivative carrying the ΔavrRps4::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ15 | pGEM-T derivative carrying the ΔhopG1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ16 | pGEM-T derivative carrying the ΔhopQ1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ17 | pGEM-T derivative carrying the ΔhopD1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ18 | pGEM-T derivative carrying the ΔhopI1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ19 | pGEM-T derivative carrying the ΔhopAV1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ20 | pGEM-T derivative carrying the ΔhopAT1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ21 | pGEM-T derivative carrying the ΔhopAF1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ22 | pGEM-T derivative carrying the ΔhopAS1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ23 | pGEM-T derivative carrying the ΔhopW1-2::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ24 | pGEM-T derivative carrying the ΔavrD1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ25 | pGEM-T derivative carrying the ΔhopAU1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ26 | pGEM-T derivative carrying the ΔhopAK1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ27 | pGEM-T derivative carrying the ΔavrB4-2::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ28 | pGEM-T derivative carrying the ΔhopAJ1::KmFRT knockout allele | Ampr Kmr | This work |
| pAZJ29 | pGEM-T derivative carrying the ΔhopF3::KmFRT knockout allele | Ampr Kmr | This work |
| pAME8 | pPnptII::avrRpt2 | Ampr Kmr | 30 |
| pAMEX | Vector for expression of N-terminal protein fusions to AvrRpt281-255 under a PnptII promoter | Ampr Kmr | 30 |
| pAME9.18 | pAMEX derivative expressing the HopW1-1::′AvrRpt2 protein fusion | Ampr Kmr | This work |
| pAME9.19 | pAMEX derivative expressing the AvrB4-2::′AvrRpt2 protein fusion | Ampr Kmr | This work |
| pAME9.20 | pAMEX derivative expressing the PSPPH1154::′AvrRpt2 protein fusion | Ampr Kmr | This work |
| pAME9.21 | pAMEX derivative expressing the avrE1::′AvrRpt2 protein fusion | Ampr Kmr | This work |
| pJRU1 | pPlac::hopW1-1/2 | Ampr Kmr | This work |
| pJRU2 | pPlac::avrB4-2 | Ampr Kmr | This work |
| pJRU3 | pPnptII::avrB4-1 | Ampr Kmr | This work |
| pJRU4 | pPnptII::avrB4-2 | Ampr Kmr | This work |
| pJRU5 | pavrB4-1 | Ampr Kmr | This work |
| pJRU6 | pPnptII::hopW1-1/2 | Ampr Kmr | This work |
TABLE 3.
Primers used in this work
| Primer | Sequence | Localization in genome | Restriction sitea |
|---|---|---|---|
| A1-hopW1-1 | 5′CCCTATAGTGAGTCGAATTCCACTGGAGTCAAGCTTACG3′ | 6121-6139c | EcoRI |
| A2-hopW1-1 | 5′GAATTCGACTCACTATAGGGTCTCAGCGGTGTAGCTTTCC3′ | 6711-6730 | EcoRI |
| B1-hopW1-1 | 5′GTTCTGCTAATGTGGACC3′ | 5607-5624 | NA |
| B2-hopW1-1 | 5′TAGGATGGGATTGCTGGG3′ | 7232-7249c | NA |
| A1-hopW1-2 | 5′CCCTATAGTGAGTCGGATCCGCCTCTATTTGCAACAGC3′ | 63160-63177c | BamHI |
| A2-hopW1-2 | 5′GGATCCGACTCACTATAGGGCTATTCCTTCTCTCAGCG3′ | 63471-63488 | BamHI |
| B1-hopW1-2 | 5′TCCAGGTCGATCATTTGC3′ | 62568-62585 | NA |
| B2-hopW1-2 | 5′CCGTGATTTCTCTTAGCC3′ | 64178-64195c | NA |
| A1-avrB4-1 | 5′CCCTATAGTGAGTCGAATTCGAGCCTTAGTGGTAGTAACC3′ | 3515337-3515356c | EcoRI |
| A2-avrB4-1 | 5′GAATTCGACTCACTATAGGGACTAGGTTCATGCATAGGC3′ | 3516388-3516406 | EcoRI |
| B1-avrB4-1 | 5′CGTGACGAGTGATCATCTGC3′ | 3514781-3514800 | NA |
| B2-avrB4-1 | 5′ATCCTTTTGCACACCGATCC3′ | 3516888-3516907c | NA |
| A1-avrB4-2 | 5′CCCTATAGTGAGTCGGATCCAGAAGCCCGCAGAATTCG3′ | 927554-927571c | BamHI |
| A2-avrB4-2 | 5′GGATCCGACTCACTATAGGGTAGATGACGAGCCTTAGG3′ | 928565-928582 | BamHI |
| B1-avrB4-2 | 5′TTCCAGCACGAAGTATCC3′ | 926946-926963 | NA |
| B2-avrB4-2 | 5′AAGCGCGAAGAACACGCTG3′ | 929015-929033c | NA |
| A1-hopAB1 | 5′CCCTATAGTGAGTCGAATTCAGACAAAGGCTTGATGCC3′ | 107146-107163c | EcoRI |
| A2-hopAB1 | 5′GAATTCGACTCACTATAGGGTCCTGATCTCATGGTTGC3′ | 108740-108757 | EcoRI |
| B1-hopAB1 | 5′TCCATCACCTGTTGAAGC3′ | 106559-106576 | NA |
| B2-hopAB1 | 5′ATCCGCAACCTCATAGAG3′ | 109299-109316c | NA |
| A1-hopAE1 | 5′CCCTATAGTGAGTCGAATTCTCTTGCCTGGCTTACCAC3′ | 4943896-4943913c | EcoRI |
| A2-hopAE1 | 5′GAATTCGACTCACTATAGGGTACTGTCGGTGGATATGG3′ | 4946706-4946723 | EcoRI |
| B1-hopAE1 | 5′AAGCCATGCAGATCCTCG3′ | 4943302-4943319 | NA |
| B2-hopAE1 | 5′TAATCAGCGCCGAACAGC3′ | 4947287-4947304c | NA |
| A1-avrB2 | 5′CCCTATAGTGAGTCGAATTCCGTTCTTACGATCGCGTAGC3′ | 102383-102402c | EcoRI |
| A2-avrB2 | 5′GAATTCGACTCACTATAGGGATTCTAGGTGGCATTGCAGG3′ | 103535-103554 | EcoRI |
| B1-avrB2 | 5′GCCGATAACCTGATCTACG3′ | 101280-101298 | NA |
| B2-avrB2 | 5′CTTAACTGAGACATCAACGGC3′ | 104481-104501c | NA |
| A1-avrD1 | 5′CCCTATAGTGAGTCGGATCCAAACAGCTGCTGATTCCC3′ | 96818-96835c | BamHI |
| A2-avrD1 | 5′GGATCCGACTCACTATAGGGTGCTGAAGCTATACAGCC3′ | 97836-97853 | BamHI |
| B1-avrD1 | 5′TCCGGATGGGGTATACTC3′ | 96266-96283 | NA |
| B2-avrD1 | 5′TGGCGGCTTTGATCTGTG3′ | 98300-98317c | NA |
| A1-hopD1 | 5′CCCTATAGTGAGTCGAATTCCAATGTGTGGTCGCAAGG3′ | 7093-7110c | EcoRI |
| A2-hopD1 | 5′GAATTCGACTCACTATAGGGATCATAGTCGACGCGACCT3′ | 9280-9298 | EcoRI |
| B1-hopD1 | 5′GCATTCAGTTCACCAGACG3′ | 6554-6572 | NA |
| B2-hopD1 | 5′CGTATAGTGACAAAGGAGG3′ | 9859-9877c | NA |
| A1-hopF3 | 5′CCCTATAGTGAGTCGAATTCAGCTTGATGCTGTGCTCC3′ | 4039619-4039636c | EcoRI |
| A2-hopF3 | 5′GAATTCGACTCACTATAGGGATGCCCATGGAAGATTCC3′ | 4040252-4040269 | EcoRI |
| B1-hopF3 | 5′TGCAACTGAACCAGCACC3′ | 4038961-4038978 | NA |
| B2-hopF3 | 5′ATCTGGCGAGCATTCTCG3′ | 4040823-4040840c | NA |
| A1-hopG1 | 5′CCCTATAGTGAGTCGAATTCAGGCAGCATGCGTATTTGC3′ | 897388-897406c | EcoRI |
| A2-hopG1 | 5′GAATTCGACTCACTATAGGGTACGTGGTCATCACAAAGC3′ | 899013-899031 | EcoRI |
| B1-hopG1 | 5′CCTTCATGCCTTGATGACTG3′ | 896841-896860 | NA |
| B2-hopG1 | 5′ACAGACCGTTGTTAGCCAG3′ | 899582-899600c | NA |
| A1-hopI1 | 5′CCCTATAGTGAGTCGAATTCTTTTCGGCGTGAGGAGAC3′ | 4988642-4988659c | EcoRI |
| A2-hopI1 | 5′GAATTCGACTCACTATAGGGATGCTTCTCCGGGAACTC3′ | 4989672-4989689 | EcoRI |
| B1-hopI1 | 5′AGATCACCGGATACATCG3′ | 4988059-4988076 | NA |
| B2-hopI1 | 5′TTCTGTAGAGCAACACGG3′ | 4990274-4990291c | NA |
| A1-hopQ1 | 5′CCCTATAGTGAGTCGAATTCATCTGCGCTTGTCCAGTC3′ | 9414-9431c | EcoRI |
| A2-hopQ1 | 5′GAATTCGACTCACTATAGGGCTGGATAGATGAACCTGC3′ | 10751-10768 | EcoRI |
| B1-hopQ1 | 5′TGTCGGAACAGATACTGC3′ | 8876-8893 | NA |
| B2-hopQ1 | 5′CGAGACATGGATGTGTGG3′ | 11381-11398c | NA |
| A1-hopR1 | 5′CCCTATAGTGAGTCGAATTCAACAAAGGAAGCGACCTG3′ | 198648-198665c | EcoRI |
| A2-hopR1 | 5′GAATTCGACTCACTATAGGGGCATCATGCCGAATAGG3′ | 204628-204644 | EcoRI |
| B1-hopR1 | 5′GCATTCTCAGGTTGTAGATGG3′ | 198090-198110 | NA |
| B2-hopR1 | 5′CGTCGAGTATCTGTATCGTTGG3′ | 205178-205199c | NA |
| A1-hopX1 | 5′CCCTATAGTGAGTCGAATTCTGTTCCTCACTGATTGCG3′ | 1508676-1508693c | EcoRI |
| A2-hopX1 | 5′GAATTCGACTCACTATAGGGACAGGTCATAGAGTTCGG3′ | 1509903-1509920 | EcoRI |
| B1-hopX1 | 5′TGAGCGTGAGAGTTACACG3′ | 1508038-1508056 | NA |
| B2-hopX1 | 5′GGTCTGACATAATCTGCG3′ | 1510496-1510513c | NA |
| A1-hopAJ1 | 5′CCCTATAGTGAGTCGGATCCCTTCATAGGTTTAGCTCGC3′ | 892773-892791c | BamHI |
| A2-hopAJ1 | 5′GGATCCGACTCACTATAGGGTGTGAAGTGAAAGTGGCC3′ | 894080-894097 | BamHI |
| B1-hopAJ1 | 5′TGACAAGCATGCCAAAGC3′ | 892244-892261 | NA |
| B2-hopAJ1 | 5′CAATCACGGAAGAAGCAC3′ | 894731-894748c | NA |
| A1-hopAK1 | 5′CCCTATAGTGAGTCGAATTCAAAAGGGCGACCGAAGTC3′ | 1652010-1652027c | EcoRI |
| A2-hopAK1 | 5′GAATTCGACTCACTATAGGGTGAACGATTCGTGATCCG3′ | 1653668-1653685 | EcoRI |
| B1-hopAK1 | 5′TTGCGTGAAGAGCAACAG3′ | 1651461-1651478 | NA |
| B2-hopAK1 | 5′CGCCAGCTATAGCAAGAC3′ | 1654216-1654233c | NA |
| A1-hopAS1 | 5′CCCTATAGTGAGTCGAATTCGATCAATACAGGTGGTGG3′ | 5376014-5376031c | EcoRI |
| A2-hopAS1 | 5′GAATTCGACTCACTATAGGGACGCTGGTCATTCAACTG3′ | 5380168-5380185 | EcoRI |
| B1-hopAS1 | 5′TGGACGCGTCTTGAATGG3′ | 5375449-5375466 | NA |
| B2-hopAS1 | 5′TGTCGGATTAGTTCAGGG3′ | 5380661-5380678c | NA |
| A1-hopAT1 | 5′CCCTATAGTGAGTCGGATCCACGCTTCTGGATCTTCGG3′ | 894814-894831c | BamHI |
| A2-hopAT1 | 5′GGATCCGACTCACTATAGGGTTGAGGATCACAAAGCGC3′ | 895177-895194 | BamHI |
| B1-hopAT1 | 5′TACATATCCTGTGCGCTG3′ | 894188-894205 | NA |
| B2-hopAT1 | 5′GGATCTGAATTCCATCGC3′ | 895766-895783c | NA |
| A1-hopAU1 | 5′CCCTATAGTGAGTCCTCGAGGGAGGGTTCCAAAGACAG3′ | 26320-26339c | XhoI |
| A2-hopAU1 | 5′CTCGAGGACTCACTATAGGGAGCCTGGAGACATTCATGC3′ | 28431-28449 | XhoI |
| B1-hopAU1 | 5′AGTACCGCAGTCCTTCAC3′ | 25800-25817 | NA |
| B2-hopAU1 | 5′CTCTGGTGATGTTTTCCG3′ | 28993-29010c | NA |
| A1-hopAV1 | 5′CCCTATAGTGAGTCGGATCCCTGCTCCAACTATTAGCC3′ | 45681-45698c | BamHI |
| A2-hopAV1 | 5′GGATCCGACTCACTATAGGGATCTCGTCGATGAAGGAC3′ | 48004-48021 | BamHI |
| B1-hopAV1 | 5′TGTTCAACTACGACCGTC3′ | 45126-45143 | NA |
| B2-hopAV1 | 5′AACATCCGGCTACTTCAG3′ | 48631-48648c | NA |
| A1-hopAW1 | 5′CCCTATAGTGAGTCGAATTCTATGCGTAGTGAACAGGG3′ | 104780-104797c | EcoRI |
| A2-hopAW1 | 5′GAATTCGACTCACTATAGGGAGAAACACTGAGTGGTCG3′ | 105491-105508 | EcoRI |
| B1-hopAW1 | 5′TTCACGTACCTCATCTGC3′ | 104148-104165 | NA |
| B2-hopAW1 | 5′ATCCAACTGCACATCAGC3′ | 106029-106046c | NA |
| A1-hopAF1 | 5′CCCTATAGTGAGTCGGATCCCCGTTGGTTATGTGATGC3′ | 1685066-1685083c | BamHI |
| A2-hopAF1 | 5′GGATCCGACTCACTATAGGGCAGCGCTTCGATTTTGCC3′ | 1686077-1686094 | BamHI |
| B1-hopAF1 | 5′CACTCAAGCCGATCTACC3′ | 1684461-1684478 | NA |
| B2-hopAF1 | 5′GCTTCATCCCCGATATCC3′ | 1686634-1686651c | NA |
| A1-avrE1 | 5′CCCTATAGTGAGTCGAATTCCTGTAGAAATGCGCGAGC3′ | 1477655-1477672c | EcoRI |
| A2-avrE1 | 5′GAATTCGACTCACTATAGGGATGTGCCACTGATGGCAG3′ | 1482889-1482906 | EcoRI |
| B1-avrE1 | 5′GAAGCTCGCGTGTCCGTC3′ | 1477068-1477085 | NA |
| B2-avrE1 | 5′CGATCATTGGCAACAGC3′ | 1483518-1483534c | NA |
| A1-avrRps4 | 5′CCCTATAGTGAGTCGGATCCAACACATCATAGCCCCTG3′ | 75967-75984c | BamHI |
| A2-avrRps4 | 5′GGATCCGACTCACTATAGCCTAATGCGCATGAGCAGGC3′ | 76681-76698 | BamHI |
| B1-avrRps4 | 5′TTTGCCTTCGCCGTACAG3′ | 74923-74940 | NA |
| B2-avrRps4 | 5′CGTCAAGACGACGGTCAG3′ | 77748-77765c | NA |
| A1-hopAY1 | 5′CCCTATAGTGAGTCGAATTCTCTCCGCTTACTGGCTTGC3′ | 110642-110660c | EcoRI |
| A2-hopAY1 | 5′GAATTCGACTCACTATAGGGCACCTGCAATTGGAGAGC3′ | 111601-111618 | EcoRI |
| B1-hopAY1 | 5′CGGTAATAATTGGCATGG3′ | 110198-110215 | NA |
| B2-hopAY1 | 5′TGCGCAACATCAACGAGG3′ | 112117-112134c | NA |
| P1-EcoRI | 5′TCAGAATTCGTGTAGGCTGGAGCTGCTTC3′ | EcoRI | |
| P2-EcoRI | 5′TCAGAATTCCATATGAATATCCTCCTTAG3′ | EcoRI | |
| P1-BamHI | 5′TCAGGATCCGTGTAGGCTGGAGCTGCTTC3′ | BamHI | |
| P2-BamHI | 5′TCAGGATCCCATATGAATATCCTCCTTAG3′ | BamHI | |
| P1-XhoI | 5′TCACTCGAGGTGTAGGCTGGAGCTGCTTC3′ | XhoI | |
| P2-XhoI | 5′TCACTCGAGCATATGAATATCCTCCTTAG3′ | XhoI | |
| qRT-avrB4-1/2 F | 5′GGCGATGTTCAATGGCTAAT3′ | 3515731-3515732; 928198-928217 | |
| qRT-avrB4-1/2 R | 5′TTTTGCAAGCTCCCATCAG3′ | 3515842-3515860c; 928070-928088c | |
| qRT-hopW1-1/2 F | 5′AATCGTCGCAGCAGGTTC3′ | 63361-63378c; 6591-6608c | |
| qRT-hopW1-1/2 R | 5′CTCTCCAACTCATGCTGAAGG3′ | 63246-63266; 6476-6496 | |
| qRT-hopAY1 F | 5′CGCTAGACCCTCGAACAGTC3′ | 110760-110779 | |
| qRT-hopAY1 R | 5′CCCTGACCTGACCCTTGTT3′ | 110937-110955c | |
| qRT-hopAH2 F | 5′TTGCTTGCCAGTCAACAGAC3′ | 3525934-3525953 | |
| qRT-hopAH2 R | 5′GCGATGTCATCAGGATTGG3′ | 3526118-3526136c | |
| qRT-hrpA F | 5′AGGGTATCAACAGCGTCAAG3′ | 1487676-1487695 | |
| qRT-hrpA R | 5′TCAGAACTGGACGACCGAGT3′ | 1487916-1487935c | |
| qRT-16S F | 5′CAATGGGCGAAAGCCTGAT3′ | 804368-804386 | |
| qRT-16S R | 5′TGCTGGCACAGAGTTAGC3′ | 804502-804519c | |
| hopW1-1/2 F | 5′GTCAAGCTTTTATGGAAAGCTACACCGC3′ | 6716-6734c; 63486-63504c | HindIII |
| hopW1-1/2 R | 5′GTCGAATTCCGATTCATTTTGCTGTTGC3′ | 6379-6397; 63149-63167 | EcoRI |
| avrB4-1 F | 5′GTCCTCGAGGAGCGGAACCGAATAAGAGG3′ | 3515319-3515338 | XhoI |
| avrB4-2 F | 5′GTCCTCGAGAGTGCTGAGGACTCGGTAGC3′ | 928612-928631c | XhoI |
| avrB4-1/2 R | 5′GTCTCTAGATTTGTTACGAATTCTGCGGG3′ | 3516364-3516383c; 927547-927566 | XbaI |
NA, not applicable.
To obtain pGEM-T-nptII-EcoRI, pGEM-T-nptII-BamHI, or pGEM-T-nptII-XhoI, a fragment containing the nptII kanamycin resistance gene, flanked by FRT (flippase recognition target) sites, was PCR amplified using the Expand high-fidelity PCR system (Roche, Germany), pKD4 (11) as a template, and primers P1 and P2 (introducing either EcoRI, BamHI, or XhoI sites at both ends of the fragment) (Table 3). The products were A/T cloned into pGEM-T (Promega, Madison, WI) and confirmed by restriction analysis.
To generate each knockout vector, we carried out two PCR amplifications of approximately 500 bp (unless otherwise indicated) of the 5′ and 3′ regions flanking the ORF to be deleted, using Pph1448a genomic DNA as a template, and primers including an EcoRI site (or BamHI or XhoI, when an EcoRI site was present on the flanking sequences) and the T7 primer sequence in such a manner as to provide homology and a cloning site between the fragments (see Fig. 2A). Each reaction was carried out at 94°C for 3 min, followed by 20 cycles at 94°C for 20s, 55°C for 30s, and 72°C for 50s, followed by 7 min at 72°C, and the reaction mixture contained 0.64 mM deoxynucleoside triphosphate (dNTP) mix, 1× buffer 2 (Expand high-fidelity PCR system [Roche]), 5% dimethyl sulfoxide (DMSO), 0.4 μM corresponding primers, 10 ng genomic DNA, and double-distilled water (H2Odd) (Nalgene, Rochester, NY). Five microliters of each gel-purified PCR product was used for a PCR, with eight cycles of polymerization at 94°C for 30 min, 52°C for 1 min, and 72°C for 1 min, finishing with 7 min at 72°C without primers or a template. The product from the previous step was used as a template in a reaction mixture (5 μl per reaction mixture) that also contained 0.64 mM dNTP mix, 1× buffer 2 (Expand high-fidelity PCR system [Roche]), 5% DMSO, 0.4 μM corresponding primers (Table 3), and H2Odd (Nalgene). The mixture was incubated at 94°C for 3 min, followed by 20 cycles at 94°C for 20 s, 53°C for 30 s, and 72°C for 1 min, finishing with 7 min at 72°C. The resulting products, the deletion alleles, were A/T cloned into pGEM-T and fully sequenced to discard mutations on flanking genes. As appropriate, EcoRI, BamHI, or XhoI fragments containing the FRT-flanked kanamycin resistance gene were obtained from pGEM-T-nptII-EcoRI, pGEM-T-nptII-BamHI, or pGEM-T-nptII-XhoI, respectively, and ligated into the EcoRI, BamHI, or XhoI fragments of the corresponding pGEM-T derivatives carrying the deletion alleles. Ligations were transformed into E. coli DH5α (16) and transformants selected directly in LB-kanamycin plates, rendering a collection of effector knockout vectors for Pph1448a (Table 2).
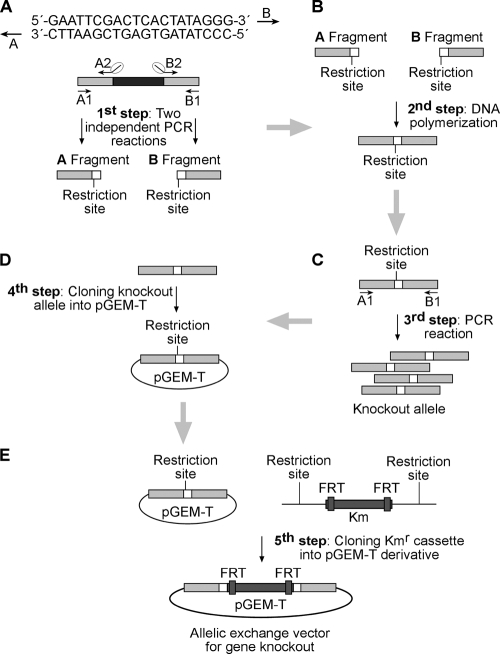
FIG. 2.
Generating allelic exchange vectors for gene knockout. (A) The flanking regions (0.5 kb) of the target gene are amplified independently. Primers A and B share a 20-nucleotide-long homologous sequence at their 5′ ends. (B) With the resulting PCR products used as both the primers and the template, a polymerization is carried out, resulting in a joint 1-kb fragment, the knockout allele. (C) Amplification of the knockout allele by use of primers A1 and B1. (D) The knockout allele is A/T cloned into pGEM-T. (E) With the use of the restriction site incorporated into primers A2 and B2, a FRT-flanked nptII fragment from the appropriate plasmid is cloned into the knockout allele, thus generating the effector knockout vector.
To generate the plasmids used in complementation experiments (Table 2), the full ORFs from avrB4-2 and hopW1-1/2 (sequences from their ORFs are 100% identical) were PCR amplified and cloned into pBBR1MCS4 (to obtain expression from a PlacZ promoter) or pAMEX (to obtain expression from a PnptII promoter). The full avrB4-1 gene, including its promoter region, was cloned into pBBR1MCS4 (to obtain expression from its native promoter) or pAMEX (to obtain expression from a PnptII promoter). The primers used, as well as the restriction sites, are listed in Table 3.
Recombinant DNA techniques were performed in accordance with standard methods (37). Genomic DNA was extracted using a Jet Flex extraction kit (Genomed, Germany), and plasmid DNA was extracted using a FavorPrep plasmid DNA extraction minikit (Favorgen Biotech Corporation, Taiwan). Routine clone analysis was carried out by quick boiling extraction (20). DNA gel purification was performed using a FavorPrep gel/PCR purification minikit (Favorgen Biotech Corporation).
Generation of knockout strains.
Allelic exchange vectors (Table 2) were transformed by electroporation into the Pph1448a strain by use of a modification of an electroporation protocol previously described for P. aeruginosa (8), using SOB medium (16) to grow Pph1448a. Transformants were plated into LB plates supplemented with kanamycin. Replica plates of the resulting colonies were carried out using LB plates supplemented with ampicillin (300 μg/ml) to determine whether each transformant was the result of plasmid integration (a single recombination event) or allelic exchange (a double recombination event). Since ampicillin selection is typically a problem in P. syringae, prospective clones were further tested for growth in liquid LB medium containing 100 μg/ml of ampicillin, with 50 μg/ml of nitrofurantoin added to avoid cross-contamination. Southern blot analysis, using nptII-FRT as a probe (a 1,495-bp fragment amplified with primers P1 and P2 from pKD4), was used to confirm that allelic exchange occurred at a single and correct position within the genome.
Double mutant strains (Table 1) were generated as follows. Plasmid pFLP2, expressing the flipase enzyme, was transformed into the corresponding single mutant strain to promote removal of the nptII gene by flipase-mediated site-specific recombination. Transformants were tested in LB plates with kanamycin to identify clones in which the kanamycin gene had been removed. Kanamycin-sensitive isolates were then grown in LB plates supplemented with 5% sucrose to select those that had lost the pFLP2 vector. A second allelic exchange vector was then transformed into the resulting kanamycin-sensitive single knockout strain, and transformants were selected and analyzed as described above.
Competitive index and disease scoring in bean plants.
CI assays with bean plants (Phaseolus vulgaris cv. Canadian wonder) were carried out as previously described (31). Eight-day-old bean plants, grown at 22°C to 28°C with a photoperiod consisting of a 16-h-light-8-h-dark cycle, were inoculated with 200 μl of a 5 × 104-CFU/ml mixed-bacterial suspension, containing equal numbers of CFU of the wild type and the mutant or gene-expressing strain, using a 2-ml syringe without a needle. Serial dilutions of the inoculum were plated onto LB agar or LB agar with the appropriate antibiotic to confirm the relative proportion of bacterial CFU between the strains, which should be close to 1. At different days postinoculation (dpi), bacteria were recovered from the infected leaves. Bacterial recovery was carried out by taking five 10-mm-diameter discs with a cork borer, which were homogenized by mechanical disruption into 1 ml of 10 mM MgCl2. Bacterial enumeration was performed by serial dilution and plating of the samples onto agar plates with cycloheximide and the appropriate antibiotic to differentiate the strains within the mixed infection. The CI is defined as the mutant-to-wild-type ratio within the output sample divided by the mutant-to-wild-type ratio within the input (inoculum) (13, 42). Competitive indices presented are the means for three replicates showing typical results from three independent experiments. Error bars represent standard errors. Each CI was analyzed using a homoscedastic 2-tailed Student t test and the null hypothesis, which states that the mean index is not significantly different from 1 or from another CI value when specified (P < 0.05).
For scoring of disease symptoms, 8-day-old bean plants were inoculated with bacterial suspensions at either 5 × 106 or 5 × 105 CFU/ml in 10 mM MgCl2, using a 2-ml syringe without a needle. Symptoms were documented at a different dpi.
Virulence and translocation assays with Arabidopsis thaliana.
Seeds of Arabidopsis ecotype Col-0 (RPS2/RPS2), an rps2 mutant derivative (rps2/rps2) (SALK_087581; European Arabidopsis Stock Center), or plants carrying the NahG transgene (12) were germinated and grown in growth chambers with 8-h-light-16-h-dark cycles at 21°C. For symptom visualization, 3- to 4-week old plants were sprayed with a bacterial suspension containing 5 × 107 CFU/ml in 10 mM MgCl2 and 0.02% Silwet L-77. Plants were then covered to keep humidity high, and symptoms were photographed at 7 dpi.
Competitive index assays for measuring growth attenuation, as well as translocation assays, were performed with Arabidopsis as previously described (30, 31). Briefly, 4- to 5-week-old plants were inoculated with a 5 × 104-CFU/ml mixed bacterial suspension, containing equal numbers of CFU of wild-type and effector-expressing strains, using a blunt syringe. Serial dilutions of the inoculum were plated onto LB agar or LB agar with kanamycin to confirm the relative proportion of bacterial CFU between the strains, which should be close to 1. At 4 days postinoculation (dpi), three 10-mm-diameter leaf discs were homogenized by mechanical disruption into 1 ml of 10 mM MgCl2. Bacterial enumeration was carried out as described for CI virulence assays with bean plants. A CI in a translocation assay is defined as the effector-expressing-strain-to-wt output ratio divided by the ratio of these strains within the inoculum. CI translocation indices were treated and presented as described for CI virulence assays.
Quantitative real-time PCR (qRT-PCR) expression analysis.
RNA was isolated from bacterial cell lysates or infected leaves by use of an RNeasy bacterial minikit (Qiagen, Germany) in accordance with the instructions provided by the manufacturer.
For extractions from infected leaves, each leaf was inoculated with 2 × 108 CFU/ml bacterial suspensions, using a blunt syringe. Two discs of 10 mm were cut from inoculation sites at 4 h postinoculation (hpi) and frozen immediately in liquid nitrogen. After macerating in 500 μl 1× RNAprotect reagent (Qiagen, Germany), samples were incubated at room temperature and centrifuged at 5,000 × g for 10 min. Pellets were resuspended in 100 μl Tris-EDTA (TE) containing 100 μg of lysozyme (Sigma) and incubated at room temperature for 10 min. DNase treatment was carried out twice, during and after the extraction process. An aliquot of RNA was tested to ensure that no trace of contaminant genomic DNA was detectable. cDNA was synthesized using the SuperScript II reverse transcriptase system (Invitrogen, CA) with random primers (Promega). After synthesis, cDNA was used as a template for quantitative real time-PCR (qRT-PCR). qRT-PCR was carried out in a RotorGene thermocycler (Qiagen) using Sybr green reagent (Qiagen) in accordance with the instructions from the manufacturer. The primers used for amplifications are detailed in Table 3.
RESULTS
Completing the Pph1448a type III effector inventory.
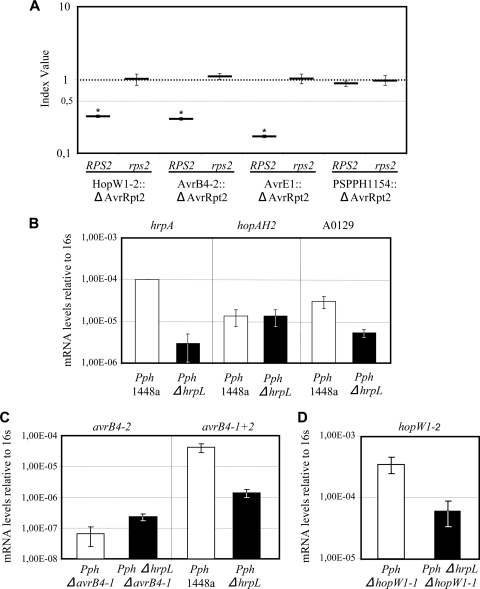
The genome of 1448a contains the genes hopW1-2 and avrB4-2, which are highly similar to those encoding the effectors HopW1-1 and AvrB4-1, respectively (7, 44). Although its HrpL-dependent expression had been established, HopW1-2 was previously discarded as not translocated (7). AvrB4-2 had not previously been tested for either T3SS-mediated translocation or HrpL-dependent expression. Nevertheless, sequence alignments showed that the predicted protein sequences of HopW1-1 and HopW1-2 are identical but that those of AvrB4-1 and AvrB4-2 differ by just 3 amino acids, thus raising the possibility of translocation for these two candidate effectors. We tested translocation of AvrB4-2 and HopW1-2, using the more sensitive AvrRpt2 CI-based translocation assay recently developed in our laboratory (30). With the use of these previously described experimental settings, interference between the HR triggered by a translocated candidate fusion to AvrRpt2 and growth of DC3000 is completely avoided, allowing detection of previously discarded candidate effectors due to the increased sensitivity of the assay (30). We generated plasmids encoding translational fusions of avrB4-2 and hopW1-2 to the sequence encoding the C-terminal domain of AvrRpt2 (AvrRpt281-255), expressed under the control of the PnptII promoter (Table 2). Particular attention was paid to primer design to avoid cross-amplification of avrB4-1 and hopW1-1 by taking advantage of the differences between their flanking regions. PtoDC3000 expressing either AvrB4-2::AvrRpt281-255 or the HopW1-2::AvrRpt281-255 protein fusion was coinoculated with PtoDC3000 into Arabidopsis Col-0 and rps2 plants. Figure 1 A shows that both CIs were significantly different from 1 in the wild type in Col-0 but not rps2 plants. Therefore, both AvrB4-2 and HopW1-2 have functional T3SS translocation signals that allow the fusion proteins to be translocated into the plant cell, thus triggering AvrRpt2-RPS2 defense responses that determine bacterial growth restriction as detected using the CI. Considering the similarities between the pair of effectors comprising HopW1-1 and HopW1-2 and that comprising AvrB4-1 and AvrB4-2, these results raised the possibility of functional redundancy between HopW1-1 and HopW1-2 and provided that AvrB4-2 was expressed within the plant, between AvrB4-1 and AvrB4-2.
FIG. 1.
(A) Graphical representation of competitive index translocation assays. Wild-type DC3000 bacteria were coinoculated into either wild-type Arabidopsis Col-0 or rps2 plants, together with DC3000 strains expressing translational fusions of candidate effector genes to ΔavrRpt2 (AvrB4-2::AvrRpt281-255, HopW1-1::AvrRpt281-255, AvrE11-326::AvrRpt281-255, or PSPPH1154::AvrRpt281-255). Four- to five-week-old Arabidopsis plants were inoculated with a 5 × 104-CFU/ml mixed bacterial suspension, containing equal numbers of CFU of wild-type and fusion-expressing bacteria. Competitive indices correspond to the means for three samples. A dashed line, corresponding to a CI of 0.5, is included for reference. Error bars represent the standard errors. Experiments were repeated twice, with similar results. Asterisks indicate results significantly different from 1, as established by Student's t test (P < 0.05). (B, C, and D) qRT-PCR expression analysis of A0129, hopAH2, and avrB4-2, carried out with Pph1448a and the ΔhrpL mutant, and expression analysis of avrB4-1 and hopW1-2, carried out with the ΔavrB4-1 and ΔavrB4-1 ΔhrpL strains and the ΔhopW1-1 and ΔhopW1-1 ΔhrpL strains, respectively, growing in planta. The expression levels of 16S and hrpA are included as controls.
We also tested the translocation of the effector candidate AvrE1, previously analyzed by Chang and colleagues (7). These authors found AvrE1 to be expressed in an HrpL-dependent manner but were not able to conclude as to its translocation, since it produced inconsistent results in their translocation assays (7). Technical difficulties in the generation of the translational fusion of the complete ORF of avrE1 to AvrRpt281-255 (5,142 bp) led us to use the first 978 bp to generate the protein fusion. Figure 1A shows that the CI of DC3000 expressing AvrE11-978::AvrRpt281-255 in mixed infection with DC3000 is significantly different from 1, thus demonstrating that AvrE1 also carries type III translocation signals.
In order to establish the complete type III secretome of Pph1448a, we also analyzed the translocation of the protein encoded by another gene, PSPPH1154, which raised our interest, as this protein was previously identified in a bioinformatics analysis as containing a eukaryotic F-box domain (4). The F-box is a short domain characteristic of the eukaryotic F-box proteins, which allows them to participate in the forming of SCF-type E3 ubiquitin (Ub) ligase complexes, thus controlling specific protein ubiquitinylation (38, 45). This pathway is essential to many developmental processes in plants, ranging from hormone signaling and flower development to stress responses. Recently, several bacterial effectors from plant pathogenic bacteria have been shown to mimic host E3 Ub ligases, possibly to alter plant defenses (5), thus raising the interesting possibility that PSPPH1154 encodes a type III effector with a similar function. However, we could not establish type III-dependent translocation for this candidate, since the value for growth of DC3000 expressing PSPPH1154::AvrRpt281-255 in mixed infection with DC3000 was not significantly different from 1 (Fig. 1A).
To fully establish these translocated candidates as effectors, we also analyzed in planta expression of AvrB4-2, A0129, and HopAH2, which had not previously been determined. We extracted RNA from bean plants 4 h after infiltration with 2 × 108 CFU/ml of either Pph1448a or a ΔhrpL mutant derivative (Table 1) and carried out qRT-PCR to detect the expression of these genes. Primers for the hrpA gene were included as a control for HrpL-dependent expression, and primers for 16S amplification (Table 3) were used for comparison purposes. As expected, expression of hrpA was induced in planta in an HrpL-dependent manner (Fig. 1B). Expression of A0129 was also found to be dependent on HrpL, although the differences between its expression in plants infected with the wild type and that in plants infected with the ΔhrpL mutant derivative were notably smaller than the differences established for hrpA (Fig. 1B). However, in planta expression of hopAH2 was found to be HrpL independent since expression in the ΔhrpL mutant was comparable to that detected in the wild-type strain (Fig. 1B).
Surprisingly, expression of avrB4-2, which lacks an hrp box (23), was found to be HrpL dependent since its expression was clearly detectable in plants infected with Pph1448a but just barely detectable in those infected with its ΔhrpL derivative (Fig. 1C). Given the strong sequence similarity between the avrB4-1 and avrB4-2 ORFs, we also tested the expression of avrB4-2 in a ΔavrB4-1 background to avoid cross-amplification. Expression of avrB4-2 in this background was found to be HrpL independent (Fig. 1C), in keeping with the absence of an hrp box in its promoter region, thus indicating that the HrpL-dependent expression detected in the wild type was due to cross-amplification of avrB4-1. We also analyzed the expression of hopW1-2, previously reported as HrpL regulated (7), in a ΔhopW1-1 background. Expression of this gene was found to be HrpL dependent, although its expression in the absence of a functional HrpL protein was not strongly reduced (Fig. 1D).
Simplifying the process of generating single and double knockout mutants.
In order to simplify and accelerate the process of generating knockout mutations in Pph1488a, we applied a series of sequential PCRs and a selectable cloning step to generate allelic exchange vectors that could be directly introduced into the target strain to produce the mutants. Each knockout vector is generated by a 5-step procedure shown in Fig. 2. Briefly, approximately 500 bp flanking the target ORF is amplified from Pph1448a genomic DNA, using primers that include a restriction site not present on the flanking sequences and the T7 primer sequence, in such a manner as to provide homology and a cloning site between the resulting fragments (Fig. 2A). These fragments are then joined through the homology provided by the T7 primer sequence in a PCR without additional primers or a template, thus generating the deletion allele (Fig. 2B). Once amplified, the deletion alleles are A/T cloned (Fig. 2C and D) and fully sequenced. Importantly, since effector genes can be carried as part of an operon or located close to other genes, accumulation of PCR-associated mutations must be kept to a minimum. For that purpose, we used a high-fidelity polymerase, DMSO, commercial H2Odd, and very few PCR cycles (see the corresponding section in Materials and Methods for details).
The knockout alleles generated are marked with an nptII kanamycin resistance gene flanked by FTR sites (Fig. 2D), which can be easily eliminated by flipase-mediated site-specific recombination (19). This allows the introduction of a second analogue vector for generation of a double mutant when required.
Allelic exchange is typically carried out by conjugation to increase the chances that the necessary recombination events take place before the nonreplicative vector is lost. However, to save the additional cloning steps required to clone the knockout allele into a conjugative vector, we used a modification of an electroporation protocol previously described for P. aeruginosa (8). In this manner, we achieved transformation efficiencies (108 to 109 CFU/μg DNA) high enough as to allow not only detection of clones having undergone plasmid integration events (single crossover) but also detection of those having undergone allelic exchange events (double crossover) in the transformation plate.
Generating knockout mutants for the Pph1448a type III effector inventory.
We set out to generate a mutant collection including single knockout strains for each of the 27 members of the complete type III effector inventory of Pph1448a. With the exception of avrE1, for which we met with technical problems when amplifying its flanking regions, we generated knockout vectors for all type III effectors and transformed these vectors into Pph1448a. On average, we obtained a frequency of transformants having undergone recombination (Kmr) that ranged between 1 and 102 CFU/μg DNA. Interestingly, when the transformants (Kmr) were tested to identify clones having undergone a double recombination event (Amps) (see Materials and Methods), the frequency of clones having sustained allelic exchange (Kmr Amps) was found to be surprisingly high (5 to 50%). However, if Km-resistant clones with the plasmid integrated (Ampr) were further grown without selective pressure, the frequency of allelic exchange was lower than the frequency of Km-resistant transformants on the initial transformation. It is possible that during transformation, the presence of the integrated plasmid determines a local increase in recombination frequencies.
The recombination frequencies achieved with this method allowed us to recover most mutants, those resulting from a double recombination event leading to allelic exchange, directly from the transformation plate. The mutants that could not be recovered this way could be obtained either after a few rounds of transformation or by doubling the bacterial concentration used in the electroporation. In one case, that of avrB2, allelic exchange was achieved only after the length of the flanking regions was increased to 1,000 bp in order to boost recombination frequencies. Four out of the 27 effector genes, hopAT1, hopAV1, hopAF1, and hopF3, have not yet been knocked out, due to the low frequency of recombination found for their loci (data not shown). A fifth effector gene, avrRps4, was not mutated either, since Southern analyses of all the transformants isolated indicate that recombination was recurrently taking place at an incorrect location within the Pph1448a genome (data not shown). Deletion of these five effector genes is currently being attempted by increasing the sizes of their flanking regions.
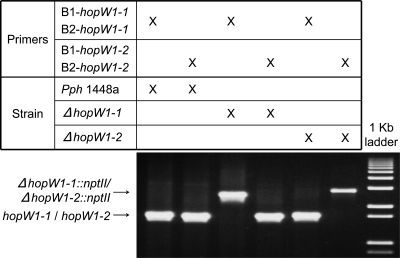
Many type III effectors are encoded within native plasmids. In Pph1448a, this is the case for 12 effectors, including HopW1-1 and HopW1-2 (23). Generation of knockout mutations in these genes poses potential problems since, although native plasmids are maintained at a very low copy number, they can still be present in more than one copy, raising the possibility of having a given strain carrying both the functional and the deletion alleles. To test this possibility, we carried out genotyping experiments using PCR to detect the presence of both types of alleles in all the selected and Southern analysis-confirmed knockout mutants of plasmid-located effector genes. Figure 3 shows the results of genotyping strains AZJ11 (ΔhopW1-1::nptII) and AZJ13 (ΔhopW1-2::nptII). Both strains carry only the knockout alleles and can therefore be used for further virulence analysis. Similar tests carried out with the rest of the mutants did, however, detect one case (ΔhopQ1), in which both alleles were present in the putative mutant strain (data not shown). Two rounds of growth of this strain, merodiploid for the hopQ1 effector gene, in LB medium supplemented with kanamycin were sufficient to obtain the final mutant strain where the wild-type allele was no longer detectable (data not shown). Transformants selected as knockout mutants of the plasmid-located avrB2 gene gave results by Southern analysis consistent with the carrying of the deletion but gave inconclusive results when genotype analyses were carried out, and therefore, these transformants were disregarded.
FIG. 3.
Genotyping analysis carried out with single knockout mutants for plasmid-located effector genes hopW1-1 and hopW1-2. PCR analyses were carried out using primers to amplify each of the two genes plus approximately 600 bp of their 5′ and 3′ flanking regions. Genomic DNA from either Pph1448a or its ΔhopW1-1 and ΔhopW1-2 mutant derivatives was used as templates. Arrows indicate the bands corresponding to the wild-type (hopW1-1/hopW1-2) or mutant (ΔhopW1-1::nptII/ΔhopW1-2::nptII) alleles.
In summary, to date, we have generated single knockout mutants for 20 of the 27 members of the Pph1448a secretome (Table 1).
Establishing functional redundancy within the Pph1448a secretome.
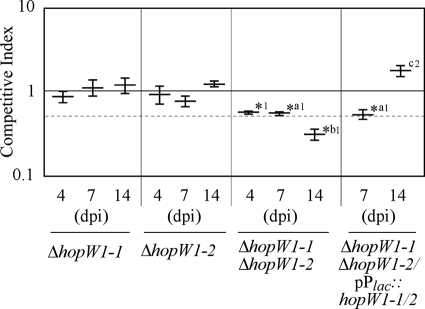
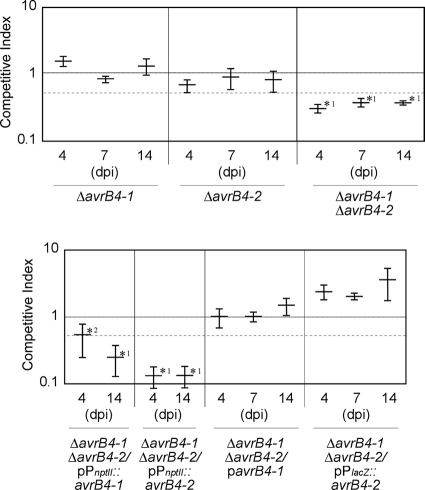
To prove the validity of our approach to the analysis of the role in virulence of the members of the Pph1448a type III secretome, we set out to establish whether the pair of effectors comprising AvrB4-1 and AvrB4-2 and that comprising HopW1-1 and HopW1-2 were functionally redundant. We generated single knockout mutations in the genes encoding all four proteins (Table 1) and analyzed the virulence of each mutant in bean plants on different days postinoculation (dpi), using CI assays carried out as described above, with inoculation doses shown to avoid complementation between coinoculated strains (31). None of the single knockout mutants showed any attenuation in bacterial growth, since the CI values obtained were not significantly different from 1 (Fig. 4 and 5).
FIG. 4.
Competitive index virulence analysis of the ΔhopW1-1 or ΔhopW1-2 single mutant, the ΔhopW1-1 ΔhopW1-2 double mutant, or the ΔhopW1-1 ΔhopW1-2 double mutant carrying a plasmid expressing the hopW1-1/2 ORF from a Plac promoter in mixed infections with Pph1448a. Eight-day-old bean plants were inoculated with 200 μl of a 5 × 104-CFU/ml mixed bacterial suspension, containing equal numbers of CFU of wild-type and mutant bacteria, and leaves were sampled at 4, 7, and 14 days postinoculation (dpi). For virulence assays, the CI is defined as the mutant-to-wt output ratio divided by the mutant-to-wt input ratio. CI values correspond to the means for three samples. A dashed line, corresponding to a CI of 0.5, is included for reference. Error bars represent the standard errors. Asterisks indicate results significantly different from 1 as established by Student's t test (P < 0.05). Mean values marked with the same letter (a, b, or c) indicate results not significantly different from each other, as established by Student's t test (P < 0.05). Mean values marked with 1 indicate results significantly different from the CI for Pph1448a carrying the empty vector (pAMEX), as established by Student's t test (P < 0.05). Mean values marked with 2 indicate a result not significantly different from the CI for Pph1448a carrying the empty vector (pAMEX), as established by a Mann-Whitney test (P < 0.05). A different test was used for this value in accordance with the specific recommendation provided by the statistics software program used.
To generate the corresponding double mutants, the kanamycin resistance gene from the ΔavrB4-2 and ΔhopW1-1 single mutant strains (AZJ16 and AZJ11) (Table 1) was removed by flipase-mediated site-specific recombination on their FRT sites (19), and the resulting strains (AZJ17 and AZJ12) (Table 1) were used as recipients for transformation with the vectors previously used in the generation of the avrB4-1 or hopW1-2 single knockout mutant. We carried out a Southern analysis of the single and double mutant strains, as well as that of the single mutants from which the kanamycin resistance gene had been removed, confirming the positions and number of insertions in each strain (data not shown). The double mutant strains thus obtained, AZJ18 (ΔavrB4-1 ΔavrB4-2) and AZJ14 (ΔhopW1-1 ΔhopW1-2) (Table 1), were analyzed for disease symptom induction and bacterial growth. A slight delay was observed in the induction of disease symptoms by the double mutant strains (Fig. 6); however, these differences were hard to establish confidently and were not always reproducible. However, when virulence in the bean was analyzed using the CI, a statistically significant attenuation was detected for both double mutant strains at all time points analyzed, as indicated by CI values significantly different from 1, with the CI values for ΔavrB4-1 ΔavrB4-2 ranging between 0.31 and 0.37 and those for ΔhopW1-1 ΔhopW1-2 ranging between 0.32 to 0.57 (Fig. 4 and 5). These results demonstrate that these pairs of effectors, that comprising HopW1-1 and HopW1-2 and that comprising AvrB4-1 and AvrB4-2, are functionally redundant and make a quantitative contribution to Pph1448a virulence.
FIG. 6.
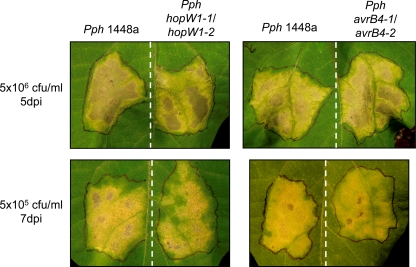
Disease symptoms in bean plants. Plants were infiltrated with the indicated doses of either the Pph1448a, the hopW1-1 hopW1-2 double mutant, or the avrB4-1 avrB4-2 double mutant strain, and the symptoms induced were documented at the indicated days postinoculation (dpi). The experiment was carried out twice, with similar results. The images show representative results.
FIG. 5.
Competitive index virulence analysis of the ΔavrB4-1 or ΔavrB4-2 single mutant, the ΔavrB4-1 ΔavrB4-2 double mutant, or the ΔavrB4-1 ΔavrB4-2 double mutant carrying plasmids expressing either AvrB4-1 or AvrB4-2 from either its own promoter, a Plac promoter, or a PnptII promoter in mixed infections with Pph1448a. Eight-day-old bean plants were inoculated with 200 μl of a 5 × 104-CFU/ml mixed bacterial suspension, containing equal numbers of CFU of wild-type and mutant bacteria, and leaves were sampled at 4, 7, and 14 days postinoculation (dpi). The CI values correspond to the means for three samples. A dashed line, corresponding to a CI of 0.5, is included for reference. Error bars represent the standard errors. Asterisks indicate results significantly different from 1 as established by Student's t test (P < 0.05). Mean values marked with 1 indicate results significantly different from the CI obtained for Pph1448a carrying the empty vector (pAMEX), as established by Student's t test (P < 0.05), whereas those marked with 2 indicate results not significantly different.
Complementing virulence attenuation of the ΔavrB4-1 ΔavrB4-2 and ΔhopW1-1 ΔhopW1-2 double mutant strains.
Given the functional redundancy of the above-mentioned pairs of effectors, the virulence attenuation detected using the CI for each of the double mutants should be complemented by expression of just one of the effector genes. This should be the case in particular for the hopW1-1 hopW1-2 double mutant strain since the nucleotide sequences of these two genes, including their promoter regions, are 100% identical. Indeed, when a plasmid carrying the common ORF region of either hopW1-1 or hopW1-2 (hopW1-1/2), expressed under the control of the medium-to-low-expression-level promoter Plac, was introduced into the ΔhopW1-1 ΔhopW1-2 double mutant strain, growth in bean plants was fully restored to wild-type levels by 14 dpi, as the CI values for the complemented strain coinoculated with the wild type were no different from 1 and were significantly different from the CI obtained for the double mutant strain without the plasmid (Fig. 4). Similarly, plasmids expressing either avrB4-1 from its own promoter or avrB4-2 from the Plac promoter were capable of fully complementing the growth of the ΔavrB4-1 ΔavrB4-2 double mutant strain at all time points analyzed, since the CIs obtained for these strains in mixed infection with the wild type were not significantly different from 1 (Fig. 5). Interestingly, if expression of either avrB4-1 or avrB4-2 was driven from the stronger PnptII promoter, no complementation of growth was detected (Fig. 5). Furthermore, the presence of the plasmid expressing avrB4-2 from the PnptII promoter resulted in a stronger attenuation of growth in the double mutant strain, since the CIs obtained for this strain were significantly smaller that the CI obtained for the double mutant strain (Fig. 5). The stability of the vector used in these assays was confirmed by establishing that the CI for the wild type expressing the empty vector in mixed infection with the wild-type strain (CI = 0.99 ± 0.09) was not significantly different from 1 and was significantly different from all significant CIs shown in Fig. 4 and 5.
Analyzing the defense responses triggered by AvrB4-1, AvrB4-2, HopW1-1, and HopW2-2 in Arabidopsis.
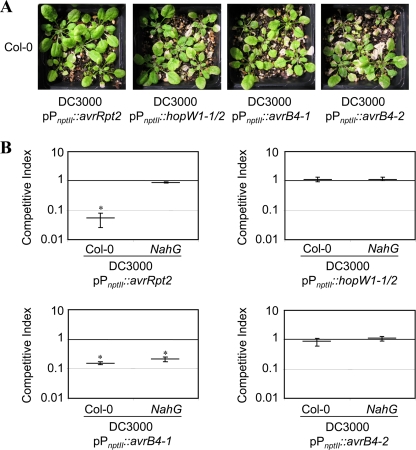
We also analyzed the capability of the AvrB4-1, AvrB4-2, HopW1-1, and HopW2-2 effectors to protect Arabidopsis against infection by PtoDC3000. Plasmids expressing HopW1-1/2, AvrB4-1, and AvrB4-2 from the PnptII promoter were transformed into PtoDC3000, and the resulting transformants were sprayed over Col-0 plants. Symptoms were observed and documented at different time points after inoculation. As controls, Col-0 plants were also sprayed with PtoDC3000 expressing AvrRpt2. The AvrRpt2-RPS2-triggered defense response determines the resistance against PtoDC3000, thus preventing the development of disease symptoms in Col-0 plants inoculated with PtoDC3000 expressing AvrRpt2 (25). Figure 7 A shows that whereas plants sprayed with PtoDC3000 expressing AvrRpt2 displayed no symptoms, plants expressing any of the four Pph effectors showed necrotic leaves. The intensity of the symptoms developed by plants sprayed with PtoDC3000 expressing AvrB4-1 in different experiments seemed lower than that developed by plants sprayed with PtoDC3000 expressing AvrB4-2 or HopW1-1/2 (Fig. 7A and data not shown), suggesting that this effector could be triggering a defense response in Arabidopsis. However, this type of assay does not allow quantification of subtle differences. In order to quantify such a difference, we carried out CI assays to determine the difference in growth of PtoDC3000 that the expression of any of these effectors may have through a putative activation of defense responses. Although expression of HopW1/2 or AvrB4-2 did not affect growth of PtoDC3000 (with CIs not significantly different from 1), expression of AvrB4-1 caused a clear attenuation of growth (with a CI significantly different from 1), suggesting that this effector may trigger a defense response in Arabidopsis (Fig. 7B), although at a lower intensity than that triggered by AvrRpt2. Interestingly, when a similar experiment was carried out with plants carrying the NahG transgene, in which, as expected, the restriction of growth determined by AvrRpt2 in Col-0 was abolished, the CI for PtoDC3000 expressing AvrB4-1 was also significantly different from 1 (Fig. 7B). The NahG transgene encodes a salicylate hydroxylase that converts salicylic acid (SA) to catechol. NahG transgenic plants accumulate extremely low levels of SA, thus hindering AvrRpt2-triggered resistance, and display enhanced susceptibility to bacterial pathogen infection (12). The fact that growth restriction determined by expression of AvrB4-1 is not abolished in NahG plants could indicate that AvrB4-1 may trigger a defense response in Arabidopsis mostly independent from SA-signaling pathways. Although the CI values for PtoDC3000 (avrB4-1) were always slightly closer to 1 in NahG than in Col-0 plants, which could indicate a small contribution of SA-signaling pathways in AvrB4-1-triggered resistance, this strain could not be confidently established, since only in some of the experiments could we detect it in a statistically significant manner (data not shown). Bacterial growth in LB was not affected by the expression of AvrB4-1 (data not shown); however, further work would be necessary to rule out an unspecific toxic effect that could affect bacterial fitness and confirm this putative SA-independent defense response.
FIG. 7.
(A) Disease symptoms caused by PtoDC3000 expressing Pph1448a effectors in Arabidopsis thaliana. Plants were sprayed with a bacterial suspension of PtoDC3000 or Pto expressing different effectors from the Pph1448a secretome, containing 5 × 107 CFU/ml in 10 mM MgCl2 and 0.02% Silwet L-77. After inoculation, plants were covered to keep humidity high, and symptoms were documented by 7 dpi. (B) Competitive index assays of PtoDC3000 bacteria coinoculated into either Arabidopsis Col-0 or NahG plants, together with DC3000 strains expressing different effector genes. Four- to five-week-old Arabidopsis plants were inoculated with a 5 × 104-CFU/ml mixed bacterial suspension, containing equal numbers of CFU of wild-type and effector-expressing bacteria, and leaves were sampled at 7 dpi. Competitive indices correspond to the means for three samples. Error bars represent the standard errors. Asterisks indicate results significantly different from 1, as established by Student's t test (P < 0.05).
DISCUSSION
We applied a highly sensitive CI-AvrRpt2-based translocation assay, recently developed in our laboratory (30), to analyze the translocation of four effector candidates from Pph1448a, two previously untested (AvrB4-2 and PSPPH1154) and two either discarded or found to be unclear for type III-mediated translocation in other assays (HopW1-2 and AvrE1) (7). Three of them were found to be translocated, including HopW1-2, the only previously discarded candidate effector not yet tested with our translocation assay. Expression within the plant was also confirmed for these candidates, which should therefore be included as validated members of the Pph1448a secretome. In planta expression was established for the translocated candidate HopAH2, although this expression was established as taking place in an HrpL-independent manner, in keeping with the absence of an hrp box in its promoter region. Interestingly, although avrB4-2 does not have a consensus hrp box upstream of its promoter sequence, we found that the expression of avrB4-2 was HrpL regulated. Chang and colleagues also found two effector-encoding operons in which HrpL regulation was experimentally demonstrated in the absence of hrp boxes (7). However, the absence of HrpL regulation detected when a ΔavrB4-1 background was used for the analysis indicates that this apparent regulation is the result of cross-amplification of HrpL-regulated avrB4-1. In the case of hopW1-2, our results show a difference in expression between wild-type and ΔhrpL mutant strains, in keeping with previous reports classifying hopW1-2 as HrpL regulated; however, this difference is small. Expression of A0129 was also found to depend on HrpL, although the level of regulation for A0129 was also lower than that measured for the hrpA gene. Our results are in keeping with A0129 being initially reported as fulfilling the three criteria for bioinformatic identification of a putative hrp box (30). Although the correlation between predicted hrp boxes and HrpL regulation is high, there are occasional examples in which genes with hrp boxes do not exhibit HrpL-dependent regulation (7, 44). Regulation by HrpL is a hallmark for most type III effector genes, but there are also exceptions to be found (e.g., hopO1-2PtoDC3000) (7). However, the level of expression required for an effector to be functional would largely depend on its function and will most likely be below the level required for the function of HrpA as a major component of the secretion pilus. Thus, this level of regulation is likely to be biologically meaningful, particularly if these effectors require a high level of expression to carry out the function of HrpA. Alternatively, these effectors may be expressed to functional levels in the absence of HrpL and respond to the presence of the activator by modulating its expression.
A0129 has a predicted catalytic triad and functional domain characteristic of cysteine proteases (http://www.sbg.bio.ic.ac.uk/∼phyre/) (24), which makes it a putative member of the C58 family of papain-like proteases (41), according to the MEROPS database for peptidases (http://merops.sanger.ac.uk/) (35). Another well-characterized member of this family, HopAR1 (previously AvrPphB and AvrPph3), defines race 3 of Pph (22, 34). However, sequence similarity between HopAR1 and the predicted protein sequence of A0129 is very limited (Fig. 8). Thus, by following the guidelines for a unified nomenclature proposed by Lindeberg and colleagues (29), we have renamed A0129 as HopAY1Pph1448a.
FIG. 8.
Amino acid alignment of A0129 and HopAR1 (Pph race 3). Squares represent residues identical between the two proteins. Arrows point to the residues that form the putative catalytic triad (cysteine, histidine, and aspartic acid).
We have developed and applied a simplified method for generating knockout mutants to the task of deleting each of the 27 members of the complete type III effector inventory of Pph1448a. We have successfully generated an effector knockout vector collection and an effector mutant collection, which already include 20 single effector mutant strains and their corresponding vectors. To our knowledge, this is the first collection of single effector mutants available for such a large type III secretome in P. syringae and represents a valuable tool for future work on the role of the type III effector in virulence. We are currently applying CI assays to this mutant collection to characterize the individual contributions of the members of the Pph1448a type III secretome to virulence.
The mutant and knockout vector collections generated in this work present the additional advantage of allowing for easy generation of any double mutant combination. We have validated this use and its application to determining functional relationships between effectors by establishing functional redundancy between two pairs of effectors, with one pair comprising AvrB4-1 and AvrB4-2 and one comprising HopW1-1 and HopW1-2, through analysis of single and double mutant strains, as well as double mutant strains expressing each of the individual effectors, using CI assays.
One interesting result, obtained while the double mutant strains were being characterized, was that the complementation of their growth defect by plasmid-based expression of one of the deleted effectors strongly depended on the promoter driving its expression. These results suggest that strong expression of some effectors, such as AvrB4-2, may result in growth attenuation within the plant. Considering the interplay between defense activation and suppression already shown to take place among effectors of a given secretome (21), increasing the expression of one of these effectors may affect bacterial growth by either favoring defense activation versus defense suppression or altering the hierarchy of translocation. In support of the relevance of such a fine equilibrium between the effectors acting within a given secretome, the best complementation results were obtained when the native promoter was used to express AvrB4-1.
The contribution of the effectors HopW1-1 and HopW1-2 to virulence has interesting implications for effector evolution since these two effectors are both 94 amino acids long, whereas their closest homologues, HopW1-1 and HopW1-2 from Pseudomonas syringae pv. maculicola ES4326, are 774 and 240 amino acids long, respectively (15, 29). Since HopW1-1PmaES4326 and HopW1-2PmaES4326 are both functional effectors, HopW1-1Pph1448a andHopW1-2Pph1448a could have easily been dismissed as inactive truncated versions, when in fact they do contribute to Pph1448a virulence.
The presence of two functionally redundant pairs of genes expressing T3SS effectors in the genome of 1448a illustrates a common evolutionary strategy for generating diversity based on gene duplication and divergent evolution. This can be an essential step both in the generation of T3SS effector families and in the adaptation of the pathogen to new hosts. In this regard, the fact that the expression of AvrB4-1 but not that of AvrB4-2 determines a reduction of bacterial growth in Arabidopsis, despite their being functionally redundant in bean plants and their differing by only 3 amino acids, supports such an evolutionary strategy.
Most effector-triggered immunities depend on SA-signaling pathways and are therefore abolished in plants expressing the NahG transgene, where the concentration of SA is kept to very low levels (12). Thus, the analysis of the plant response to AvrB4-1 carries considerable interest and could lead to a better understanding of the plant responses that shape the P. syringae-plant interaction.
Acknowledgments
We are very grateful to T. Duarte for technical assistance and E. R. Bejarano and J. Ruiz-Albert for their helpful suggestions.
The work was supported by project grants BIO2006-00673 (from the Ministerio de Educación y Ciencia and BIO2009-11516 (from the Ministerio de Ciencia e Innovación [Spain]) to C. R. Beuzón and P06-CVI-2088 (from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía) to E. R. Bejarano. The work was cofunded by Fondos Europeos de Desarrollo Regional (FEDER).
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., A. O. Charkowski, W. L. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. U. S. A. 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angot, A., N. Peeters, E. Lechner, F. Vailleau, C. Baud, L. Gentzbittel, E. Sartorel, P. Genschik, C. Boucher, and S. Genin. 2006. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U. S. A. 103:14620-14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angot, A., A. Vergunst, S. Genin, and N. Peeters. 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badel, J. L., R. Shimizu, H. S. Oh, and A. Collmer. 2006. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19:99-111. [DOI] [PubMed] [Google Scholar]
- 7.Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu, S. Gombar, S. R. Grant, F. M. Ausubel, and J. L. Dangl. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. U. S. A. 102:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 9.Cohn, J. R., and G. B. Martin. 2005. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44:139-154. [DOI] [PubMed] [Google Scholar]
- 10.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney, T. P., S. Uknes, B. Vernooij, L. Friedrich, K. Weymann, D. Negrotto, T. Gaffney, M. Gut-Rella, H. Kessmann, E. Ward, and J. Ryals. 1994. A central role of salicylic acid in plant disease resistance. Science 266:1247-1250. [DOI] [PubMed] [Google Scholar]
- 13.Freter, R., B. Allweiss, P. C. O'Brien, S. A. Halstead, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect. Immun. 34:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant, S. R., E. J. Fisher, J. H. Chang, B. M. Mole, and J. L. Dangl. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60:425-449. [DOI] [PubMed] [Google Scholar]
- 15.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies of transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hauck, P., R. Thilmony, and S. Y. He. 2003. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. U. S. A. 100:8577-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, P., L. Shan, N. C. Lin, G. B. Martin, B. Kemmerling, T. Nurnberger, and J. Sheen. 2006. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125:563-575. [DOI] [PubMed] [Google Scholar]
- 19.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma, D. L. Arnold, M. J. Gibbon, J. Murillo, J. D. Taylor, and A. Vivian. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. U. S. A. 96:10875-10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenner, C., E. Hitchin, J. Mansfield, K. Walters, P. Betteridge, D. Teverson, and J. Taylor. 1991. Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol. Plant Microbe Interact. 4:553-562. [PubMed] [Google Scholar]
- 23.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. Deboy, A. S. Durkin, M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel, B. N., A. F. Bent, D. Dahlbeck, R. W. Innes, and B. J. Staskawicz. 1993. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5:865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kvitko, B. H., D. H. Park, A. C. Velasquez, C. F. Wei, A. B. Russell, G. B. Martin, D. J. Schneider, and A. Collmer. 2009. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X., H. Lin, W. Zhang, Y. Zou, J. Zhang, X. Tang, and J. M. Zhou. 2005. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. U. S. A. 102:12990-12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, N. C., and G. B. Martin. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant Microbe Interact. 18:43-51. [DOI] [PubMed] [Google Scholar]
- 29.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 30.Macho, A. P., J. Ruiz-Albert, P. Tornero, and C. R. Beuzón. 2009. Identification of new type III effectors and analysis of the plant response by competitive index. Mol. Plant Pathol. 10:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macho, A. P., A. Zumaquero, I. Ortiz-Martín, and C. R. Beuzón. 2007. Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol. Plant Pathol. 8:437-450. [DOI] [PubMed] [Google Scholar]
- 32.Mudgett, M. B., O. Chesnokova, D. Dahlbeck, E. T. Clark, O. Rossier, U. Bonas, and B. J. Staskawicz. 2000. Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. U. S. A. 97:13324-13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz-Martin, I., A. P. Macho, L. Lambersten, C. Ramos, and C. R. Beuzon. 2006. Suicide vectors for antibiotic marker exchange and rapid generation of multiple knockout mutants by allelic exchange in Gram-negative bacteria. J. Microbiol. Methods 67:395-407. [DOI] [PubMed] [Google Scholar]
- 34.Puri, N., C. Jenner, M. Bennett, R. Stewart, J. Mansfield, N. Lyons, and J. Taylor. 1997. Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol. Plant Microbe Interact. 10:247-256. [DOI] [PubMed] [Google Scholar]
- 35.Rawlings, N. D., and A. J. Barrett. 2000. MEROPS: the peptidase database. Nucleic Acids Res. 28:323-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronald, P. C., J. M. Salmeron, F. M. Carland, and B. J. Staskawicz. 1992. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174:1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 39.Shan, L., P. He, J. Li, A. Heese, S. C. Peck, T. Nurnberger, G. B. Martin, and J. Sheen. 2008. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan, L., P. He, J. M. Zhou, and X. Tang. 2000. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant Microbe Interact. 13:592-598. [DOI] [PubMed] [Google Scholar]
- 41.Shindo, T., and R. A. Van der Hoorn. 2008. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 9:119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teverson, D. M. 1991. Genetics of pathogenicity and resistance in the halo-blight disease of beans in Africa. Ph.D. thesis. University of Birmingham, Birmingham, United Kingdom.
- 44.Vencato, M., F. Tian, J. R. Alfano, C. R. Buell, S. Cartinhour, G. A. DeClerck, D. S. Guttman, J. Stavrinides, V. Joardar, M. Lindeberg, P. A. Bronstein, J. W. Mansfield, C. R. Myers, A. Collmer, and D. J. Schneider. 2006. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant Microbe Interact. 19:1193-1206. [DOI] [PubMed] [Google Scholar]
- 45.Vierstra, R. D. 2003. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8:135-142. [DOI] [PubMed] [Google Scholar]
- 46.Xiang, T., N. Zong, Y. Zou, Y. Wu, J. Zhang, W. Xing, Y. Li, X. Tang, L. Zhu, J. Chai, and J. M. Zhou. 2008. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18:74-80. [DOI] [PubMed] [Google Scholar]
- 47.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]