Abstract
Topoisomerases form a covalent enzyme-DNA intermediate after initial DNA cleavage. Trapping of the cleavage complex formed by type IIA topoisomerases initiates the bactericidal action of fluoroquinolones. It should be possible also to identify novel antibacterial lead compounds that act with a similar mechanism on type IA bacterial topoisomerases. The cellular response and repair pathways for trapped topoisomerase complexes remain to be fully elucidated. The RuvAB and RecG proteins could play a role in the conversion of the initial protein-DNA complex to double-strand breaks and also in the resolution of the Holliday junction during homologous recombination. Escherichia coli strains with ruvA and recG mutations are found to have increased sensitivity to low levels of norfloxacin treatment, but the mutations had more pronounced effects on survival following the accumulation of covalent complexes formed by mutant topoisomerase I defective in DNA religation. Covalent topoisomerase I and DNA gyrase complexes are converted into double-strand breaks for SOS induction by the RecBCD pathway. SOS induction following topoisomerase I complex accumulation is significantly lower in the ruvA and recG mutants than in the wild-type background, suggesting that RuvAB and RecG may play a role in converting the initial single-strand DNA-protein cleavage complex into a double-strand break prior to repair by homologous recombination. The use of a ruvB mutant proficient in homologous recombination but not in replication fork reversal demonstrated that the replication fork reversal function of RuvAB is required for SOS induction by the covalent complex formed by topoisomerase I.
DNA topoisomerases can modulate DNA superhelicity and help overcome topological barriers in cellular processes by cleaving the DNA backbone phosphodiester linkage to allow topological changes in DNA substrates. The ends of the cleaved DNA are covalently linked to an active-site tyrosine on the topoisomerase proteins in cleavage complex intermediates. Covalent protein-DNA complexes exist only transiently during catalysis because the cleaved DNA is rapidly religated. The stabilization of covalent complexes formed by human topoisomerase I or II due to the action of certain anticancer drugs results in the apoptotic death of cancer cells. Quinolone antibiotics are highly bactericidal because they cause the accumulation of covalent complexes formed by bacterial DNA gyrase and topoisomerase IV enzymes. Although a similar topoisomerase poison inhibitor remains to be identified for bacterial type IA topoisomerases, bacterial topoisomerase I complex accumulation due to mutations that inhibit DNA religation has also been shown to cause rapid bacterial cell death (4, 36). The requirement of a DNA cleavage step in the mechanism of action of topoisomerases increases the vulnerability of cells to conditions that would trap the covalent protein-DNA complex. These conditions include the presence of DNA intercalators, toxic metabolites, and DNA lesions, as well as protein thiolation (9, 28-31, 38). Response to and repair of the trapped covalent topoisomerase-DNA complex are thus needed for cell survival. In eukaryotes, 3′-tyrosyl DNA phosphodiesterase (TDP1) and 5′-tyrosyl DNA phosphodiesterase (TDP2), which can cleave the covalent linkage between topoisomerases and DNA, have been identified (8, 15, 27). Tyrosyl DNA phosphodiesterases have not been identified in bacteria. Repair of covalent bacterial topoisomerase-DNA complexes may require the action of endonucleases to remove the DNA-bound topoisomerase proteins, similar to the Rad1-Rad10 repair pathway characterized in yeast (37). In Escherichia coli, covalent topoisomerase I and DNA gyrase complexes have been shown to be processed into double-strand DNA breaks (DSB), which are then repaired via the RecBCD-mediated RecA homologous recombination pathway with induction of the SOS regulon (24, 34). The RuvABC and RecG activities could play significant roles in the response to the covalent topoisomerase complexes. They are both capable of resolving the Holliday junctions following DSB formation in the later stages of homologous recombination repair (11). SbcCD has been shown previously to remove protein from a protein-bound DNA end with nucleolytic activity to create a DSB (7). In addition, it is also possible that RuvAB and RecG might act at arrested forks to process replication forks blocked by the covalently bound topoisomerase proteins and generate DSB substrates for RecBCD (1, 32). Previous studies have not clearly elucidated the roles of RuvABC and RecG in the response to covalent topoisomerase complexes. We examine here the effects of mutations in the ruvA and recG genes on both bacterial survival and SOS induction following the accumulation of covalent topoisomerase I or gyrase complexes with cleaved DNA.
MATERIALS AND METHODS
E. coli strains and growth media.
The E. coli strains and plasmids used in this study are listed in Table 1. E. coli cells were grown in Luria-Bertani (LB) broth and, when appropriate, with an antibiotic (ampicillin at 100 μg/ml, chloramphenicol at 20 μg/ml, kanamycin at 50 μg/ml, or spectinomycin at 60 μg/ml) at 37°C for cell viability and luciferase assays. Mueller-Hinton broth (MHB) was used for culture dilutions for MIC tests. Strain BW27784 was used to measure cell death upon the induction of mutant bacterial topoisomerase I enzymes deficient in DNA rejoining. The mutant topoisomerases included a mutant Yersinia pestis topoisomerase I expressed from plasmid pAYTOP-G122S (34) and a mutant E. coli topoisomerase I expressed from plasmid pETOP-G116S (4). In these plasmids, the expression of the mutant topoisomerase is under the control of the BAD promoter. The use of BW27784 and its derivatives allows uniform controlled induction of expression from the BAD promoter dependent on increasing concentrations of arabinose (18). Plasmids pAYTOP and pETOP were used to express wild-type recombinant Y. pestis and E. coli topoisomerase I proteins, respectively. For strain maintenance and overnight culture of transformed strains, 2% glucose was included in the LB medium to suppress the potentially lethal expression of the recombinant mutant topoisomerase proteins. The relevant E. coli gene mutations being studied were introduced into BW27784 by P1-mediated generalized transduction according to standard procedures and selected by the associated antibiotic resistance markers. The presence of the mutations in the resulting strains was confirmed by PCR.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or description | Source, reference, or construction |
|---|---|---|
| Strains | ||
| BW27784 | Δ(araBAD)567 Δ(rhaBAD)568 | Yale E. coli Genetic Stock Center; 18 |
| Δ(araFGH) Φ(ΔaraEpPCP18-araE) | ||
| JW1850-2 | ruvA786(del)::kan; Keio collection | Yale E. coli Genetic Stock Center; 2 |
| JW3627-1 | recG756(del)::kan; Keio collection | Yale E. coli Genetic Stock Center; 2 |
| JW1852-1 | ruvC789(del)::kan; Keio collection | Yale E. coli Genetic Stock Center; 2 |
| JHS4 | BW27784ruvA786(del)::kan | P1(JW1850-2) × BW27784, Kanr |
| JHS5 | BW27784 recG756(del)::kan | P1(JW3627-1) × BW27784, Kanr |
| JHS7 | BW27784ruvC789(del)::kan | P1(JW1852-1) × BW27784, Kanr |
| JJC671 | AB1157ruvA60::Tn10 (ruvB polar) [pGB2-RuvAB] | Bénédicte Michel; 23 |
| JHS6 | BW27784 ruvA60::Tn10 (ruvB polar) | P1(JJC671) × BW27784, Tetr |
| Plasmids | ||
| pETOP | With E. coli topA under control of BAD promoter, high copy number | 4 |
| pETOP-G116S | pETOP with G116S mutation | Site-directed mutagenesis of pETOP |
| pAYTOP | With Y. pestis topA under control of BAD promoter, medium copy number | 5 |
| pAYTOP-G122S | pAYTOP with G122S mutation | 34 |
| pDinlux | SOS reporter plasmid with dinD1::luxCADBE fusion | 5 |
| pGB2 | Cloning vector | Bénédicte Michel; 6 |
| pGB2-RuvAB | Contains entire ruvAB operon | Bénédicte Michel; 32 |
| pGB2-RuvA+ RuvB-Y184H | RuvAB proficient in recombination but not RFR | Bénédicte Michel; 20 |
Determination of cell killing by mutant topoisomerase I.
Overnight cultures of transformants of mutant topoisomerase clones grown in LB medium with 2% glucose plus antibiotics were diluted (1:100) in LB medium with antibiotics and incubated in a shaker at 37°C. When cells reached exponential phase (A600 = 0.4), the cultures were divided into noninduced (no arabinose) and induced (0.00002 to 0.0002% arabinose) samples. After an additional 2 h at 37°C with shaking, serial dilutions of the cultures were carried out with LB medium and the diluted cultures were plated on LB medium plates containing appropriate antibiotics and 2% glucose. The viable counts were recorded after overnight incubation at 37°C. For Western blot analysis of mutant topoisomerase I expression, cells were collected at 2 h after arabinose addition and lysed by boiling in sodium dodecyl sulfate (SDS) gel sample buffer for 5 min. The lysates were electrophoresed in a 10% SDS gel before transfer and blotting with mouse monoclonal antibodies against E. coli topoisomerase I.
Determination of cell viability following norfloxacin treatment.
Exponential-phase cultures in LB medium (A600 = 0.4) were treated with norfloxacin (50 to 200 ng/ml) and incubated with shaking at 37°C for 2 h. Cultures were diluted and plated on LB medium plates, and the ratio of the number of surviving colonies from the treated culture to the number of surviving colonies from the untreated control culture was calculated.
Fluoroquinolone susceptibility.
The broth macrodilution method was used to measure MICs for BW27784 and its mutant derivatives treated with norfloxacin. The inoculum was added to a 0.85% saline solution and adjusted to equal the turbidity of a McFarland 0.5 standard using a spectrophotometer. The inoculum was then further diluted in MHB (1:200). One-milliliter volumes of the diluted inoculum were added to tubes with 1-ml volumes of MHB containing 2-fold serial dilutions of norfloxacin (0.015 to 32 μg/ml). The tubes were incubated overnight at 37°C, and the presence or absence of growth was recorded. The MIC was determined as the lowest concentration of antibiotic that completely inhibited bacterial growth.
Ciprofloxacin Etests were done with wild-type and mutant E. coli strains on MHB plates. The inoculum was adjusted in saline as described above and then swabbed onto plates to form a bacterial lawn. Etest strips (AB BIODISK) were applied, and the plates were incubated at 37°C for 18 h before reading of the MIC according to the directions outlined by the manufacturer.
Luciferase assay to measure SOS induction.
To measure SOS induction from topoisomerase I cleavage complex accumulation, overnight cultures of strains transformed with pAYTOP-G122S and pDinlux (5) were diluted in LB medium containing appropriate antibiotics. Cultures were grown to exponential phase and then distributed into a 96-well Microlite 1 plate (Thermo Scientific). Arabinose (0.00001 to 0.0001%) was added to cultures to induce mutant topoisomerase I expression from plasmid pAYTOP-G122S. SOS induction is measured as an increase in the luciferase signal from the plasmid pDinlux following arabinose addition and normalized to the luciferase signal from a control culture not treated with arabinose. The pDinlux plasmid contains the gene fusion luxCADBE from Vibrio fischeri under the control of the SOS-inducible dinD1 promoter. Luminescence was measured at 37°C for 35 cycles at 10 min per cycle with 30 s of shaking before and during the measurements. The Perkin-Elmer 7000 BioAssay Reader was used.
To measure SOS induction following topoisomerase I cleavage complex accumulation, luminescence measurements were conducted with strains carrying pDinlux after treatment with norfloxacin (15 to 200 ng/ml).
RESULTS
Effects of null mutations of recA and recG on viability following quinolone treatment or induction of topoisomerase I cleavage complexes.
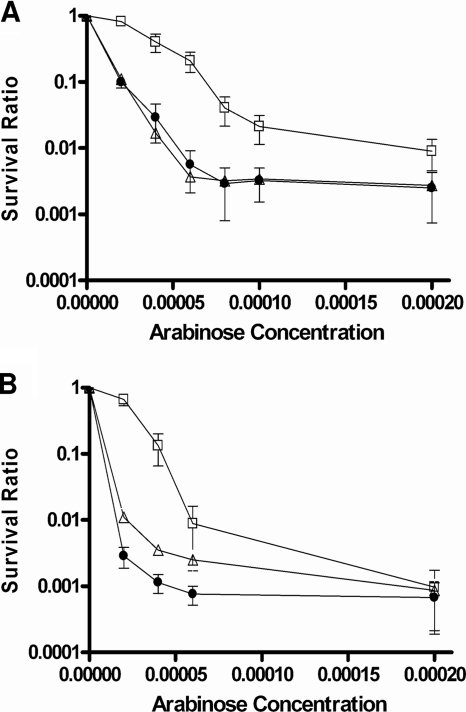
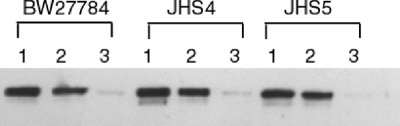
Null mutations in the ruvA and recG genes present in the Keio collection of E. coli mutants (2) were transduced into E. coli strain BW27784 to examine the effects of the mutations on viability upon the induction of mutant topoisomerase I cleavage complexes with arabinose. In strain BW27784, the expression level of induced recombinant topoisomerase proteins can be correlated with the arabinose concentration (18). The results (Fig. 1 A) showed that hypersensitivity to induction of mutant recombinant Y. pestis topoisomerase I YTOP-G122S from plasmid pAYTOP-G122S could be observed in both the ruvA and recG mutants (up to 57-fold for ruvA mutant JHS4 and up to 37-fold for recG mutant JHS5 compared to BW27784 at 0.00006% arabinose). A similar mutant E. coli topoisomerase I, ETOP-G116S, was expressed under the control of the BAD promoter in plasmid pETOP-G116S present at a higher copy number than pAYTOP-G122S. A greater effect on viability was observed after induction of the BAD promoter with arabinose (Fig. 1B), also with hypersensitivity for the ruvA and recG mutants (up to 60-fold for ruvA and up to 230-fold for recG compared to the wild type at 0.00002% arabinose). This hypersensitivity was not due to an increased level of mutant topoisomerase protein being induced in the ruvA and recG mutant strains, as demonstrated by Western blotting with antibodies against E. coli topoisomerase I (Fig. 2). ETOP-G116S accumulated at lower levels than wild-type ETOP in BW27784, but the levels were not further affected by the ruvA or recG mutation.
FIG. 1.
Effects of ruvA and recG mutations on relative cell viability following induction of mutant topoisomerase I with arabinose. YTOP-G122S (A) and ETOP-G116S (B), encoded by plasmids pAYTOP-G122S and pETOP-G116S, respectively, were induced in strains BW27784 (squares), JHS4 (triangles), and JHS5 (circles) with the indicated concentrations of arabinose. Viable counts at 2 h after the addition of arabinose were normalized against the viable counts of uninduced cultures to obtain survival ratios. The results represent the average and standard deviation of at least three experiments.
FIG. 2.
Expression of wild-type and mutant recombinant topoisomerase I in the ruvA and recG mutant backgrounds following arabinose induction. Shown is a Western blot analysis of total proteins of BW27784, JHS4, and JHS5 with pETOP induced for 2 h with 0.00004% arabinose (lane 1), pETOP-G116S induced with 0.00004% arabinose (lane 2), or pETOP-G116S with no arabinose present (lane 3), using monoclonal antibodies against E. coli topoisomerase I.
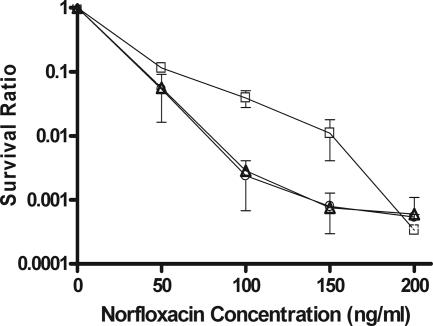
The hypersensitivity of the ruvA and recG mutants to type II topoisomerase cleavage complex accumulation was also seen, but to a lesser degree, when viability was compared after treatment with different concentrations of norfloxacin (Fig. 3). With norfloxacin at 100 and 150 ng/ml, the ruvA and recG mutations resulted in ∼14- to 16-fold lower viability. With norfloxacin at 200 ng/ml, there was no difference in the survival ratio normalized to the viable counts in untreated cultures.
FIG. 3.
Effects of ruvA and recG null mutations on cell survival after norfloxacin treatment. Viable counts of BW27784 (squares), JHS4 (triangles), and JHS5 (circles) were measured at 2 h after the addition of norfloxacin and normalized against the viable counts of untreated cultures to obtain survival ratios. The results represent the average and standard deviation of at least three experiments.
Because the effects of the ruvA and recG mutations on viability following norfloxacin treatment were relatively modest, additional experiments were conducted to evaluate the effect of the ruvA and recG mutations on growth inhibition by quinolones. MICs of norfloxacin and ciprofloxacin were measured by the broth macrodilution method, as well as with Etest strips. The results (Table 2) showed that MICs were 2- to 4-fold lower in the ruvA and recG mutants.
TABLE 2.
Effects of ruvA and recG mutations on fluoroquinolone MICs
| Strain | MICa (ng/ml) |
|
|---|---|---|
| Ciprofloxacin | Norfloxacin | |
| BW27784 | 12 | 60 |
| JHS4 | 3 | 30 |
| JHS5 | 6 | 30 |
Ciprofloxacin MICs were determined with Etest strips. Identical results were obtained in two independent experiments. Norfloxacin MICs were measured by the broth macrodilution method. The results shown were identical in three experiments.
RuvA and RecG are required for SOS induction following topoisomerase I cleavage complex accumulation but not for SOS induction by quinolones.
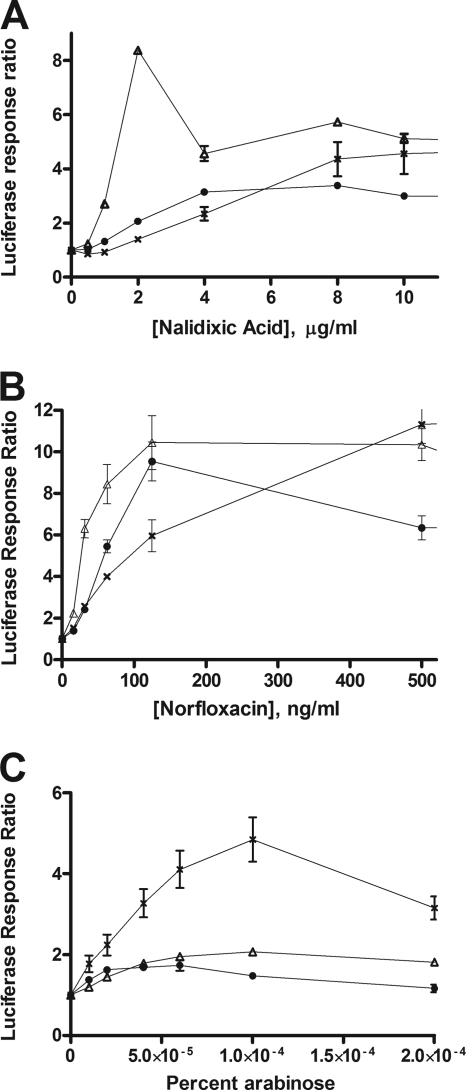
Covalent topoisomerase I and DNA gyrase complexes are converted into double-stranded DNA breaks for RecA loading via the RecBCD pathway (24, 34), inducing the SOS regulon. The luciferase activity from transcription of the dinD1::luxCADBE fusion was used here to monitor SOS induction at multiple time points at different levels of topoisomerase cleavage complex accumulation. In a previous study, the effects of the ruvA and recG mutations on SOS induction were measured with nalidixic acid treatment at a single concentration of 10 μg/ml with a β-galactosidase assay for the transcription of dinD::lacZ (26). Induction of lacZ was evident for the ruvA and recG mutants that were studied but at a lower level than in the wild-type background and with a large variation in the induced β-galactosidase units for the recG mutant (26). This previous result is consistent with our results obtained here with multiple concentrations of nalidixic acid and norfloxacin (Fig. 4 A and B). The ruvA and recG mutations resulted in small increases in dinD promoter activity in the absence of any treatment (data not shown), as reported in the previous study. Accumulation of gyrase cleavage complexes was initiated by the addition of quinolones. Induction of the SOS response could clearly be seen in the ruvA and recG mutant strains. The degree of induction relative to that in the wild type varied, depending on the concentration of the quinolone drug used.
FIG. 4.
SOS induction in ruvA and recG null mutants following treatment with quinolones or topoisomerase I cleavage complex accumulation. The luciferase signal at 250 min after the addition of nalidixic acid (A), norfloxacin (B), or arabinose (C) to induce mutant topoisomerase from pAYTOP-G122S was divided by the luciferase signal from an untreated culture to obtain luciferase response ratios. The results represent the average and standard deviation of at least three experiments. Symbols: ×, BW27784; triangles, JHS4; circles, JHS5.
In contrast, a null mutation in either the ruvA or the recG gene consistently diminished the SOS induction following mutant topoisomerase I cleavage complex accumulation initiated by the addition of arabinose (Fig. 4C). At concentrations of arabinose not greater than 0.00004%, the effect of the mutant YTOP-G122S protein on viability was relatively modest (Fig. 1A). The survival ratios of JHS4 and JHS5 at these low arabinose concentrations were not lower than that of BW27784 at 0.0001% arabinose. In BW27784, robust luciferase activity could still be observed at 0.0001% arabinose. Therefore, the absence of a luciferase signal in the ruvA and recG mutant strains at all of the arabinose concentrations tested was unlikely to be due to lack of ATP from loss of viability.
Effect of the ruvC mutation on cell viability and SOS induction following mutant topoisomerase I complex accumulation.
We hypothesize that the replication fork reversal (RFR) function of RuvAB and RecG is responsible for processing of topoisomerase I cleavage complexes to generate the DSB substrate for RecBCD. This model predicts that a null mutation in ruvC would not eliminate the SOS response following mutant topoisomerase I complex accumulation. However, because the RuvABC complex is required for homologous recombination repair following RecA loading, the ruvC mutation would lead to a decrease in cell viability. These predictions were tested by the construction of strain JHS7 carrying the deletion mutation in ruvC. The results obtained with JHS7 transformed with pAYTOP-G122S following arabinose induction of mutant topoisomerase I were in agreement with the model (Fig. 5). The ruvC mutation decreased cell viability significantly (Fig. 5A). SOS induction following mutant topoisomerase I cleavage complex accumulation in the ruvC mutant was similar to that in wild-type BW27784 at low arabinose concentrations (Fig. 5B). At high arabinose concentrations, the luciferase response ratio began to decrease in wild-type BW27784, and more rapidly in JHS7 with the ruvC mutation, due to the loss of viability. The SOS response was not seen when wild-type topoisomerase I was induced from pAYTOP in JHS7 (Fig. 5B).
FIG. 5.
Effect of ruvC mutation on viability and SOS response following topoisomerase I cleavage complex accumulation. Following induction of wild-type or mutant topoisomerase, relative viability (A) or SOS induction, as indicated by luciferase response ratio, at 250 min (B) was determined. Symbols: ×, BW27784 with pAYTOP-G122S; circles, JHS7 with wild-type pAYTOP; squares, JHS7 with pAYTOP-G122S.
The RFR function of RuvAB is required for processing of the topoisomerase I cleavage complex into a DSB.
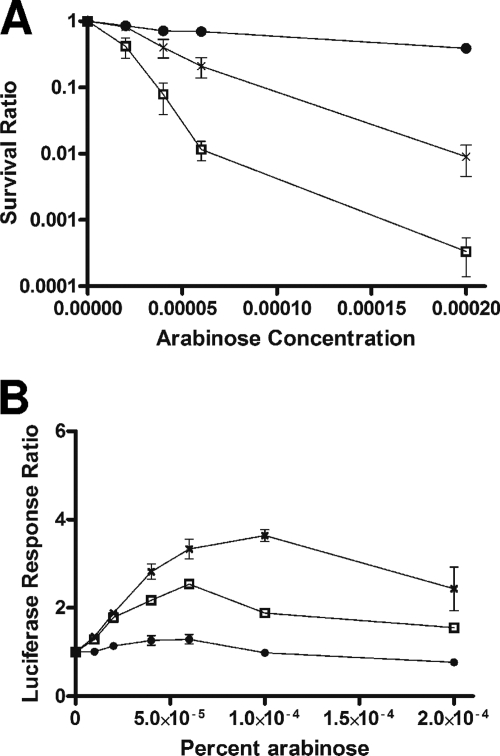
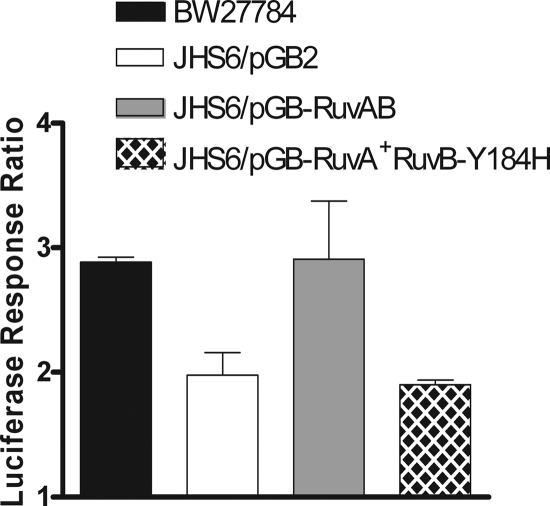
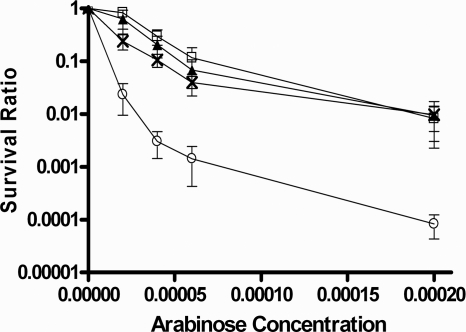
ruvA and ruvB mutants specifically impaired in RFR have been studied previously (3, 20). To further test our hypothesis, the ruvA60::Tn10 (ruvB polar) mutation (23) was transduced into BW27784, resulting in strain JHS6. This ruvA60::Tn10 (ruvB polar) mutation also prevents the expression of ruvB downstream, thus requiring both RuvA and RuvB activities for complementation (23). Plasmid pGB-ruvAB encodes wild-type RuvA and RuvB for both RFR and homologous recombination functions. Plasmid pGB-RuvA+ RuvB-Y184H encodes wild-type RuvA along with a mutant RuvB protein that when expressed in the ruvA60::Tn10 (ruvB polar) genetic background would result in RuvAB activity that is deficient in RFR at arrested forks while maintaining Holliday junction resolution function during homologous recombination (20). The effect of the ruvA60::Tn10 (ruvB polar) mutation in strain JHS6 on SOS induction and viability following topoisomerase I cleavage complex accumulation was similar to those observed for the Keio ruvA null mutation in JHS4. When JHS6 was transformed with vector pGB2 with no RuvAB function, SOS induction after topoisomerase I cleavage complex accumulation was diminished (Fig. 6) along with viability (Fig. 7). Plasmid pGB-RuvAB, but not pGB-RuvA+ RuvB-Y184H, was able to restore the SOS induction after topoisomerase I cleavage complex accumulation (Fig. 6). This result showed that the RFR function is involved in the conversion of the topoisomerase I-associated DNA break into the DSB substrate for RecBCD-dependent loading of RecA. Plasmid pGB-RuvA+ RuvB-Y184H restored the viability of JHS6 to a level close to that obtained with plasmid pGB-RuvAB following topoisomerase I cleavage accumulation (Fig. 7). The ruvA60::Tn10 (ruvB polar) mutation in JHS6 did not decrease the SOS induction ratio in response to norfloxacin (data not shown), similar to the results obtained with JHS4 (Fig. 4B), confirming that RuvAB activity was not required to generate the DSB repair substrate for RecBCD.
FIG. 6.
Effect of RFR-deficient ruvB allele on SOS induction in response to topoisomerase I cleavage complex accumulation. All of the strains here contain plasmids pAYTOP-G116S and pDinlux. The luciferase signal from an induced culture was measured at 250 min after the addition of 0.00004% arabinose and divided by the luciferase signal from a noninduced culture not treated with arabinose to obtain the luciferase response ratio. The results represent the average and standard deviation from at least three experiments.
FIG. 7.
Effect of RFR-deficient ruvB allele on viability following topoisomerase I cleavage complex accumulation. All of the strains here contained plasmid pAYTOP-G122S to measure complementation of RuvAB activity for survival after induction of mutant topoisomerase I by arabinose. Symbols: squares, BW27784 control; circles, JHS6/pGB2; triangles, JHS6/pGB-RuvAB; ×, JHS6/pGB-RuvA+ RuvB-Y184H.
DISCUSSION
Mutations affecting either RuvABC or RecG activity have been shown to result in increased sensitivity to nalidixic acid and ciprofloxacin in previous studies (25, 35). This can be attributed to the Holliday junction resolution function of RuvABC and RecG during homologous recombination initiated by the RecBCD pathway. Either RuvABC or RecG can act in separate pathways to resolve the Holliday junction following RecA-mediated homologous recombination. Double mutants with defects in both RuvABC and RecG exhibit extremely poor growth phenotypes, even in the absence of DNA damage, due to the requirement of a homologous recombination function during exponential cell growth (16). While their functions in homologous recombination should account for the effects of the ruvA and recG null mutations on viability following gyrase cleavage complex accumulation, it is not quite clear from previous studies if the replication fork restart function of RuvAB and RecG acting at arrested forks (1) is involved in generating the DSB repair substrate for RecA loading by RecBCD following quinolone treatment. As we saw no drastic decrease in SOS induction following quinolone treatment in the ruvA and recG mutants, our results suggest that RuvAB and RecG do not play a role in generating the DSB repair substrate for RecBCD during the response to quinolone treatment. Instead of converting the type IIA topoisomerase cleavage complex into a DSB, RuvAB has been shown in vitro to reverse the DNA strand breakage and displace the ternary complex formed by quinolone-bound topoisomerase IV and DNA (33). Since the DNA strand breakage made by the mutant recombinant topoisomerase I was not reversible, this could account for the difference in outcome from interaction with RuvAB. However, the reversible complexes formed by gyrase and topoisomerase IV are also converted into an irreversible complex in the cell killing pathway. Processing of the irreversible topoisomerase-quinolone-DNA complex to convert the DSB made by the type IIA topoisomerases into substrates for RecBCD is likely to involve factors other than RuvAB.
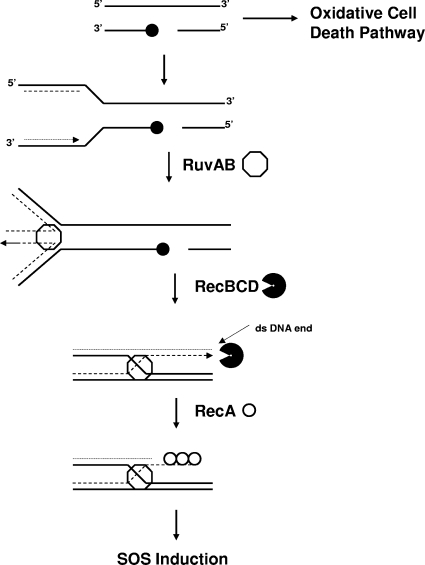
SOS induction following topoisomerase I cleavage complex accumulation in E. coli also occurs via the RecBCD pathway, with the SOS induction shown to be abolished completely by a recB null mutation in our previous study (34). In contrast to the response to quinolone treatment, processing by RuvAB and RecG at replication forks arrested by covalent topoisomerase I complex accumulation is likely to be required to generate the DSB repair substrate for RecBCD and subsequent RecA-mediated SOS induction. A model of the RFR action of RuvAB following topoisomerase I cleavage complex trapping is shown in Fig. 8. RecG could potentially initiate replication fork regression (1), although it cannot be ruled out that it may have additional roles in response to the topoisomerase-mediated DNA damage. The effect of the individual ruvA or recG null mutation on SOS induction and viability is less than that observed previously for recA and recB null mutations (34), probably because RuvA and RecG could act independently of each other for similar repair functions.
FIG. 8.
Model of RFR by RuvAB for SOS induction following topoisomerase I cleavage complex trapping (adapted from reference 3). Symbols: filled circles, topoisomerase I; octagons, RuvAB; incised circles, RecBCD; unfilled circles, RecA.
The arrest of replication forks by type I topoisomerase cleavage complex accumulation has been proposed as part of the mechanism of action of the anticancer drug camptothecin that targets human topoisomerase I (14). The RFR function for repair could account for at least part of the decreased viability observed in the ruvA and recG mutants following topoisomerase I cleavage complex accumulation. However, the RuvAB RFR deficiency had virtually no effect on viability following topoisomerase I cleavage complex accumulation compared to the loss of homologous recombination function from the null mutation (Fig. 7). In a recently reported study (22), the RFR function deficiency of the RuvB-Y184H mutant was found to similarly decrease the constitutive SOS induction from RecA4142 loading onto reversed replication forks with no effect on UV sensitivity.
The results of our studies suggest that topoisomerase I cleavage complex accumulation can trigger the oxidative cell killing pathway (21) in a mechanism not affected by DNA repair at the arrested forks. RecA is likely to be involved in response to the oxidative cell killing pathway. Other bactericidal antibiotics, such as aminoglycosides, which do not act on DNA replication can also involve oxidative damage in at least part of their bactericidal action (10, 19). The pathway from topoisomerase cleavage complex accumulation to changes in metabolism that result in the production of superoxide and other reactive oxygen species remains to be fully elucidated.
Certain synthetic peptides have been shown to trap Holliday junction intermediates in E. coli and inhibit the activities of RuvABC and RecG (12, 13, 17). Administration of such DNA repair inhibitors in combination with drugs that trap topoisomerase cleavage complexes could improve the efficiency of antibacterial therapy and limit the development of drug resistance.
Acknowledgments
We thank Bénédicte Michel for generously providing strains and helpful suggestions. We gratefully acknowledge the use of E. coli strains made available by National BioResource Project (NIG, Japan). We also thank Anca Segall and Thirunavukkarasu Annamalai for comments and suggestions.
This work was supported by Public Health Service grant R01 AI 069313 from the National Institutes of Health.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Atkinson, J., and P. McGlynn. 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37:3475-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baharoglu, Z., A. S. Bradley, M. Le Masson, I. Tsaneva, and B. Michel. 2008. ruvA mutants that resolve Holliday junctions but do not reverse replication forks. PLoS Genet. 4:e1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, B., S. Shukla, S. Vasunilashorn, S. Mukhopadhyay, and Y. C. Tse-Dinh. 2005. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J. Biol. Chem. 280:38489-38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, B., I. Liu, and Y. C. Tse-Dinh. 2007. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 59:640-645. [DOI] [PubMed] [Google Scholar]
- 6.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 7.Connelly, J. C., E. S. de Leau, and D. R. F. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair 2:795-807. [DOI] [PubMed] [Google Scholar]
- 8.Cortes Ledesma, F., S. F. El Khamisy, M. C. Zuma, K. Osborn, and K. W. Caldecott. 2009. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461:674-678. [DOI] [PubMed] [Google Scholar]
- 9.Deweese, J. E., and N. Osheroff. 2009. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 37:738-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwyer, D. J., M. A. Kohanski, and J. J. Collins. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12:482-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grove, J. I., L. Harris, C. Buckman, and R. G. Lloyd. 2008. DNA double strand break repair and crossing over mediated by RuvABC resolvase and RecG translocase. DNA Repair (Amsterdam) 7:1517-1530. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson, C. W., and A. M. Segall. 2006. DNA repair, a novel antibacterial target: Holliday junction-trapping peptides induce DNA damage and chromosome segregation defects. Mol. Microbiol. 59:1129-1148. [DOI] [PubMed] [Google Scholar]
- 13.Gunderson, C. W., J. L. Boldt, R. N. Authement, and A. M. Segall. 2009. Peptide wrwycr inhibits the excision of several prophages and traps Holliday junctions inside bacteria. J. Bacteriol. 191:2169-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiang, Y. H., M. G. Lihou, and L. F. Liu. 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49:5077-5082. [PubMed] [Google Scholar]
- 15.Interthal, H., J. J. Pouliot, and J. J. Champoux. 2001. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. U. S. A. 98:12009-12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishioka, K., H. Iwasaki, and H. Shinagawa. 1997. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet. Syst. 72:91-99. [DOI] [PubMed] [Google Scholar]
- 17.Kepple, K. V., N. Patel, P. Salamon, and A. M. Segall. 2008. Interactions between branched DNAs and peptide inhibitors of DNA repair. Nucleic Acids Res. 36:5319-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 19.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 20.Le Masson, M., Z. Baharoglu, and B. Michel. 2008. ruvA and ruvB mutants specifically impaired for replication fork reversal. Mol. Microbiol. 70:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, I. F., T. Annamali, J. H. Sutherland, and Y. C. Tse-Dinh. 2009. Hydroxyl radicals are involved in cell killing by bacterial topoisomerase I cleavage complex. J. Bacteriol. 191:5315-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long, J. E., S. C. Massoni, and S. J. Sandler. 2010. RecA4142 causes SOS constitutive expression by loading onto reversed replication forks in Escherichia coli K-12. J. Bacteriol. 192:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel, B., G. D. Recchia, M. Penel-Colin, S. D. Ehrlich, and D. J. Sherratt. 2000. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol. 37:180-191. [DOI] [PubMed] [Google Scholar]
- 24.Newmark, K. G., E. K. O'Reilly, J. R. Pohlhaus, and K. N. Kreuzer. 2005. Genetic analysis of the requirements for SOS induction by nalidixic acid in Escherichia coli. Gene 356:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otsuji, N., H. Iyehara, and Y. Hideshima. 1974. Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J. Bacteriol. 117:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pohlhaus, J. R., D. T. Long, E. O'Reilly, and K. N. Kreuzer. 2008. The epsilon subunit of DNA polymerase III Is involved in the nalidixic acid-induced SOS response in Escherichia coli. J. Bacteriol. 190:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pouliot, J. J., K. C. Yao, C. A. Robertson, and H. A. Nash. 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286:552-555. [DOI] [PubMed] [Google Scholar]
- 28.Pourquier, P., M. A. Bjornsti, and Y. Pommier. 1998. Induction of topoisomerase I cleavage complexes by the vinyl chloride adduct 1,N6-ethenoadenine. J. Biol. Chem. 273:27245-27249. [DOI] [PubMed] [Google Scholar]
- 29.Pourquier, P., A. A. Pilon, G. Kohlhagen, A. Mazumder, A. Sharma, and Y. Pommier. 1997. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J. Biol. Chem. 272:26441-26447. [DOI] [PubMed] [Google Scholar]
- 30.Pourquier, P., L. M. Ueng, J. Fertala, D. Wang, H. J. Park, J. M. Essigmann, M. A. Bjornsti, and Y. Pommier. 1999. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7,8-Dihydro-8-oxoguanine and 5-hydroxycytosine. J. Biol. Chem. 274:8516-8523. [DOI] [PubMed] [Google Scholar]
- 31.Sabourin, M., and N. Osheroff. 2000. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Res. 28:1947-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 33.Shea, M. E., and H. Hiasa. 2003. The RuvAB branch migration complex can displace topoisomerase IV.quinolone.DNA ternary complexes. J. Biol. Chem. 278:48485-48490. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland, J. H., B. Cheng, I. F. Liu, and Y. C. Tse-Dinh. 2008. SOS induction by stabilized topoisomerase IA cleavage complex occurs via the RecBCD pathway. J. Bacteriol. 190:3399-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamae, C., A. Liu, K. Kim, D. Sitz, J. Hong, E. Becket, A. Bui, P. Solaimani, K. P. Tran, H. Yang, and J. H. Miller. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190:5981-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tse-Dinh, Y. C. 2009. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 37:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. U. S. A. 99:13669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, H., Y. Mao, A. Y. Chen, N. Zhou, E. J. LaVoie, and L. F. Liu. 2001. Stimulation of topoisomerase II-mediated DNA damage via a mechanism involving protein thiolation. Biochemistry 40:3316-3323. [DOI] [PubMed] [Google Scholar]